PAI1 Regulates Cell Morphology and Migration Markers in Trastuzumab-Resistant HER2-Positive Breast Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture, Transfection, Drug Treatment
2.2. Combination Therapy Experiment
2.3. Quantitative Real-Time PCR
2.4. Gene Expression Heatmap
2.5. Western Blot
2.6. Immunofluorescence
2.7. Statistical Analysis
3. Results
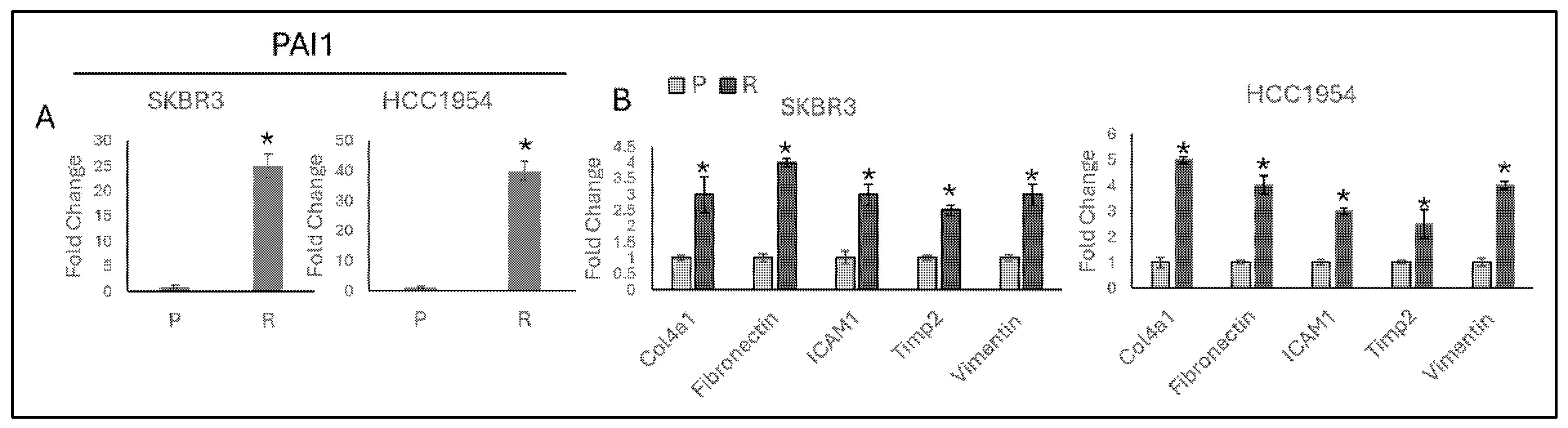
3.1. TGF-β and Migration Responsive Genes Regulated by Acquired Resistance in HER2-Positive Trastuzumab Resistant Cell Lines
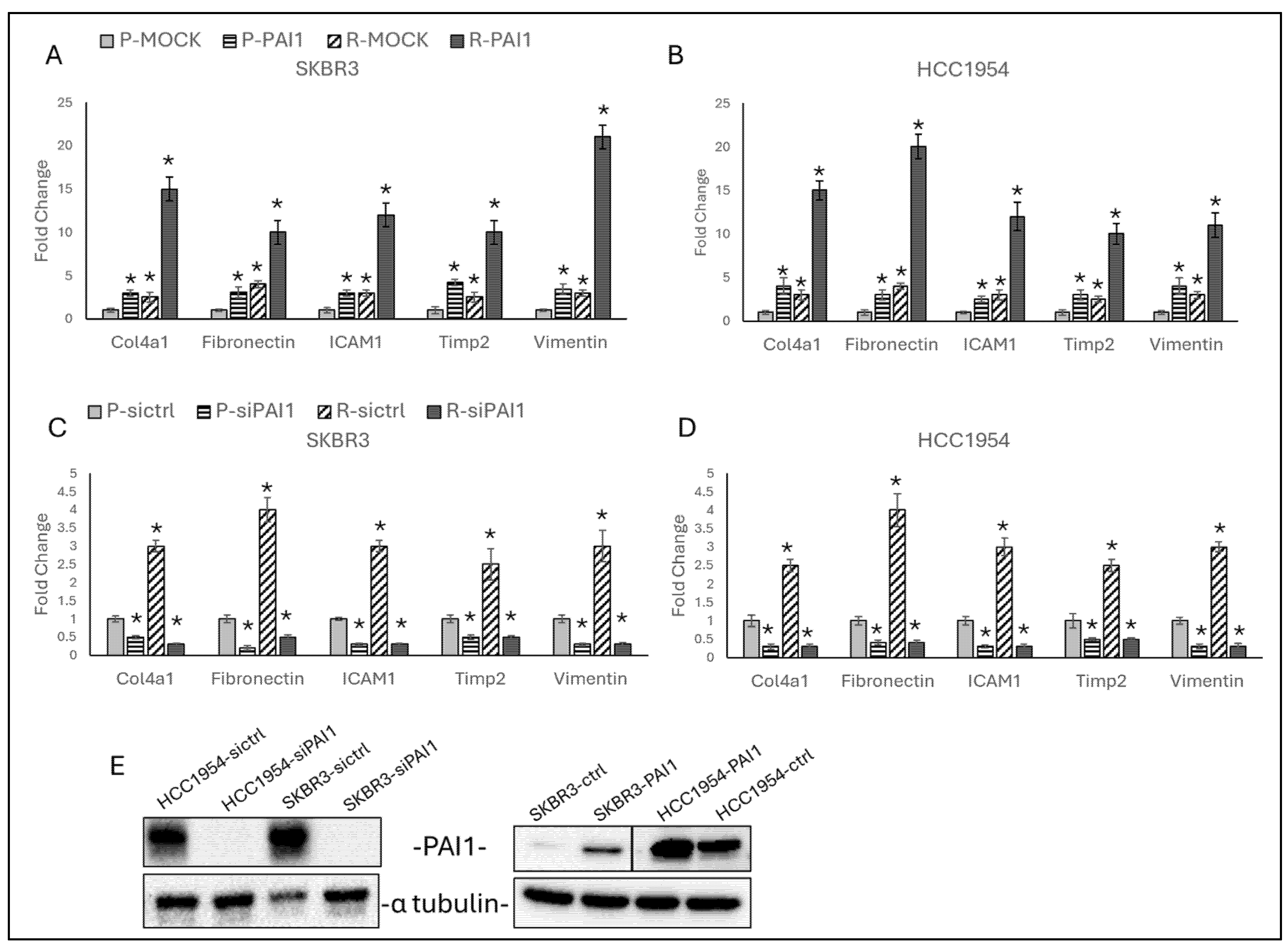
3.2. PAI1 Regulates Migration Responsive Gene Expressions in Parental and Resistant HER2-Positive Cell Lines
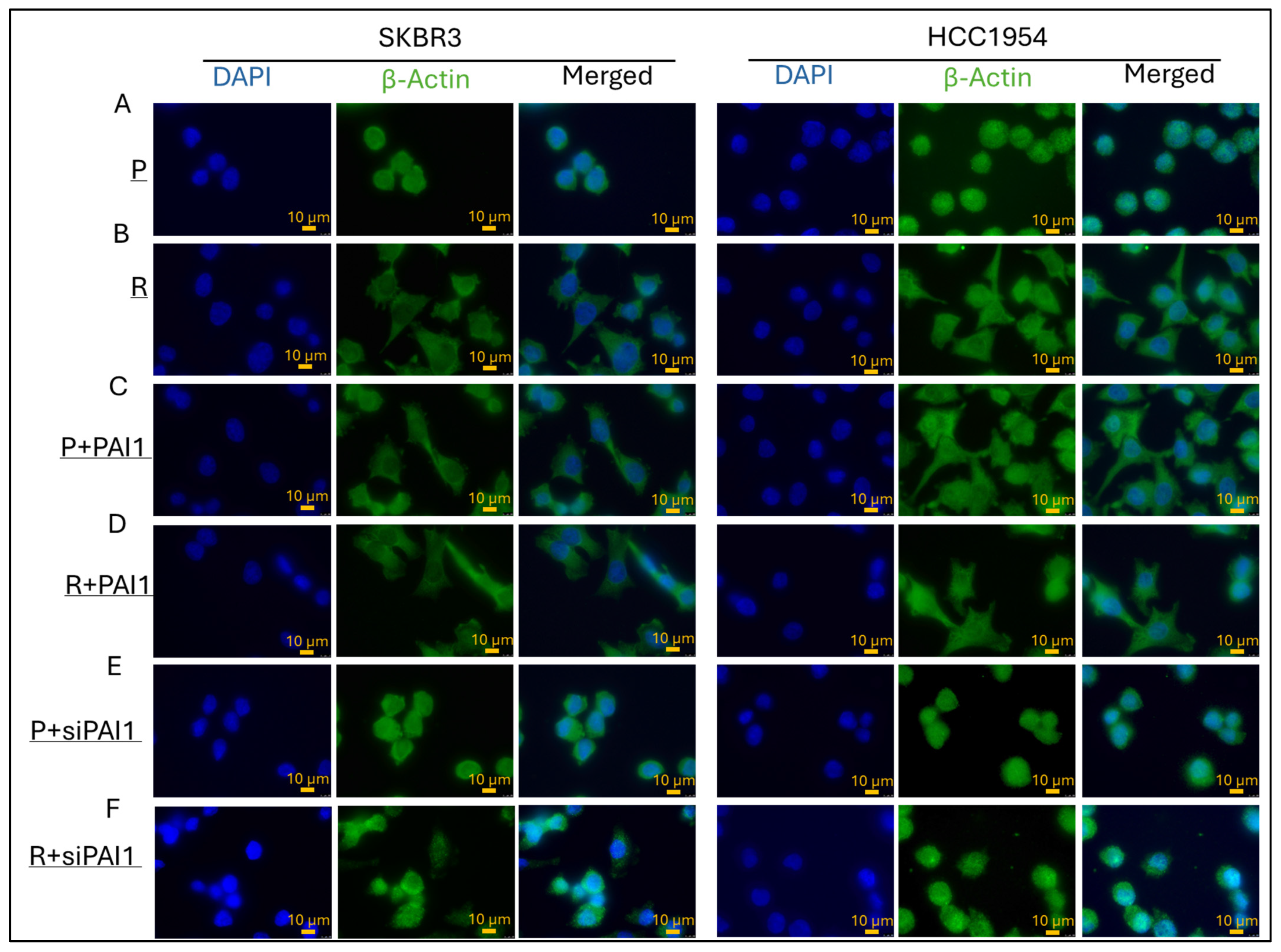
3.3. PAI1 Regulates Cell Morphology in Parental and Resistant HER2-Positive Cell Lines
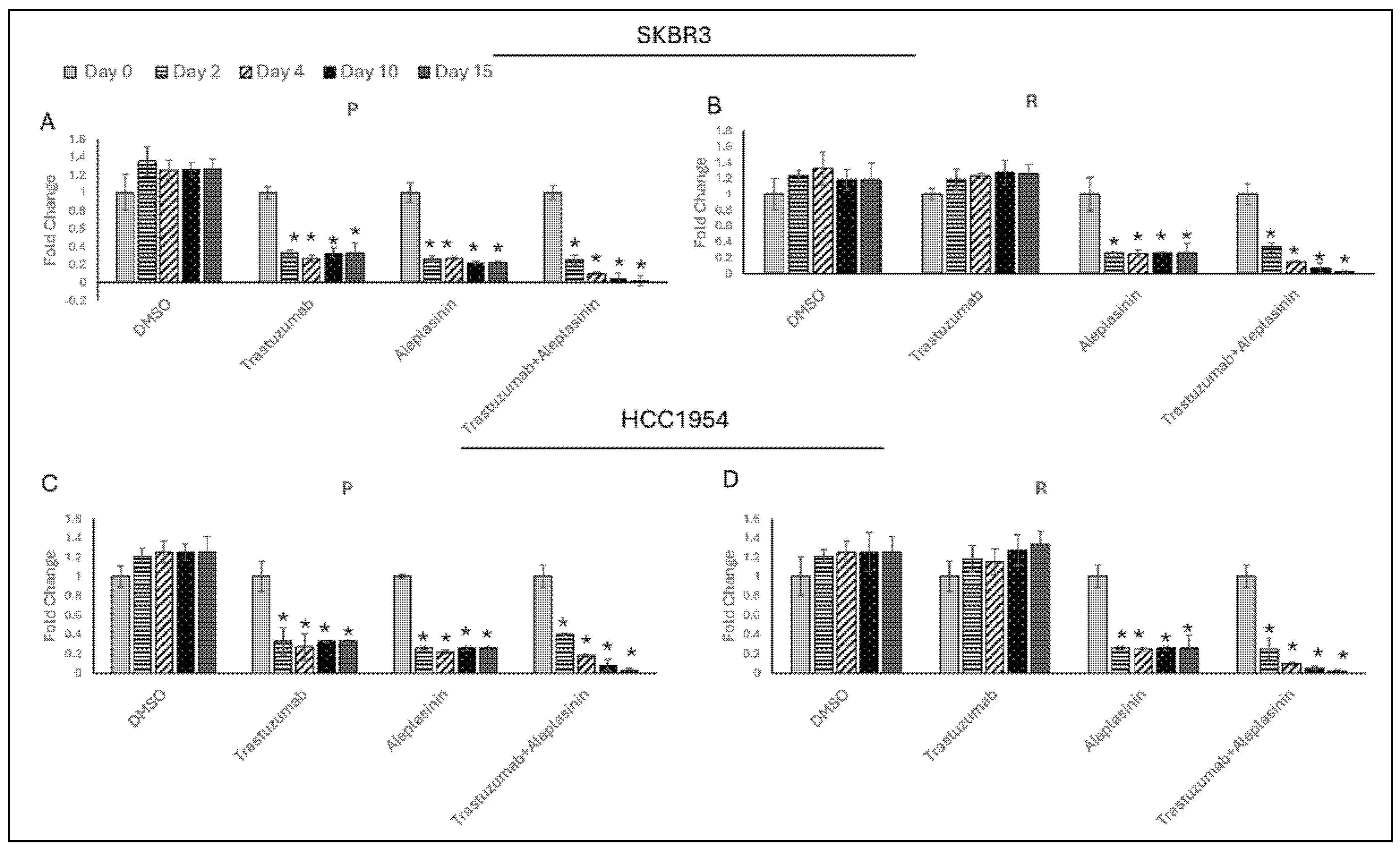
3.4. Synergistic Effect of Trastuzumab+Aleplasinin on HER2-Positive Breast Cancer Cells
3.5. Trastzumab+Aleplasinin Combination Decrease PAI1 Expression
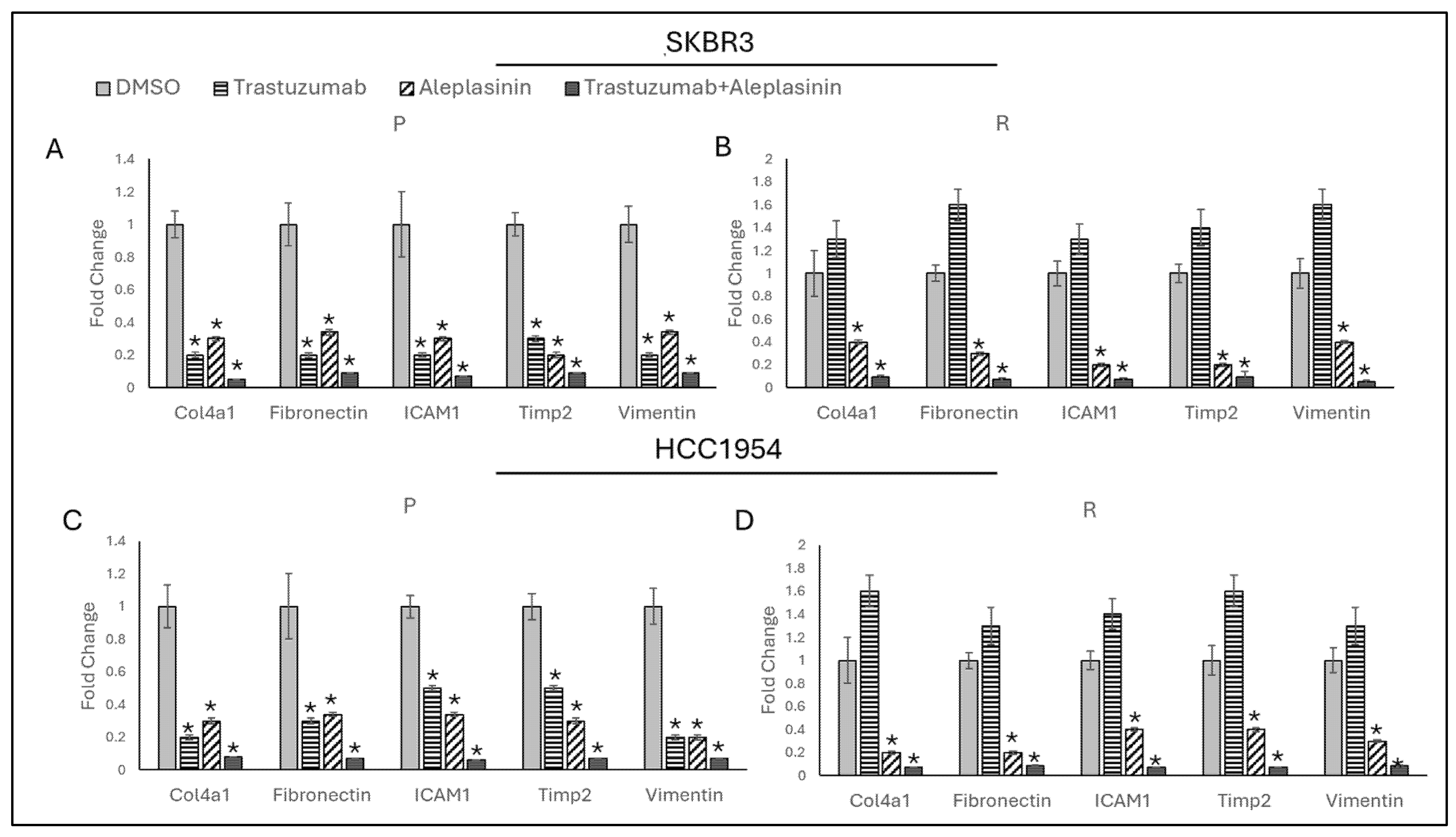
3.6. Trastzumab+Aleplasinin Combination Decrease Migration Responsive Marker Expressions
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Murthy, R.K.; Loi, S.; Okines, A.; Paplomata, E.; Hamilton, E.; Hurvitz, S.A.; Lin, N.U.; Borges, V.; Abramson, V.; Anders, C.; et al. Tucatinib, Trastuzumab, and Capecitabine for HER2-Positive Metastatic Breast Cancer. N. Engl. J. Med. 2020, 382, 597–609. [Google Scholar] [CrossRef]
