Obesity and Environmental Risk Factors Significantly Modify the Association between Ischemic Stroke and the Hero Chaperone C19orf53
Abstract
:1. Introduction
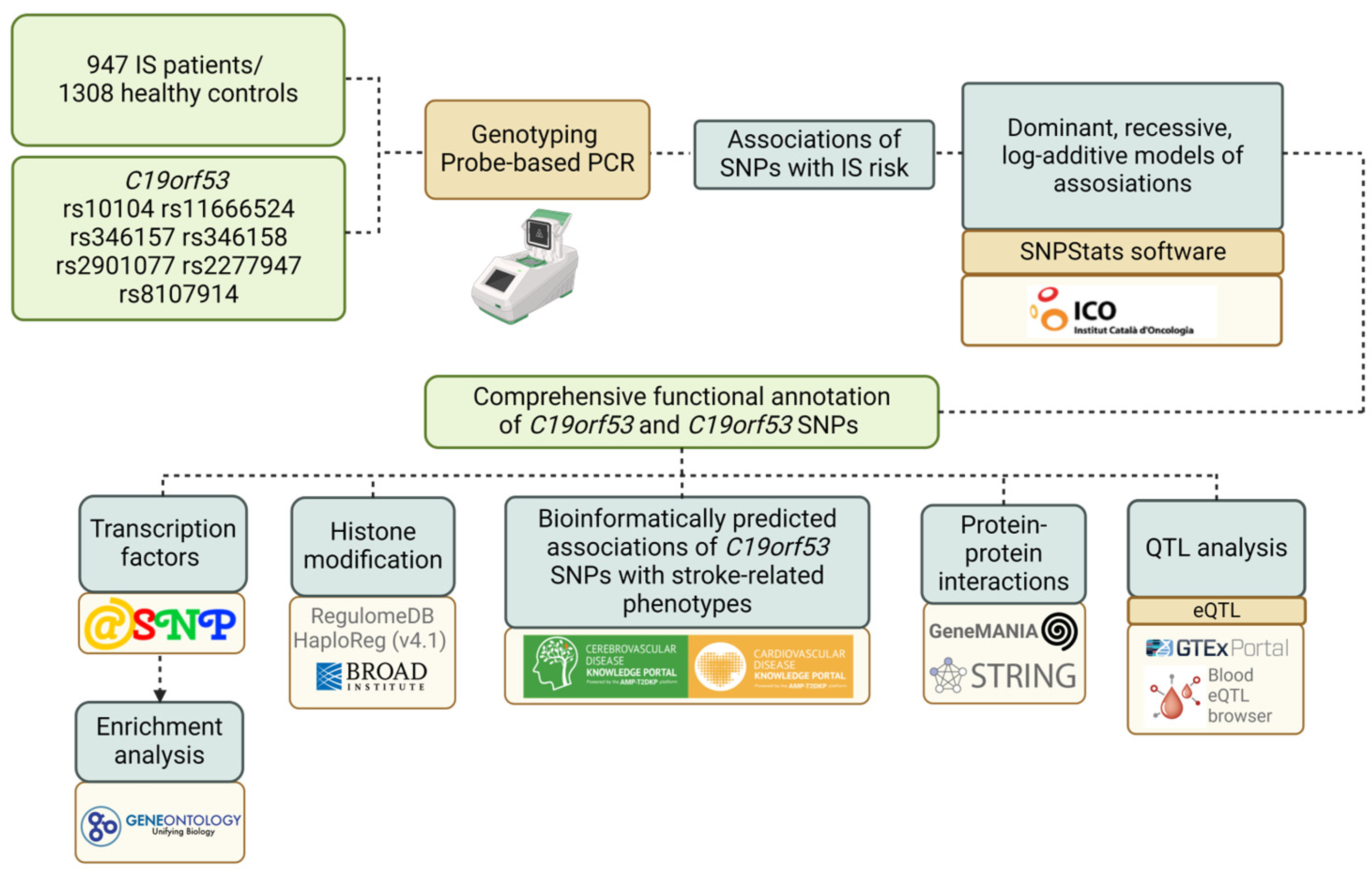
2. Materials and Methods
2.1. Experimental Design
2.2. Genetic Analysis
2.3. Statistical and Bioinformatic Analysis
- The GTExportal (http://www.gtexportal.org/ (accessed on 14 May 2024)) was employed to analyze the expression levels of the studied genes in the brain, whole blood, and blood vessels as well as to examine the expression quantitative trait loci (eQTLs) [30].
- For the examination of C19orf53 SNPs binding to quantitative expression trait loci (eQTL) in peripheral blood, the eQTLGen resource available at https://www.eqtlgen.org/ (accessed on 14 May 2024) was employed [31].
- HaploReg (v4.2), a bioinformatics tool available at https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php (accessed on 14 May 2024), was utilized to assess the associations between C19orf53 SNPs and specific histone modifications indicative of promoters and enhancers. These modifications included acetylation of lysine residues at positions 27 and 9 of the histone H3 protein as well as mono-methylation at position 4 (H3K4me1) and tri-methylation at position 4 (H3K4me3) of the histone H3 protein. Additionally, the tool was applied to investigate the positioning of SNPs in DNase hypersensitive regions, regulatory motif sites, and locations binding to regulatory proteins [32].
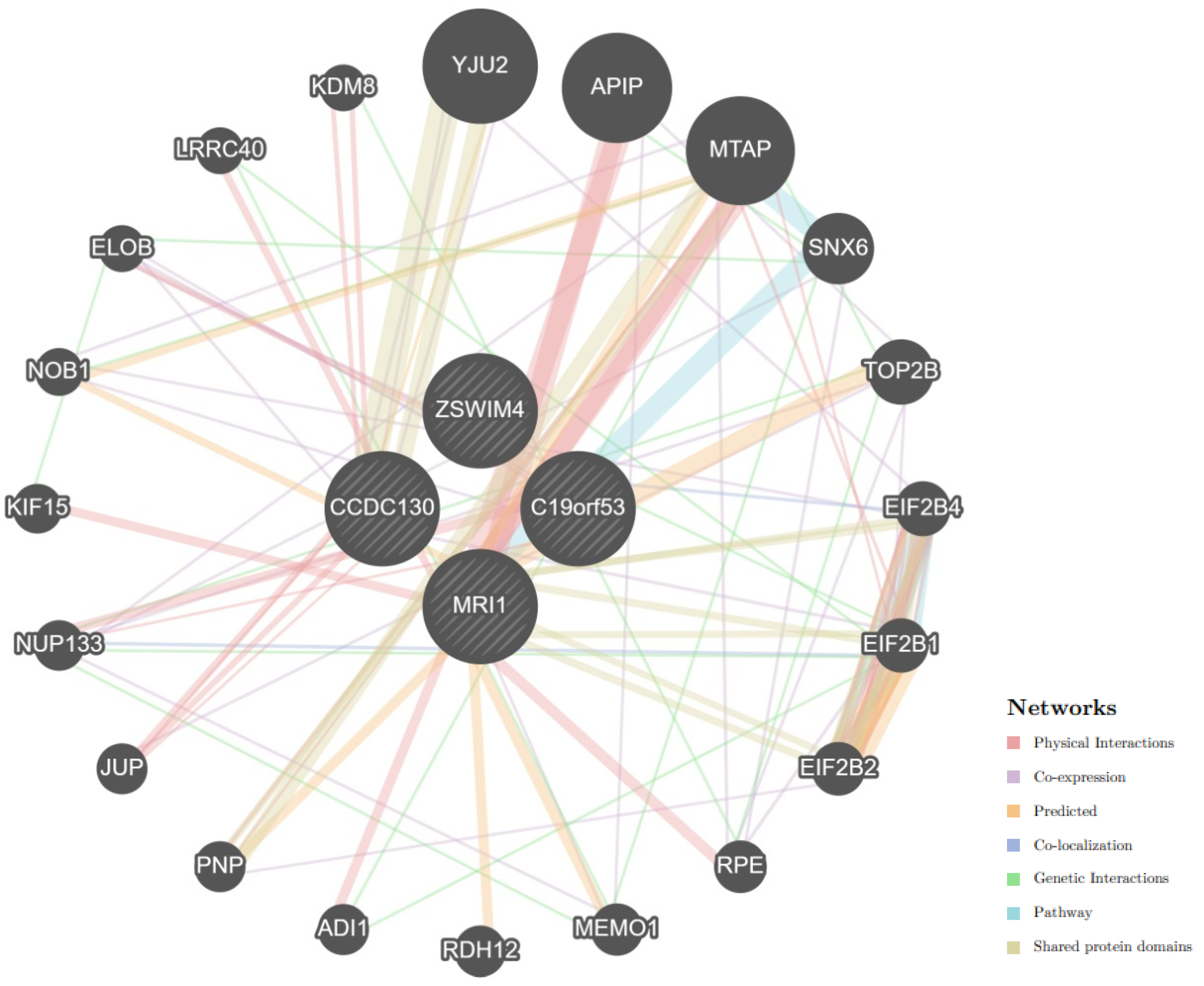
- Bioinformatic tool STRING database (https://string-db.org/ (accessed on 14 May 2024)) was employed to analyze the primary interaction partners of C19orf53. Additionally, the analysis of biological processes and molecular functions related to interactions with key functionally related proteins was conducted using the STRING database [33].
- The mechanisms of interactions between C19orf53 and cis-eQTL-associated genes were analyzed using the GeneMANIA tool [34].
- The atSNP Function Prediction online tool (http://atsnp.biostat.wisc.edu/search (accessed on 15 May 2024)) was utilized to assess how C19orf53 SNPs affect the gene’s affinity for transcription factors (TFs) based on the presence of reference or alternative alleles [35]. This tool evaluated the impact of SNPs on TF-DNA interactions using a positional weight matrix to calculate the degree of influence.
- The Gene Ontology online tool (http://geneontology.org/ (accessed on 15 May 2024)) allowed us to analyze the involvement of transcription factors associated with the reference and SNP alleles in overrepresented biological processes related to IS pathogenesis [36]. This tool helped identify biological functions regulated by TFs linked to C19orf53 SNPs.
- The Cerebrovascular Disease Knowledge Portal (CDKP) (https://cd.hugeamp.org/ (accessed on 16 May 2024)) and Cardiovascular Disease Knowledge Portal (https://cvd.hugeamp.org/ (accessed on 16 May 2024)) integrate and analyze genetic association data from major consortia to study cardio- and cerebrovascular diseases, aiding in the exploration of associations between C19orf53 SNPs and atherosclerosis-related conditions and risk factors for IS (like low density lipids, body mass index, total cholesterol) [37].
3. Results
3.1. C19orf53 SNPs and the Ischemic Stroke Risk: An Analysis of Associations
3.2. C19orf53 SNPs and Clinical Parameters
3.3. The Role of Bioinformatic Tools in Substantiating the Results
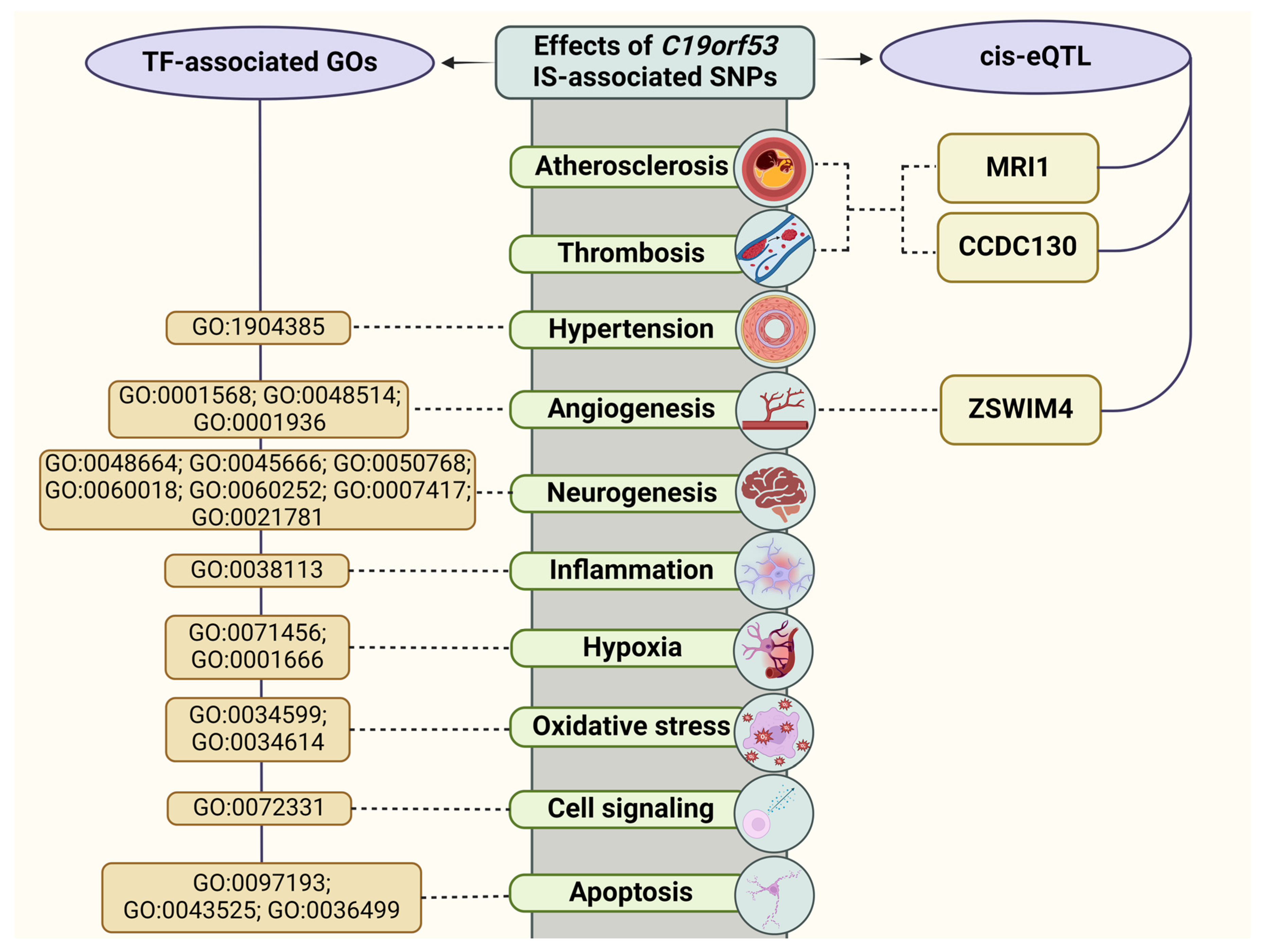
3.3.1. QTL-Effects
3.3.2. Histone Modifications
3.3.3. Analysis of Transcription Factors
3.3.4. Bioinformatic Analysis of the Associations of C19orf53 SNPs with IS-Related Phenotypes
3.3.5. Protein–Protein Interactions
4. Discussion
4.1. C19orf53 SNPs and the IS Risk: Underlying Mechanisms
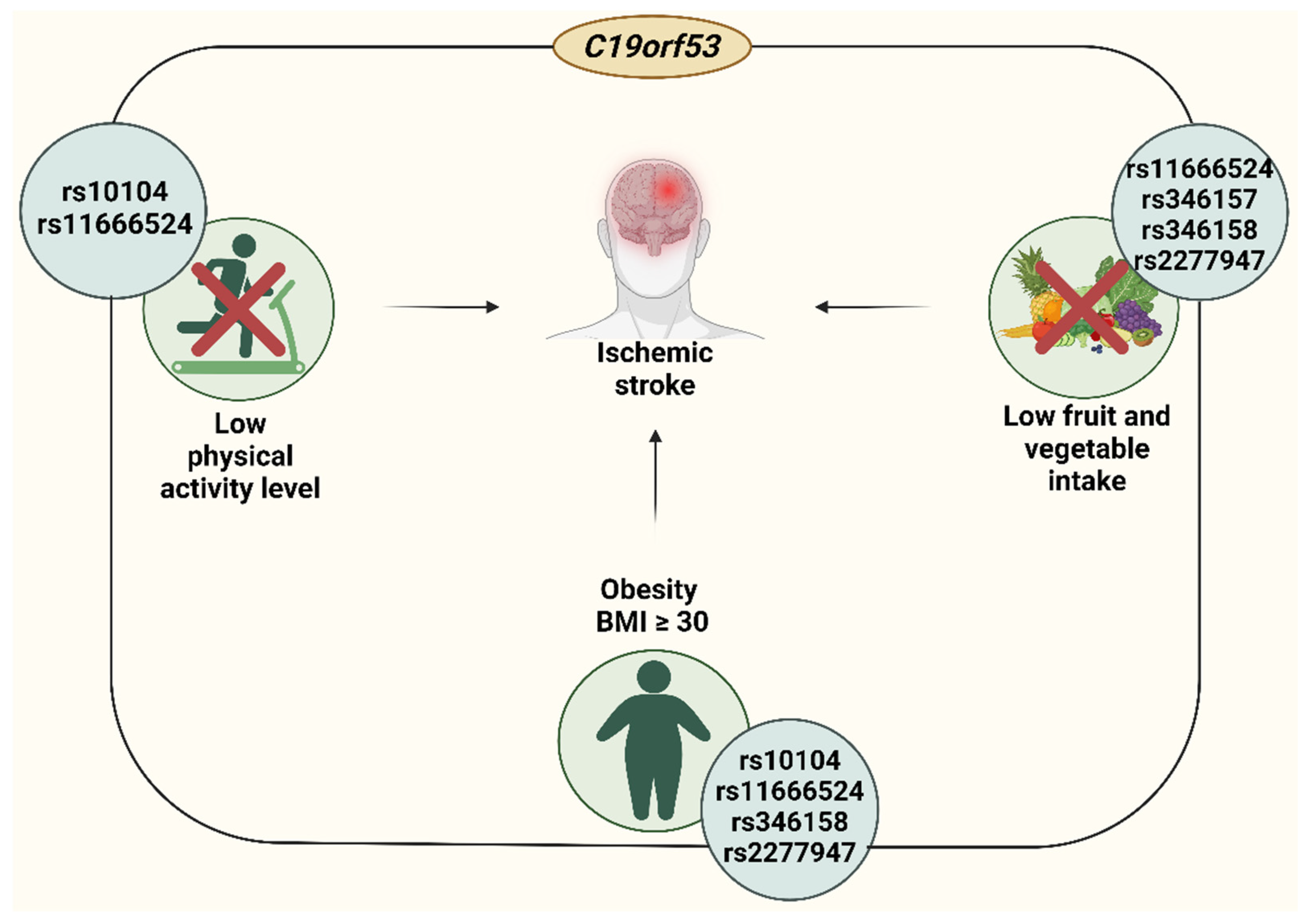
4.2. Insuficient Fruit and Vegetable Intake-Linked Associations of C19orf53
4.3. Low Physical Activity-Related Correlates of C19orf53
4.4. Obesity-Related Correlates of C19orf53
5. Conclusions
6. Limitations of the Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saini, V.; Guada, L.; Yavagal, D.R. Global Epidemiology of Stroke and Access to Acute Ischemic Stroke Interventions. Neurology 2021, 97, S6–S16. [Google Scholar] [CrossRef] [PubMed]
- Koyama, R.; Shichita, T. Glial Roles in Sterile Inflammation after Ischemic Stroke. Neurosci. Res. 2023, 187, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Candelario-Jalil, E.; Dijkhuizen, R.M.; Magnus, T. Neuroinflammation, Stroke, Blood-Brain Barrier Dysfunction, and Imaging Modalities. Stroke 2022, 53, 1473–1486. [Google Scholar] [CrossRef] [PubMed]
- Giffard, R.G.; Yenari, M.A. Many Mechanisms for Hsp70 Protection From Cerebral Ischemia. J. Neurosurg. Anesthesiol. 2004, 16, 53. [Google Scholar] [CrossRef]
- Kobzeva, K.A.; Soldatova, M.O.; Stetskaya, T.A.; Soldatov, V.O.; Deykin, A.V.; Freidin, M.B.; Bykanova, M.A.; Churnosov, M.I.; Polonikov, A.V.; Bushueva, O.Y. Association between HSPA8 Gene Variants and Ischemic Stroke: A Pilot Study Providing Additional Evidence for the Role of Heat Shock Proteins in Disease Pathogenesis. Genes 2023, 14, 1171. [Google Scholar] [CrossRef]
- Truettner, J.S.; Hu, K.; Liu, C.L.; Dietrich, W.D.; Hu, B. Subcellular Stress Response and Induction of Molecular Chaperones and Folding Proteins after Transient Global Ischemia in Rats. Brain Res. 2009, 1249, 9–18. [Google Scholar] [CrossRef]
- Venediktov, A.A.; Bushueva, O.Y.; Kudryavtseva, V.A.; Kuzmin, E.A.; Moiseeva, A.V.; Baldycheva, A.; Meglinski, I.; Piavchenko, G.A. Closest Horizons of Hsp70 Engagement to Manage Neurodegeneration. Front. Mol. Neurosci. 2023, 16, 1230436. [Google Scholar] [CrossRef]
- Jiang, J.; Cyr, D.; Babbitt, R.W.; Sessa, W.C.; Patterson, C. Chaperone-Dependent Regulation of Endothelial Nitric-Oxide Synthase Intracellular Trafficking by the Co-Chaperone/Ubiquitin Ligase CHIP. J. Biol. Chem. 2003, 278, 49332–49341. [Google Scholar] [CrossRef]
- Bushueva, O.; Solodilova, M.; Ivanov, V.; Polonikov, A. Gender-Specific Protective Effect of the −463G>A Polymorphism of Myeloperoxidase Gene against the Risk of Essential Hypertension in Russians. J. Am. Soc. Hypertens. 2015, 9, 902–906. [Google Scholar] [CrossRef]
- Bushueva, O. Single Nucleotide Polymorphisms in Genes Encoding Xenobiotic Metabolizing Enzymes Are Associated with Predisposition to Arterial Hypertension. Res. Results Biomed. 2020, 6, 447–456. [Google Scholar] [CrossRef]
- Madrigal-Matute, J.; de Bruijn, J.; van Kuijk, K.; Riascos-Bernal, D.F.; Diaz, A.; Tasset, I.; Martín-Segura, A.; Gijbels, M.J.J.; Sander, B.; Kaushik, S.; et al. Protective Role of Chaperone-Mediated Autophagy against Atherosclerosis. Proc. Natl. Acad. Sci. USA 2022, 119, e2121133119. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Metzler, B.; Jahangiri, M.; Mandal, K. Molecular Chaperones and Heat Shock Proteins in Atherosclerosis. Am. J. Physiol.-Heart Circ. Physiol. 2012, 302, H506–H514. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, A.; Kotani, K.; Bushueva, O.; Taniguchi, N.; Lazarenko, V. The Cardio-Ankle Vascular Index and Ankle-Brachial Index in Young Russians. J. Atheroscler. Thromb. 2015, 22, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Staron, M.; Wu, S.; Hong, F.; Stojanovic, A.; Du, X.; Bona, R.; Liu, B.; Li, Z. Heat-Shock Protein Gp96/Grp94 Is an Essential Chaperone for the Platelet Glycoprotein Ib-IX-V Complex. Blood J. Am. Soc. Hematol. 2011, 117, 7136–7144. [Google Scholar] [CrossRef]