- Baselga, J.; Swain, S.M. CLEOPATRA: A phase III evaluation of pertuzumab and trastuzumab for HER2-positive metastatic breast cancer. Clin. Breast Cancer 2010, 10, 489–491. [Google Scholar] [CrossRef]
- Swain, S.M.; Miles, D.; Kim, S.B.; Im, Y.H.; Im, S.A.; Semiglazov, V.; Ciruelos, E.; Schneeweiss, A.; Loi, S.; Monturus, E.; et al. Pertuzumab, trastuzumab, and docetaxel for HER2-positive metastatic breast cancer (CLEOPATRA): End-of-study results from a double-blind, randomised, placebo-controlled, phase 3 study. Lancet Oncol. 2020, 21, 519–530. [Google Scholar] [CrossRef]
- Luque-Cabal, M.; Garcia-Teijido, P.; Fernandez-Perez, Y.; Sanchez-Lorenzo, L.; Palacio-Vazquez, I. Mechanisms Behind the Resistance to Trastuzumab in HER2-Amplified Breast Cancer and Strategies to Overcome It. Clin. Med. Insights Oncol. 2016, 10, 21–30. [Google Scholar] [CrossRef]
- Modi, S.; Saura, C.; Yamashita, T.; Park, Y.H.; Kim, S.B.; Tamura, K.; Andre, F.; Iwata, H.; Ito, Y.; Tsurutani, J.; et al. Trastuzumab Deruxtecan in Previously Treated HER2-Positive Breast Cancer. N. Engl. J. Med. 2020, 382, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Boz Er, A.B.; Er, I. Targeting ITGβ3 to Overcome Trastuzumab Resistance through Epithelial–Mesenchymal Transition Regulation in HER2-Positive Breast Cancer. Int. J. Mol. Sci. 2024, 25, 8640. [Google Scholar] [CrossRef]