- Nosaka, M.; Ishida, Y.; Kuninaka, Y.; Ishigami, A.; Taruya, A.; Shimada, E.; Hashizume, Y.; Yamamoto, H.; Kimura, A.; Furukawa, F. Relationship between Intrathrombotic Appearance of HSP27 and HSP70 and Thrombus Ages in a Murine Model of Deep Vein Thrombosis. Sci. Rep. 2023, 13, 22416. [Google Scholar] [CrossRef]
- Tang, Y.; Liao, S.; Liu, G.; Xiong, X.; Liu, H.; Li, F.; Tan, Z.; Kong, X.; Yin, Y.; Tan, B. Advanced Single-Cell Pooled CRISPR Screening Identifies C19orf53 Required for Cell Proliferation Based on mTORC1 Regulators. Cell Biol. Toxicol. 2022, 38, 43–68. [Google Scholar] [CrossRef]
- Zhang, Q.; Burdette, J.E.; Wang, J.-P. Integrative Network Analysis of TCGA Data for Ovarian Cancer. BMC Syst. Biol. 2014, 8, 1338. [Google Scholar] [CrossRef]
- Kong, W.; Hayashi, T.; Fiches, G.; Xu, Q.; Li, M.Z.; Que, J.; Liu, S.; Zhang, W.; Qi, J.; Santoso, N. Diversified Application of Barcoded PLATO (PLATO-BC) Platform for Identification of Protein Interactions. Genom. Proteom. Bioinform. 2019, 17, 319–331. [Google Scholar] [CrossRef]
- Tsuboyama, K.; Osaki, T.; Matsuura-Suzuki, E.; Kozuka-Hata, H.; Okada, Y.; Oyama, M.; Ikeuchi, Y.; Iwasaki, S.; Tomari, Y. A Widespread Family of Heat-Resistant Obscure (Hero) Proteins Protect against Protein Instability and Aggregation. PLoS Biol. 2020, 18, e3000632. [Google Scholar] [CrossRef] [PubMed]
- Kobzeva, K.A.; Shilenok, I.V.; Belykh, A.E.; Gurtovoy, D.E.; Bobyleva, L.A. C9orf16 (BBLN) Gene, Encoding a Member of Hero Proteins, Is a Novel Marker in Ischemic Stroke Risk. Res. Results Biomed. 2022, 8, 278–292. [Google Scholar] [CrossRef]
- Belykh, A.E.; Soldatov, V.O.; Stetskaya, T.A.; Kobzeva, K.A.; Soldatova, M.O.; Polonikov, A.V.; Deykin, A.V.; Churnosov, M.I.; Freidin, M.B.; Bushueva, O.Y. Polymorphism of SERF2, the Gene Encoding a Heat-Resistant Obscure (Hero) Protein with Chaperone Activity, Is a Novel Link in Ischemic Stroke. IBRO Neurosci. Rep. 2023, 14, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Shilenok, I.; Kobzeva, K.; Stetskaya, T.; Freidin, M.; Soldatova, M.; Deykin, A.; Soldatov, V.; Churnosov, M.; Polonikov, A.; Bushueva, O. SERPINE1 mRNA Binding Protein 1 Is Associated with Ischemic Stroke Risk: A Comprehensive Molecular–Genetic and Bioinformatics Analysis of SERBP1 SNPs. Int. J. Mol. Sci. 2023, 24, 8716. [Google Scholar] [CrossRef] [PubMed]
- Lam, A.Y.W.; Tsuboyama, K.; Tadakuma, H.; Tomari, Y. DNAJA2 and Hero11 Mediate Similar Conformational Extension and Aggregation Suppression of TDP-43. RNA 2024, 8, rna.080165.124. [Google Scholar] [CrossRef] [PubMed]
- Stetskaya, T.A.; Kobzeva, K.A.; Zaytsev, S.M.; Shilenok, I.V.; Komkova, G.V.; Goryainova, N.V.; Bushueva, O.Y. HSPD1 Gene Polymorphism Is Associated with an Increased Risk of Ischemic Stroke in Smokers. Res. Results Biomed. 2024, 10, 175–186. [Google Scholar] [CrossRef]
- Bushueva, O.Y.; Bulgakova, I.V.; Ivanov, V.P.; Polonikov, A.V. Association of Flavin Monooxygenase Gene E158K Polymorphism with Chronic Heart Disease Risk. Bull. Exp. Biol. Med. 2015, 159, 776–778. [Google Scholar] [CrossRef]
- Vialykh, E.K.; Solidolova, M.A.; Bushueva, O.I.; Bulgakova, I.V.; Polonikov, A.V. [Catalase gene polymorphism is associated with increased risk of cerebral stroke in hypertensive patients]. Zhurnal Nevrol. Psikhiatrii Im. S.S. Korsakova 2012, 112, 3–7. [Google Scholar]
- Polonikov, A.; Vialykh, E.; Vasil’eva, O.; Bulgakova, I.; Bushueva, O.; Illig, T.; Solodilova, M. Genetic Variation in Glutathione S-Transferase Genes and Risk of Nonfatal Cerebral Stroke in Patients Suffering from Essential Hypertension. J. Mol. Neurosci. 2012, 47, 511–513. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and Modifications of Primer Design Program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef]
- Johnson, J.L.; Abecasis, G.R. GAS Power Calculator: Web-Based Power Calculator for Genetic Association Studies. BioRxiv 2017. [Google Scholar] [CrossRef]
- THE GTEX CONSORTIUM. The GTEx Consortium Atlas of Genetic Regulatory Effects across Human Tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Võsa, U.; Claringbould, A.; Westra, H.-J.; Bonder, M.J.; Deelen, P.; Zeng, B.; Kirsten, H.; Saha, A.; Kreuzhuber, R.; Kasela, S. Unraveling the Polygenic Architecture of Complex Traits Using Blood eQTL Metaanalysis. BioRxiv 2018, 447367. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg: A Resource for Exploring Chromatin States, Conservation, and Regulatory Motif Alterations within Sets of Genetically Linked Variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef] [PubMed]
- von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and Predicted Protein–Protein Associations, Integrated and Transferred across Organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA Update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef]
- Shin, S.; Hudson, R.; Harrison, C.; Craven, M.; Keleş, S. atSNP Search: A Web Resource for Statistically Evaluating Influence of Human Genetic Variation on Transcription Factor Binding. Bioinformatics 2019, 35, 2657–2659. [Google Scholar] [CrossRef]
- Consortium, G.O. The Gene Ontology Resource: 20 Years and Still GOing Strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Crawford, K.M.; Gallego-Fabrega, C.; Kourkoulis, C.; Miyares, L.; Marini, S.; Flannick, J.; Burtt, N.P.; von Grotthuss, M.; Alexander, B.; Costanzo, M.C.; et al. Cerebrovascular Disease Knowledge Portal. Stroke 2018, 49, 470–475. [Google Scholar] [CrossRef]