- Bang, Y.J.; Giaccone, G.; Im, S.A.; Oh, D.Y.; Bauer, T.M.; Nordstrom, J.L.; Li, H.; Chichili, G.R.; Moore, P.A.; Hong, S.; et al. First-in-human phase 1 study of margetuximab (MGAH22), an Fc-modified chimeric monoclonal antibody, in patients with HER2-positive advanced solid tumors. Ann. Oncol. 2017, 28, 855–861. [Google Scholar] [CrossRef]
- Vivekanandhan, S.; Knutson, K.L. Resistance to Trastuzumab. Cancers 2022, 14, 5115. [Google Scholar] [CrossRef]
- Shi, Q.; Huang, F.; Wang, Y.; Liu, H.; Deng, H.; Chen, Y.G. HER2 phosphorylation induced by TGF-beta promotes mammary morphogenesis and breast cancer progression. J. Cell Biol. 2024, 223, e202307138. [Google Scholar] [CrossRef]
- Ranganathan, P.; Agrawal, A.; Bhushan, R.; Chavalmane, A.K.; Kalathur, R.K.; Takahashi, T.; Kondaiah, P. Expression profiling of genes regulated by TGF-beta: Differential regulation in normal and tumour cells. BMC Genom. 2007, 8, 98. [Google Scholar] [CrossRef]
- Yan, X.; Xiong, X.; Chen, Y.G. Feedback regulation of TGF-beta signaling. Acta Biochim. Biophys. Sin. (Shanghai) 2018, 50, 37–50. [Google Scholar] [CrossRef]
- Samarakoon, R.; Higgins, S.P.; Higgins, C.E.; Higgins, P.J. The TGF-beta1/p53/PAI-1 Signaling Axis in Vascular Senescence: Role of Caveolin-1. Biomolecules 2019, 9, 341. [Google Scholar] [CrossRef]