- Lin, C.-H.; Kuo, Y.-W.; Kuo, C.-Y.; Huang, Y.-C.; Hsu, C.-Y.; Hsu, H.-L.; Lin, Y.-H.; Wu, C.-Y.; Huang, Y.-C.; Lee, M.; et al. Shortened Activated Partial Thromboplastin Time Is Associated With Acute Ischemic Stroke, Stroke Severity, and Neurological Worsening. J. Stroke Cerebrovasc. Dis. 2015, 24, 2270–2276. [Google Scholar] [CrossRef]
- Song, M.-S.; Choi, Y.J.; Kim, H.; Nam, M.J.; Lee, C.; Han, K.; Jung, J.-H.; Park, Y.-G.; Kim, D.-H.; Park, J.-H. Relationship between Blood Pressure Levels and Ischemic Stroke, Myocardial Infarction, and Mortality in Very Elderly Patients Taking Antihypertensives: A Nationwide Population-Based Cohort Study. BMC Geriatr. 2021, 21, 620. [Google Scholar] [CrossRef]
- Hoshino, T.; Ishizuka, K.; Toi, S.; Mizuno, T.; Nishimura, A.; Takahashi, S.; Wako, S.; Kitagawa, K. Atherogenic Dyslipidemia and Residual Vascular Risk After Stroke or Transient Ischemic Attack. Stroke 2022, 53, 79–86. [Google Scholar] [CrossRef]
- Khoshnam, S.E.; Winlow, W.; Farzaneh, M.; Farbood, Y.; Moghaddam, H.F. Pathogenic Mechanisms following Ischemic Stroke. Neurol. Sci. 2017, 38, 1167–1186. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-Q.; Chen, W.; Yan, M.-H.; Lai, J.-J.; Tang, N.; Wu, L. Ischemic Preconditioning Protects Brain from Ischemia/Reperfusion Injury by Attenuating Endoplasmic Reticulum Stress-Induced Apoptosis through PERK Pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 5736–5744. [Google Scholar] [PubMed]
- Bushueva, O.; Barysheva, E.; Markov, A.; Belykh, A.; Koroleva, I.; Churkin, E.; Polonikov, A.; Ivanov, V.; Nazarenko, M. DNA Hypomethylation of the MPO Gene in Peripheral Blood Leukocytes Is Associated with Cerebral Stroke in the Acute Phase. J. Mol. Neurosci. 2021, 71, 1914–1932. [Google Scholar] [CrossRef]
- Soldatov, V.O. Hypoxia-Inducible Factor: Basic Biology and Involvement in Cardiovascular Pathology. Asian J. Pharm. (AJP) 2018, 12. [Google Scholar] [CrossRef]
- Doyle, K.P.; Simon, R.P.; Stenzel-Poore, M.P. Mechanisms of Ischemic Brain Damage. Neuropharmacology 2008, 55, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Jariel-Encontre, I.; Bossis, G.; Piechaczyk, M. Ubiquitin-Independent Degradation of Proteins by the Proteasome. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2008, 1786, 153–177. [Google Scholar] [CrossRef]
- Thiebaut, A.M.; Hedou, E.; Marciniak, S.J.; Vivien, D.; Roussel, B.D. Proteostasis During Cerebral Ischemia. Front. Neurosci. 2019, 13. [Google Scholar] [CrossRef]
- Guiraud, S.P.; Montoliu, I.; Da Silva, L.; Dayon, L.; Galindo, A.N.; Corthésy, J.; Kussmann, M.; Martin, F.-P. High-Throughput and Simultaneous Quantitative Analysis of Homocysteine–Methionine Cycle Metabolites and Co-Factors in Blood Plasma and Cerebrospinal Fluid by Isotope Dilution LC–MS/MS. Anal. Bioanal. Chem. 2017, 409, 295–305. [Google Scholar] [CrossRef]
- Zheng, J.; Xiao, H.; Duan, Y.; Song, B.; Zheng, C.; Guo, Q.; Li, F.; Li, T. Roles of Amino Acid Derivatives in the Regulation of Obesity. Food Funct. 2021, 12, 6214–6225. [Google Scholar] [CrossRef]
- Forde, N.; Duffy, G.B.; McGettigan, P.A.; Browne, J.A.; Mehta, J.P.; Kelly, A.K.; Mansouri-Attia, N.; Sandra, O.; Loftus, B.J.; Crowe, M.A.; et al. Evidence for an Early Endometrial Response to Pregnancy in Cattle: Both Dependent upon and Independent of Interferon Tau. Physiol. Genom. 2012, 44, 799–810. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Z.; Zeng, Y.; Zhou, L.; Long, Q.; Hassan, I.U.; Zhang, Y.; Qi, X.; Cai, D.; Mao, B.; et al. ZSWIM4 Regulates Embryonic Patterning and BMP Signaling by Promoting Nuclear Smad1 Degradation. EMBO Rep 2024, 25, 646–671. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Yin, G.; Hu, Y.; Shi, S.; Jiang, J.; Song, X.; Zhang, Z.; Wei, Z.; Tang, C.; Lyu, H. Coicis Semen Protects against Focal Cerebral Ischemia-Reperfusion Injury by Inhibiting Oxidative Stress and Promoting Angiogenesis via the TGFβ/ALK1/Smad1/5 Signaling Pathway. Aging 2020, 13, 877–893. [Google Scholar] [CrossRef] [PubMed]
- González-Núñez, M.; Muñoz-Félix, J.M.; López-Novoa, J.M. The ALK-1/Smad1 Pathway in Cardiovascular Physiopathology. A New Target for Therapy? Biochim. Biophys. Acta 2013, 1832, 1492–1510. [Google Scholar] [CrossRef] [PubMed]
- Azad, A.K.; Chakrabarti, S.; Xu, Z.; Davidge, S.T.; Fu, Y. Coiled-Coil Domain Containing 3 (CCDC3) Represses Tumor Necrosis Factor-α/Nuclear Factor κB-Induced Endothelial Inflammation. Cell. Signal. 2014, 26, 2793–2800. [Google Scholar] [CrossRef]
- Kobayashi, S.; Fukuhara, A.; Taguchi, T.; Matsuda, M.; Tochino, Y.; Otsuki, M.; Shimomura, I. Identification of a New Secretory Factor, CCDC3/Favine, in Adipocytes and Endothelial Cells. Biochem. Biophys. Res. Commun. 2010, 392, 29–35. [Google Scholar] [CrossRef]
- Kobayashi, S.; Kita, S.; Okuzaki, D.; Fujishima, Y.; Otsuki, M.; Kato, H.; Nishizawa, Y.; Miyashita, K.; Yokoyama, C.; Fukuhara, A. Favine/CCDC3 Deficiency Accelerated Atherosclerosis and Thrombus Formation Is Associated with Decreased MEF2C-KLF2 Pathway. iScience 2022, 25, 105252. [Google Scholar] [CrossRef]
- Selhub, J.; Troen, A.M. Sulfur Amino Acids and Atherosclerosis: A Role for Excess Dietary Methionine. Ann. N. Y. Acad. Sci. 2016, 1363, 18–25. [Google Scholar] [CrossRef]
- Vinknes, K.J.; Refsum, H.; Turner, C.; Khaw, K.-T.; Wareham, N.J.; Forouhi, N.G.; Imamura, F. Plasma Sulfur Amino Acids and Risk of Cerebrovascular Diseases: A Nested Case-Control Study in the EPIC-Norfolk Cohort. Stroke 2021, 52, 172–180. [Google Scholar] [CrossRef]