- Milenkovic, J.; Milojkovic, M.; Jevtovic Stoimenov, T.; Djindjic, B.; Miljkovic, E. Mechanisms of plasminogen activator inhibitor 1 action in stromal remodeling and related diseases. Biomed. Pap. Med. Fac. Univ. Palacky. Olomouc Czech Repub. 2017, 161, 339–347. [Google Scholar] [CrossRef]
- Dovnik, N.F.; Takac, I. Prognostic significance of uPA/PAI-1 level, HER2 status, and traditional histologic factors for survival in node-negative breast cancer patients. Radiol. Oncol. 2017, 51, 65–73. [Google Scholar] [CrossRef]
- Kokot, A.; Gadakh, S.; Saha, I.; Gajda, E.; Lazniewski, M.; Rakshit, S.; Sengupta, K.; Mollah, A.F.; Denkiewicz, M.; Gorczak, K.; et al. Unveiling the Molecular Mechanism of Trastuzumab Resistance in SKBR3 and BT474 Cell Lines for HER2 Positive Breast Cancer. Curr. Issues Mol. Biol. 2024, 46, 2713–2740. [Google Scholar] [CrossRef]
- Li, H.; Yuan, W.; Bin, S.; Wu, G.; Li, P.; Liu, M.; Yang, J.; Li, X.; Yang, K.; Gu, H. Overcome trastuzumab resistance of breast cancer using anti-HER2 chimeric antigen receptor T cells and PD1 blockade. Am. J. Cancer Res. 2020, 10, 688–703. [Google Scholar]
- Boz Er, A.B. Integrin β3 Reprogramming Stemness in HER2-Positive Breast Cancer Cell Lines. Biology 2024, 13, 429. [Google Scholar] [CrossRef]
- Chou, T.-C. The combination index (CI < 1) as the definition of synergism and of synergy claims. Synergy 2018, 7, 49–50. [Google Scholar] [CrossRef]