- Sikora, M.; Bretes, E.; Perła-Kaján, J.; Utyro, O.; Borowczyk, K.; Piechocka, J.; Głowacki, R.; Wojtasz, I.; Kaźmierski, R.; Jakubowski, H. Homocysteine Thiolactone and Other Sulfur-Containing Amino Acid Metabolites Are Associated with Fibrin Clot Properties and the Risk of Ischemic Stroke. Sci. Rep. 2024, 14, 11222. [Google Scholar] [CrossRef]
- Paterson, P.G.; Lyon, A.W.; Kamencic, H.; Andersen, L.B.; Juurlink, B.H. Sulfur Amino Acid Deficiency Depresses Brain Glutathione Concentration. Nutr. Neurosci. 2001, 4, 213–222. [Google Scholar] [CrossRef]
- Solodilova, M.; Drozdova, E.; Azarova, I.; Klyosova, E.; Bykanova, M.; Bushueva, O.; Polonikova, A.; Churnosov, M.; Polonikov, A. The Discovery of GGT1 as a Novel Gene for Ischemic Stroke Conferring Protection against Disease Risk in Non-Smokers and Non-Abusers of Alcohol. J. Stroke Cerebrovasc. Dis. 2024, 33, 107685. [Google Scholar] [CrossRef] [PubMed]
- Elshorbagy, A.K.; Kozich, V.; Smith, A.D.; Refsum, H. Cysteine and Obesity: Consistency of the Evidence across Epidemiologic, Animal and Cellular Studies. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-H.; Kim, D.; Kim, S.T.; Jeong, S.; Kim, J.L.; Shim, S.M.; Heo, A.J.; Song, X.; Guo, Z.S.; Bartlett, D.L. PARK7 Modulates Autophagic Proteolysis through Binding to the N-Terminally Arginylated Form of the Molecular Chaperone HSPA5. Autophagy 2018, 14, 1870–1885. [Google Scholar] [CrossRef] [PubMed]
- Rudnick, N.D.; Griffey, C.J.; Guarnieri, P.; Gerbino, V.; Wang, X.; Piersaint, J.A.; Tapia, J.C.; Rich, M.M.; Maniatis, T. Distinct Roles for Motor Neuron Autophagy Early and Late in the SOD1G93A Mouse Model of ALS. Proc. Natl. Acad. Sci. USA 2017, 114, E8294–E8303. [Google Scholar] [CrossRef] [PubMed]
- Gurkar, A.U.; Chu, K.; Raj, L.; Bouley, R.; Lee, S.-H.; Kim, Y.-B.; Dunn, S.E.; Mandinova, A.; Lee, S.W. Identification of ROCK1 Kinase as a Critical Regulator of Beclin1-Mediated Autophagy during Metabolic Stress. Nat. Commun. 2013, 4, 2189. [Google Scholar] [CrossRef]
- Azarova, I.E.; Gureeva, A.V.; Postnikova, M.I.; Makarenko, V.V.; Klyosova, E.Y.; Polonikov, A.V. The Link of Single Nucleotide Polymorphism Rs4880 of the SOD2 Gene to the Development of Microvascular Complications of Type 2 Diabetes Mellitus. Res. Results Biomed. 2023, 9, 461–473. [Google Scholar]
- Patel, H.; Alkhawam, H.; Madanieh, R.; Shah, N.; Kosmas, C.E.; Vittorio, T.J. Aerobic vs Anaerobic Exercise Training Effects on the Cardiovascular System. World J. Cardiol. 2017, 9, 134. [Google Scholar] [CrossRef]
- Szyller, J.; Bil-Lula, I. Heat Shock Proteins in Oxidative Stress and Ischemia/Reperfusion Injury and Benefits from Physical Exercises: A Review to the Current Knowledge. Oxidative Med. Cell. Longev. 2021, 2021, 6678457. [Google Scholar] [CrossRef]
- Kochanowicz, A.; Sawczyn, S.; Niespodziński, B.; Mieszkowski, J.; Kochanowicz, K.; Żychowska, M. Cellular Stress Response Gene Expression During Upper and Lower Body High Intensity Exercises. PLoS ONE 2017, 12, e0171247. [Google Scholar] [CrossRef]
- Kalmar, B.; Greensmith, L. Induction of Heat Shock Proteins for Protection against Oxidative Stress. Adv. Drug Deliv. Rev. 2009, 61, 310–318. [Google Scholar] [CrossRef]
- Yenari, M.A.; Liu, J.; Zheng, Z.; Vexler, Z.S.; Lee, J.E.; Giffard, R.G. Antiapoptotic and Anti-inflammatory Mechanisms of Heat-shock Protein Protection. Ann. N. Y. Acad. Sci. 2005, 1053, 74–83. [Google Scholar] [PubMed]
- Chen, Y.; Voegeli, T.S.; Liu, P.P.; Noble, E.G.; Currie, R.W. Heat Shock Paradox and a New Role of Heat Shock Proteins and Their Receptors as Anti-Inflammation Targets. Inflamm. Allergy-Drug Targets (Former. Curr. Drug Targets-Inflamm. Allergy)(Discontin.) 2007, 6, 91–100. [Google Scholar]
- Cotman, C.W.; Berchtold, N.C.; Christie, L.-A. Exercise Builds Brain Health: Key Roles of Growth Factor Cascades and Inflammation. Trends Neurosci. 2007, 30, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.M.; Montgomery, M.H.; Gregory, E.J.; Berman, N.E. Exercise Preconditioning Improves Traumatic Brain Injury Outcomes. Brain Res. 2015, 1622, 414–429. [Google Scholar] [CrossRef]
- Carro, E.; Nuñez, A.; Busiguina, S.; Torres-Aleman, I. Circulating Insulin-like Growth Factor I Mediates Effects of Exercise on the Brain. J. Neurosci. 2000, 20, 2926–2933. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, L.; Yang, X.; Wan, Y.; Jia, J. The Effects of Exercise Preconditioning on Cerebral Blood Flow Change and Endothelin-1 Expression after Cerebral Ischemia in Rats. J. Stroke Cerebrovasc. Dis. 2014, 23, 1696–1702. [Google Scholar] [CrossRef]
- Otsuka, S.; Sakakima, H.; Sumizono, M.; Takada, S.; Terashi, T.; Yoshida, Y. The Neuroprotective Effects of Preconditioning Exercise on Brain Damage and Neurotrophic Factors after Focal Brain Ischemia in Rats. Behav. Brain Res. 2016, 303, 9–18. [Google Scholar] [CrossRef]
- Pikula, A.; Beiser, A.S.; Chen, T.C.; Preis, S.R.; Vorgias, D.; DeCarli, C.; Au, R.; Kelly-Hayes, M.; Kase, C.S.; Wolf, P.A. Serum Brain–Derived Neurotrophic Factor and Vascular Endothelial Growth Factor Levels Are Associated with Risk of Stroke and Vascular Brain Injury: Framingham Study. Stroke 2013, 44, 2768–2775. [Google Scholar] [CrossRef]
- Strazzullo, P.; D’Elia, L.; Cairella, G.; Garbagnati, F.; Cappuccio, F.P.; Scalfi, L. Excess Body Weight and Incidence of Stroke: Meta-Analysis of Prospective Studies with 2 Million Participants. Stroke 2010, 41, e418–e426. [Google Scholar] [CrossRef]
- Wang, X.; Huang, Y.; Chen, Y.; Yang, T.; Su, W.; Chen, X.; Yan, F.; Han, L.; Ma, Y. The Relationship between Body Mass Index and Stroke: A Systemic Review and Meta-Analysis. J. Neurol. 2022, 269, 6279–6289. [Google Scholar] [CrossRef]
- Shiozawa, M.; Kaneko, H.; Itoh, H.; Morita, K.; Okada, A.; Matsuoka, S.; Kiriyama, H.; Kamon, T.; Fujiu, K.; Michihata, N.; et al. Association of Body Mass Index with Ischemic and Hemorrhagic Stroke. Nutrients 2021, 13, 2343. [Google Scholar] [CrossRef] [PubMed]

| Baseline and Clinical Characteristics | IS Patients (n = 947) | Controls (n = 1308) | p-Value | |
|---|---|---|---|---|
| Age, Me [Q1; Q3] | 63 [55; 70] | 59 [53; 66] | <0.001 | |
| Gender | Males, N (%) | 524 (55.33%) | 614 (46.9%) | <0.001 |
| Females, N (%) | 423 (44.67%) | 694 (53.1%) | ||
| Smoking | Yes, N (%) | 469 (49.52%) | 413 (31.6%) | <0.001 |
| No, N (%) | 478 (50.48%) | 895 (68.4%) | ||
| Low physical activity | Yes, N (%) | 354 (39.91%) | ND | |
| No, N (%) | 533 (60.09%) | |||
| Low fruit/vegetable consumption | Yes, N (%) | 416 (46.9%) | ND | |
| No, N (%) | 471 (53.1%) | |||
| Coronary artery disease | Yes, N (%) | 116 (13.08%) | - | |
| No, N (%) | 771 (86.92%) | - | ||
| Obesity | Yes, N (%) | 61 (6.4%) | ND | |
| No, N (%) | 530 (56%) | |||
| ND, N (%) | 356 (37.6%) | |||
| Family history of cerebrovascular diseases | Yes, N (%) | 312 (35.29%) | ND | |
| No, N (%) | 572 (64.71%) | ND | ||
| Age at onset of stroke, Me [Q1; Q3] | 61 [54; 70] (n = 904) | - | ||
| Number of strokes including event in question | 1, N (%) | 800 (88.59%) | - | |
| 2, N (%) | 89 (9.86%) | - | ||
| 3, N (%) | 14 (1.55%) | - | ||
| Stroke localization | Right/left middle cerebral artery basin, N (%) | 750 (83.24%) | - | |
| Vertebrobasilar basin, N (%) | 151 (16.76%) | - | ||
| Area of lesion in stroke, mm2, Me [Q1; Q3] | 105 [30; 468] (n = 883) | - | ||
| Total cholesterol, mmol/L, Me [Q1; Q3] | 5.2 [4.4; 5.9] (n = 608) | ND | ||
| Triglycerides, mmol/L, Me [Q1; Q3] | 1.3 [1.1; 1.8] (n = 601) | ND | ||
| Glucose level, mmol/L, Me [Q1; Q3] | 4.8 [4.3; 5.5] (n = 892) | ND | ||
| Prothrombin time, seconds, Me [Q1; Q3] | 10.79 [10.14; 11.70] (n = 880) | ND | ||
| International normalized ratio, Me [Q1; Q3] | 1 [0.94; 1.09] (n = 596) | ND | ||
| Activated partial thromboplastin time, seconds, Me [Q1; Q3] | 32.7 [29; 37] (n = 599) | ND | ||
| SNP | N | Dominant Model | Recessive Model | Log-Additive Model | N | Dominant Model | Recessive Model | Log-Additive Model | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) 1 | P (Pbonf) | OR (95% CI) 1 | P (Pbonf) | OR (95% CI) 1 | P (Pbonf) | OR (95% CI) 1 | P (Pbonf) | OR (95% CI) 1 | P (Pbonf) | OR (95% CI) 1 | P (Pbonf) | |||
| BMI ≥ 30 | BMI < 30 | |||||||||||||
| rs10104 C19orf53 (A/G) | 1286 | 1.52 [0.90–2.55] | 0.12 | 4.70 [2.25–9.82] | 0.0003 (0.0009) | 1.82 [1.22–2.72] | 0.004 (0.01) | 1745 | 0.94 [0.76–1.16] | 0.57 (1.0) | 1.60 [1.02–2.51] | 0.046 (1.0) | 1.02 [0.86–1.22] | 0.79 (1.0) |
| rs11666524 C19orf53 (G/A) | 1330 | 1.54 [0.92–2.58] | 0.098 | 3.88 [1.88–8.02] | 0.001 (0.003) | 1.76 [1.19–2.60] | 0.006 (0.02) | 1792 | 0.94 [0.76–1.15] | 0.55 (1.0) | 1.64 [1.09–2.48] | 0.02 (0.06) | 1.04 [0.88–1.23] | 0.68 (1.0) |
| rs346158 C19orf53 (T/C) | 1301 | 1.40 [0.84–2.35] | 0.2 | 3.73 [1.81–7.69] | 0.002 (0.006) | 1.65 [1.11–2.44] | 0.015 (0.045) | 1769 | 0.90 [0.73–1.11] | 0.32 (0.96) | 1.60 [1.06–2.40] | 0.03 (0.09) | 1.01 [0.85–1.19] | 0.94 (1.0) |
| rs2277947 C19orf53 (G/A) | 1242 | 1.60 [0.95–2.70] | 0.079 | 4.11 [1.98–8.53] | 0.0008 (0.002) | 1.82 [1.22–2.70] | 0.004 (0.01) | 1699 | 0.98 [0.80–1.21] | 0.87 (1.0) | 1.60 [1.05–2.45] | 0.03 (0.09) | 1.06 [0.90–1.26] | 0.48 (1.0) |
| Low physical activity level (f+) | Normal physical activity level (f−) | |||||||||||||
| rs10104 C19orf53 (A/G) | 1597 | 0.86 [0.67–1.09] | 0.21 (0.63) | 1.91 [1.19–3.07] | 0.009 (0.03) | 1.00 [0.82–1.22] | 0.99 (2.3) | 1774 | 0.93 [0.76–1.15] | 0.5 (1.5) | 1.14 [0.71–1.83] | 0.6 (1.8) | 0.97 [0.81–1.15] | 0.7 (2.1) |
| rs11666524 C19orf53 (G/A) | 1623 | 0.85 [0.67–1.08] | 0.18 (0.5) | 1.80 [1.13–2.84] | 0.016 (0.048) | 0.99 [0.81–1.20] | 0.91 (2.7) | 1798 | 0.97 [0.79–1.20] | 0.79 (2.4) | 1.29 [0.83–2.00] | 0.26 (0.8) | 1.02 [0.86–1.21] | 0.84 (2.5) |
| Low fruit/vegetable intake (f+) | Normal fruit/vegetable intake (f−) | |||||||||||||
| rs11666524 C19orf53 (G/A) | 1739 | 0.97 [0.78–1.21] | 0.81 (2.4) | 1.81 [1.20–2.75] | 0.006 (0.02) | 1.08 [0.91–1.29] | 0.37 (1.1) | 1682 | 0.86 [0.69–1.09] | 0.21 (0.6) | 1.13 [0.68–1.86] | 0.64 (1.9) | 0.92 [0.76–1.11] | 0.38 (1.1) |
| rs346157 C19orf53 (A/G) | 1752 | 0.91 [0.73–1.13] | 0.4 (1.2) | 1.42 [1.08–1.86] | 0.012 (0.036) | 1.06 [0.91–1.24] | 0.43 (1.3) | 1699 | 1.04 [0.82–1.31] | 0.76 (2.3) | 1.14 [0.85–1.53] | 0.4 (1.2) | 1.06 [0.90–1.24] | 0.51 (1.5) |
| rs346158 C19orf53 (T/C) | 1740 | 0.96 [0.77–1.19] | 0.71 (2.1) | 1.93 [1.29–2.89] | 0.0017 (0.005) | 1.09 [0.92–1.30] | 0.33 (1.0) | 1687 | 0.86 [0.68–1.08] | 0.18 (0.5) | 1.07 [0.65–1.76] | 0.79 (2.4) | 0.91 [0.75–1.10] | 0.31 (0.9) |
| rs2277947 C19orf53 (G/A) | 1668 | 1.02 [0.82–1.27] | 0.86 (2.6) | 1.81 [1.18–2.76] | 0.0074 (0.02) | 1.12 [0.93–1.33] | 0.23 (0.7) | 1604 | 0.90 [0.71–1.14] | 0.38 (1.1) | 1.24 [0.75–2.04] | 0.4 (1.2) | 0.96 [0.79–1.17] | 0.68 (2.0) |
| SNP | Assessed Allele | Other Allele | Gene | p-Value | FDR | Z-Score | N Samples |
|---|---|---|---|---|---|---|---|