- Sun, Y.; Vandenbriele, C.; Kauskot, A.; Verhamme, P.; Hoylaerts, M.F.; Wright, G.J. A Human Platelet Receptor Protein Microarray Identifies the High Affinity Immunoglobulin E Receptor Subunit alpha (FcepsilonR1alpha) as an Activating Platelet Endothelium Aggregation Receptor 1 (PEAR1) Ligand. Mol. Cell. Proteom. 2015, 14, 1265–1274. [Google Scholar] [CrossRef]
- Jun, F.; Hong, J.; Liu, Q.; Guo, Y.; Liao, Y.; Huang, J.; Wen, S.; Shen, L. Epithelial membrane protein 3 regulates TGF-beta signaling activation in CD44-high glioblastoma. Oncotarget 2017, 8, 14343–14358. [Google Scholar] [CrossRef]
- Czekay, R.P.; Wilkins-Port, C.E.; Higgins, S.P.; Freytag, J.; Overstreet, J.M.; Klein, R.M.; Higgins, C.E.; Samarakoon, R.; Higgins, P.J. PAI-1: An Integrator of Cell Signaling and Migration. Int. J. Cell Biol. 2011, 2011, 562481. [Google Scholar] [CrossRef]
- Ciriza, J.; Garcia-Ojeda, M.E. Expression of migration-related genes is progressively upregulated in murine Lineage-Sca-1+c-Kit+ population from the fetal to adult stages of development. Stem Cell Res. Ther. 2010, 1, 14. [Google Scholar] [CrossRef]
- Lappalainen, P.; Kotila, T.; Jegou, A.; Romet-Lemonne, G. Biochemical and mechanical regulation of actin dynamics. Nat. Rev. Mol. Cell Biol. 2022, 23, 836–852. [Google Scholar] [CrossRef]
- Chou, T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef]
- Wei, X.; Li, S.; He, J.; Du, H.; Liu, Y.; Yu, W.; Hu, H.; Han, L.; Wang, C.; Li, H.; et al. Tumor-secreted PAI-1 promotes breast cancer metastasis via the induction of adipocyte-derived collagen remodeling. Cell Commun. Signal 2019, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Oh, D.Y. HER2-targeted therapies beyond breast cancer—An update. Nat. Rev. Clin. Oncol. 2024, 21, 675–700. [Google Scholar] [CrossRef]


| Cell Lines | p-IC50 (μM) | R-IC50 (μM) |
|---|---|---|
| SKBR3 | 7.5 ± 0.6 | 6.4 ± 0.6 |
| HCC1954 | 10.2 ± 0.8 | 12.5 ± 1.1 |
| Gene Name | Sequence | Product Length |
|---|---|---|
| WWP1 | F: GCAGCTCATCTCCAACCATAG | 201 bp |
| R: GAGACGGAGATGAAGGTGTG | ||
| CARM1 | F: CAGTTTTATGGCTACCTGTCCC | 150 bp |
| R: AAACGACAGGATCCCAGAGC | ||
| RASGM1 | F: AAATCAATGCCCGTGACTGG | 166 bp |
| R: TCAGAGAACGATATCCTCCGG | ||
| THBS1 | F: GCTCTACCAGTGTCCTCCTC | 213 bp |
| R: TCTCTTCAGTCACTTTGCGG | ||
| KCDT1 | F: AGGAGTGTTGGAGGAAGCAG | 193 bp |
| R: CAACTGCTCGAACTTCCAGC | ||
| SGCA | F: CAGGTCATTGAGGTCACAGC | 183 bp |
| R: CCCCAAGGCTGAGAGGAAG | ||
| MCAM | F: ACTGGTTTTCTGTCCACAAGG | 203 bp |
| R: GATGCGGTACTCCTGGGAC | ||
| FXR2 | F: GGGGATGAAGTGGAGGTTTATTC | 177 bp |
| R: AAGGGGATTGGGATTAACTGG | ||
| MTMR3 | F: TTCCCAGGAAGCAGCTGATC | 249 bp |
| R: ACTGACACCTGATAACTTTGCAG | ||
| SOCS3 | F: GAGAGCGGCTTCTACTGGAG | 162 bp |