| rs10104 C19orf53 | |||||||
| rs10104 | G | A | C19orf53 | 3.27 × 10−310 | 0 | −46.0132 | 27007 |
| rs10104 | G | A | MRI1 | 3.27 × 10−310 | 0 | −38.6135 | 26793 |
| rs10104 | G | A | CCDC130 | 1.10 × 10−19 | 0 | 9.0787 | 27007 |
| rs10104 | G | A | ZSWIM4 | 8.02 × 10−11 | 0 | 6.5002 | 21718 |
| rs11666524 C19orf53 | |||||||
| rs11666524 | A | G | C19orf53 | 3.27 × 10−310 | 0 | −46.2734 | 27279 |
| rs11666524 | A | G | MRI1 | 3.27 × 10−310 | 0 | −39.214 | 27065 |
| rs11666524 | A | G | CCDC130 | 6.37 × 10−20 | 0 | 9.1379 | 27279 |
| rs11666524 | A | G | ZSWIM4 | 5.99 × 10−11 | 0 | 6.5441 | 21990 |
| rs346157 C19orf53 | |||||||
| rs346157 | G | A | C19orf53 | 2.14 × 10−214 | 0 | −31.2513 | 27274 |
| rs346157 | G | A | MRI1 | 9.14 × 10−150 | 0 | −26.0647 | 27060 |
| rs346157 | G | A | CCDC130 | 2.22 × 10−36 | 0 | 12.5961 | 27274 |
| rs346158 C19orf53 | |||||||
| rs346158 | C | T | C19orf53 | 3.27 × 10−310 | 0 | −44.6143 | 26890 |
| rs346158 | C | T | MRI1 | 3.27 × 10−310 | 0 | −38.1044 | 26676 |
| rs346158 | C | T | CCDC130 | 9.10 × 10−18 | 0 | 8.5849 | 26890 |
| rs346158 | C | T | ZSWIM4 | 3.91 × 10−11 | 0 | 6.6077 | 21601 |
| rs2277947 C19orf53 | |||||||
| rs2277947 | A | G | C19orf53 | 3.27 × 10−310 | 0 | −46.393 | 27395 |
| rs2277947 | A | G | MRI1 | 3.27 × 10−310 | 0 | −39.1083 | 27181 |
| rs2277947 | A | G | CCDC130 | 5.30 × 10−20 | 0 | 9.1579 | 27395 |
| rs2277947 | A | G | ZSWIM4 | 3.56 × 10−11 | 0 | 6.6213 | 22106 |
| SNP | GTEx Portal Data (https://gtexportal.org) | |||
|---|---|---|---|---|
| Gene Expressed | p-Value | Effect (NES) | Tissue | |
| rs10104 C19orf53 (A/G) | C19orf53 | 2.7 × 10−18 | ↓ (−0.23) | Artery—Aorta |
| MRI1 | 3.0 × 10−5 | ↓ (−0.27) | Artery—Aorta | |
| C19orf53 | 8.9 × 10−18 | ↓ (−0.17) | Artery—Tibial | |
| MRI1 | 1.0 × 10−5 | ↓ (−0.23) | Artery—Tibial | |
| C19orf53 | 9.4 × 10−7 | ↓ (−0.33) | Brain—Amygdala | |
| C19orf53 | 5.1 × 10−8 | ↓ (−0.34) | Brain—Anterior cingulate cortex (BA24) | |
| C19orf53 | 2.5 × 10−18 | ↓ (−0.36) | Brain—Caudate (basal ganglia) | |
| C19orf53 | 2.8 × 10−6 | ↓ (−0.27) | Brain—Cerebellum | |
| C19orf53 | 4.9 × 10−15 | ↓ (−0.42) | Brain—Cortex | |
| MRI1 | 1.7 × 10−5 | ↓ (−0.39) | Brain—Cortex | |
| C19orf53 | 1.2 × 10−15 | ↓ (−0.38) | Brain—Frontal Cortex (BA9) | |
| C19orf53 | 6.2 × 10−12 | ↓ (−0.31) | Brain—Hippocampus | |
| C19orf53 | 3.6 × 10−7 | ↓ (−0.24) | Brain—Hypothalamus | |
| C19orf53 | 3.0 × 10−15 | ↓ (−0.33) | Brain—Nucleus accumbens (basal ganglia) | |
| C19orf53 | 1.3 × 10−8 | ↓ (−0.3) | Brain—Putamen (basal ganglia) | |
| C19orf53 | 6.4 × 10−12 | ↓ (−0.29) | Pituitary | |
| C19orf53 | 2.5 × 10−11 | ↓ (−0.15) | Adipose—Subcutaneous | |
| MRI1 | 9.1 × 10−6 | ↓ (−0.21) | Adipose—Subcutaneous | |
| C19orf53 | 2.3 × 10−9 | ↓ (−0.15) | Adipose—Visceral (Omentum) | |
| rs11666524 C19orf53 (G/A) | C19orf53 | 5.5 × 10−18 | ↓ (−0.23) | Artery—Aorta |
| MRI1 | 2.1 × 10−5 | ↓ (−0.27) | Artery—Aorta | |
| C19orf53 | 1.7 × 10−17 | ↓ (−0.17) | Artery—Tibial | |
| MRI1 | 9.3 × 10−6 | ↓ (−0.23) | Artery—Tibial | |
| C19orf53 | 9.3 × 10−7 | ↓ (−0.33) | Brain—Amygdala | |
| C19orf53 | 5.6 × 10−8 | ↓ (−0.34) | Brain—Anterior cingulate cortex (BA24) | |
| C19orf53 | 7.8 × 10−19 | ↓ (−0.36) | Brain—Caudate (basal ganglia) | |
| C19orf53 | 2.7 × 10−6 | ↓ (−0.27) | Brain—Cerebellum | |
| C19orf53 | 8.7 × 10−16 | ↓ (−0.42) | Brain—Cortex | |
| MRI1 | 2.8 × 10−5 | ↓ (−0.38) | Brain—Cortex | |
| C19orf53 | 1.0 × 10−15 | ↓ (−0.37) | Brain—Frontal Cortex (BA9) | |
| C19orf53 | 1.4 × 10−10 | ↓ (−0.3) | Brain—Hippocampus | |
| C19orf53 | 4.3 × 10−7 | ↓ (−0.23) | Brain—Hypothalamus | |
| C19orf53 | 1.9 × 10−14 | ↓ (−0.32) | Brain—Nucleus accumbens (basal ganglia) | |
| C19orf53 | 8.6 × 10−9 | ↓ (−0.3) | Brain—Putamen (basal ganglia) | |
| C19orf53 | 8.5 × 10−12 | ↓ (−0.29) | Pituitary | |
| C19orf53 | 1.6 × 10−11 | ↓ (−0.16) | Adipose—Subcutaneous | |
| MRI1 | 6.1 × 10−6 | ↓ (−0.21) | Adipose—Subcutaneous | |
| C19orf53 | 5.1 × 10−9 | ↓ (−0.14) | Adipose—Visceral (Omentum) | |
| rs346157 C19orf53 (A/G) | C19orf53 | 6.2 × 10−10 | ↓ (−0.15) | Artery—Aorta |
| C19orf53 | 3.2 × 10−10 | ↓ (−0.11) | Artery—Tibial | |
| C19orf53 | 2.1 × 10−7 | ↓ (−0.27) | Brain—Cortex | |
| C19orf53 | 3.9 × 10−6 | ↓ (−0.22) | Brain—Frontal Cortex (BA9) | |
| C19orf53 | 2.5 × 10−5 | ↓ (−0.17) | Brain—Nucleus accumbens (basal ganglia) | |
| C19orf53 | 5.9 × 10−8 | ↓ (−0.2) | Pituitary | |
| C19orf53 | 9.6 × 10−8 | ↓ (−0.11) | Adipose—Subcutaneous | |
| rs346158 C19orf53 (T/C) | C19orf53 | 9.5 × 10−14 | ↓ (−0.19) | Artery—Aorta |
| MRI1 | 6.3 × 10−5 | ↓ (−0.25) | Artery—Aorta | |
| C19orf53 | 4.1 × 10−13 | ↓ (−0.14) | Artery—Tibial | |
| MRI1 | 5.2 × 10−5 | ↓ (−0.2) | Artery—Tibial | |
| C19orf53 | 1.1 × 10−5 | ↓ (−0.3) | Brain—Amygdala | |
| C19orf53 | 1.7 × 10−7 | ↓ (−0.32) | Brain—Anterior cingulate cortex (BA24) | |
| C19orf53 | 2.8 × 10−13 | ↓ (−0.31) | Brain—Caudate (basal ganglia) | |
| C19orf53 | 5.0 × 10−10 | ↓ (−0.36) | Brain—Cortex | |
| MRI1 | 8.9 × 10−7 | ↓ (−0.47) | Brain—Cortex | |
| C19orf53 | 3.2 × 10−15 | ↓ (−0.38) | Brain—Frontal Cortex (BA9) | |
| C19orf53 | 3.8 × 10−8 | ↓ (−0.26) | Brain—Hippocampus | |
| C19orf53 | 7.8 × 10−6 | ↓ (−0.21) | Brain—Hypothalamus | |
| C19orf53 | 3.8 × 10−10 | ↓ (−0.28) | Brain—Nucleus accumbens (basal ganglia) | |
| C19orf53 | 1.0 × 10−5 | ↓ (−0.25) | Brain—Putamen (basal ganglia) | |
| C19orf53 | 8.4 × 10−9 | ↓ (−0.25) | Pituitary | |
| C19orf53 | 4.4 × 10−10 | ↓ (−0.14) | Adipose—Subcutaneous | |