| R: CTGGATGCGCAGGTTCTTG | ||
| SLC2A4 | F: AGAGCCACCCCAGGAAAAG | 225 bp |
| R: CGGAGAGGACTGGTCACTG | ||
| MMP2 | F: CAGGATCATTGGCTACACACC | 151 bp |
| R: CCAGCGGCCAAAGTTGATC | ||
| MMP9 | F: TGGATCCAAAACTACTCGGAAG | 250 bp |
| R: CATCGTCGAAATGGGCGTC | ||
| PAI1 | F: AGACCGATTATTGACCGACC | 210 bp |
| R: CCAGTTGTGAGATCCGCTAC | ||
| TIMP1 | F: GACCACCTTATACCAGCGTTATG | 183 bp |
| R: CCATCCTGCAGTTTTCCAGC | ||
| TGIF1 | F: AGGTCTGTAACTGGTTCATCAAC | 231 bp |
| R: GAGACAGTGGCCTCCCTAG | ||
| COL6A1 | F: AAGTCTTCTCGGTGGCCATC | 198 bp |
| R: CGAAGGAGCAGCACACTTG | ||
| AAK1 | F: TGAGGGATCTTTCAGGGCAC | 208 bp |
| R: GGCAACAGCTTCACAGGTATC | ||
| MAN1A1 | F: ACAGTGGGGTCAACATCATG | 155 bp |
| R: TGCGGATCAAATGAGTCTCG | ||
| NT5C2 | F: TGCTGTGTACAAGTCCCCAG | 184 bp |
| R: RGAGGTTTCCATAGGCATCGAC | ||
| IRS1 | F: TCTTCTTCATCGAGGTGGGC | 170 bp |
| R: CCCCGACGATTGGCTCTTAC | ||
| KCNIP3 | F: GGGCTTTAAGAATGAGTGTCCC | 151 bp |
| R: AAAGTGGATGGCCCCGTTC | ||
| C4BPA | F: TCTACAAACGATGCAGACACC | 236 bp |
| R: GCTGTGCCTTCCATTCCTG | ||
| PSG1 | F: TCACCTTACACCTGGAGACTC | 159 bp |
| R: AGCTGTGAGTCATAGGGAGG | ||
| ALAD | F: AGAGTTCCCAAGGACGAGC | 172 bp |
| R: ATGCTCCGTTTTCACTCAGG | ||
| FGD1 | F: AGAAGTTTGAAAGAGAGCCTGTG | 214 bp |
| R: CTATCAATGCCGCTGTCCCG | ||
| TFAP2B | F: GTCCAGTCAGTTGAAGATGCC | 165 bp |
| R: GACGGAGCAAAACACCTCG | ||
| AHCYL1 | F: CGCTGGTCTGCTTGTAACATC | 175 bp |
| R: CCCCATCATCCAGGATCATG | ||
| HSPG2 | F: AGATGGTTTATTTCCGAGCCC | 181 bp |
| R: AGCTCCTTGATGAACACCAC | ||
| IL1RAP | F: CGACTATCACTTGGTATATGGGC | 164 bp |
| R: CCTGGTGAGATGAAACGTACG | ||
| EIF3S6 | F: GGACAAGCATGGTTTTAGGC | 186 bp |
| R: TGCATTAAGATTTCAGAGGCCAG | ||
| HSP47 | F: CGCCATGTTCTTCAAGCCAC | 244 bp |
| R: CTTTTCAAGGCGCTCGAGAG | ||
| MIDORI | F: ACAGCGACGTCAGGTTCAC | 169 bp |
| R: GCATCTTCTTCTCGACACCTG | ||
| GAPDH | F: ACAGTTGCCATGTAGACC | 152 bp |
| R: TTGAGCACAGGGTACTTTA |
| Cell Lines | Drug Combo CI Value | Description of Effect |
|---|---|---|
| SKBR3-P | 1.06 | Nearly additive |
| SKBR3-R | 0.00 | Very strong synergism |
| HCC1954-P | 0.92 | Nearly additive |
| HCC1954-R | 0.49 | Synergism |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boz Er, A.B.; Er, I. PAI1 Regulates Cell Morphology and Migration Markers in Trastuzumab-Resistant HER2-Positive Breast Cancer Cells. Life 2024, 14, 1040. https://doi.org/10.3390/life14081040
Boz Er AB, Er I. PAI1 Regulates Cell Morphology and Migration Markers in Trastuzumab-Resistant HER2-Positive Breast Cancer Cells. Life. 2024; 14(8):1040. https://doi.org/10.3390/life14081040
Chicago/Turabian StyleBoz Er, Asiye Busra, and Idris Er. 2024. "PAI1 Regulates Cell Morphology and Migration Markers in Trastuzumab-Resistant HER2-Positive Breast Cancer Cells" Life 14, no. 8: 1040. https://doi.org/10.3390/life14081040