| MRI1 | 5.9 × 10−7 | ↓ (−0.23) | Adipose—Subcutaneous | |
| C19orf53 | 2.4 × 10−7 | ↓ (−0.13) | Adipose—Visceral (Omentum) | |
| rs2277947 C19orf53 (G/A) | C19orf53 | 5.5 × 10−18 | ↓ (−0.23) | Artery—Aorta |
| MRI1 | 2.1 × 10−5 | ↓ (−0.27) | Artery—Aorta | |
| C19orf53 | 1.7 × 10−17 | ↓ (−0.17) | Artery—Tibial | |
| MRI1 | 9.3 × 10−6 | ↓ (−0.23) | Artery—Tibial | |
| C19orf53 | 9.3 × 10−7 | ↓ (−0.33) | Brain—Amygdala | |
| C19orf53 | 9.9 × 10−8 | ↓ (−0.34) | Brain—Anterior cingulate cortex (BA24) | |
| C19orf53 | 5.2 × 10−18 | ↓ (−0.36) | Brain—Caudate (basal ganglia) | |
| C19orf53 | 3.0 × 10−6 | ↓ (−0.27) | Brain—Cerebellum | |
| C19orf53 | 1.1 × 10−14 | ↓ (−0.41) | Brain—Cortex | |
| MRI1 | 1.0 × 10−5 | ↓ (−0.4) | Brain—Cortex | |
| C19orf53 | 4.7 × 10−15 | ↓ (−0.37) | Brain—Frontal Cortex (BA9) | |
| C19orf53 | 1.4 × 10−10 | ↓ (−0.3) | Brain—Hippocampus | |
| C19orf53 | 4.3 × 10−7 | ↓ (−0.23) | Brain—Hypothalamus | |
| C19orf53 | 1.9 × 10−14 | ↓ (−0.32) | Brain—Nucleus accumbens (basal ganglia) | |
| C19orf53 | 2.4 × 10−8 | ↓ (−0.3) | Brain—Putamen (basal ganglia) | |
| C19orf53 | 8.5 × 10−12 | ↓ (−0.29) | Pituitary | |
| C19orf53 | 4.8 × 10−11 | ↓ (−0.15) | Adipose—Subcutaneous | |
| MRI1 | 5.1 × 10−6 | ↓ (−0.21) | Adipose—Subcutaneous | |
| C19orf53 | 6.2 × 10−9 | ↓ (−0.14) | Adipose—Visceral (Omentum) | |
| SNP (Ref/Alt Allele) | Tissue Marks | Brain Regions | Peripheral Blood | Adipose | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (1) | (2) | (3) | (4) | (5) | (6) | (7) | (8) | (9) | ||
| rs10104 C19orf53 (A/G) | H3K4me1 | Enh | Enh | Enh | Enh | - | Enh | Enh | Enh | Enh |
| H3K4me3 | Pro | Pro | Pro | Pro | Pro | Pro | Pro | Pro | Pro | |
| H3K27ac | Enh | Enh | Enh | Enh | Enh | Enh | Enh | Enh | Enh | |
| H3K9ac | - | Pro | Pro | Pro | Pro | Pro | Pro | Pro | Pro | |
| DNase | - | - | - | - | - | - | - | DNase | - | |
| rs11666524 C19orf53 (G/A) | H3K4me1 | Enh | Enh | Enh | Enh | Enh | Enh | Enh | Enh | Enh |
| H3K4me3 | Pro | Pro | Pro | Pro | - | - | - | Pro | Pro | |
| H3K27ac | Enh | Enh | Enh | Enh | Enh | - | - | Enh | Enh | |
| H3K9ac | - | Pro | Pro | Pro | - | Pro | Pro | Pro | Pro | |
| DNase | - | - | - | - | - | - | - | - | - | |
| rs346157 C19orf53 (A/G) | H3K4me1 | - | Enh | Enh | - | Enh | Enh | Enh | Enh | - |
| H3K4me3 | - | - | - | - | - | - | - | Pro | Pro | |
| H3K27ac | Enh | Enh | Enh | Enh | Enh | - | - | Enh | Enh | |
| H3K9ac | - | Pro | - | Pro | - | - | - | Pro | - | |
| DNase | - | - | - | - | - | - | - | - | - | |
| rs346158 C19orf53 (T/C) | H3K4me1 | - | - | Enh | - | - | Enh | Enh | Enh | - |
| H3K4me3 | - | - | - | - | - | - | - | Pro | Pro | |
| H3K27ac | Enh | Enh | Enh | Enh | Enh | - | Enh | Enh | Enh | |
| H3K9ac | - | Pro | - | Pro | - | - | - | Pro | - | |
| DNase | - | - | - | - | - | - | - | - | - | |
| rs2277947 C19orf53 (G/A) | H3K4me1 | Enh | Enh | Enh | Enh | Enh | - | Enh | Enh | Enh |
| H3K4me3 | - | - | - | - | - | - | - | Pro | Pro | |
| H3K27ac | - | - | - | - | - | - | - | Enh | Enh | |
| H3K9ac | - | - | - | - | - | - | - | Pro | Pro | |
| DNase | - | - | - | - | - | - | - | - | - | |
| Tag SNPs | Phenotype | p-Value | Beta (OR) | Sample Size |
|---|---|---|---|---|
| rs10104 C19orf53 (A/G) | 1 All ischemic stroke | 0.015 | OR▲1.0173 | 1,047,650 |
| 1 Modified Rankin scale score 0–2 vs. 3–6 adj stroke severity | 0.04 | OR▲1.1224 | 5666 | |
| 2 Atrial fibrillation or flutter | 0.014 | OR▲1.0291 | 130,776 | |
| 2 Triglyceride-to-HDL ratio | 0.023 | Beta▲0.0058 | 579,558 | |
| 2 Serum ApoB | 0.028 | Beta▼−0.0048 | 450,527 | |
| rs11666524 C19orf53 (G/A) | 1 All ischemic stroke | 0.0004 | OR▲1.0214 | 794,817 |
| 1 Modified Rankin scale score 0–2 vs. 3–6 adj stroke severity | 0.029 | OR▲1.1324 | 5666 | |
| 1 Any stroke | 0.046 | OR▲1.0178 | 851,850 | |
| 2 Atrial fibrillation or flutter | 0.01 | OR▲1.0293 | 130,776 | |
| 2 Triglyceride-to-HDL ratio | 0.029 | Beta▲0.0075 | 418,488 | |
| 2 Serum ApoB | 0.03 | Beta▼−0.0047 | 436,068 | |
| rs346157 C19orf53 (A/G) | 1 All ischemic stroke | 0.005 | OR▲1.0707 | 38,850 |
| 1 Cerebral white matter hyperintensity volume | 0.033 | Beta▲0.0267 | 11,226 | |
| 2 Dyslipidemia | 0.01 | OR▲1.0360 | 56,375 | |
| 2 Obese vs. controls | 0.040 | OR▲1.0896 | 4752 | |
| rs346158 C19orf53 (T/C) | 1 Any stroke | 0.006 | OR▲1.0870 | 12,406 |
| 1 All ischemic stroke | 0.003 | OR▲1.0202 | 1,042,380 | |
| 1 Modified Rankin scale score 0–2 vs. 3–6 adj stroke severity | 0.012 | OR▲1.1511 | 5666 | |
| 1 Modified Rankin scale score 0–2 vs. 3–6 | 0.04 | OR▲1.1030 | 5802 | |
| 2 Triglyceride-to-HDL ratio | 0.01 | Beta▲0.0085 | 418,488 | |
| 2 Atrial fibrillation or flutter | 0.015 | OR▲1.0289 | 130,776 | |
| 2 Serum ApoB | 0.04 | Beta▼−0.0045 | 436,068 | |
| rs2277947 C19orf53 (G/A) | 1 All ischemic stroke | 0.005 | OR▲1.0193 | 1,062,920 |
| 1 Modified Rankin scale score 0–2 vs. 3–6 adj stroke severity | 0.03 | OR▲1.1308 | 5666 | |
| 2 Atrial fibrillation or flutter | 0.01 | OR▲1.0292 | 130,776 | |
| 2 Triglyceride-to-HDL ratio | 0.02 | Beta▲0.0079 | 418,488 | |
| 2 Serum ApoB | 0.035 | Beta▼−0.0047 | 436,068 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shilenok, I.; Kobzeva, K.; Deykin, A.; Pokrovsky, V.; Patrakhanov, E.; Bushueva, O. Obesity and Environmental Risk Factors Significantly Modify the Association between Ischemic Stroke and the Hero Chaperone C19orf53. Life 2024, 14, 1158. https://doi.org/10.3390/life14091158
Shilenok I, Kobzeva K, Deykin A, Pokrovsky V, Patrakhanov E, Bushueva O. Obesity and Environmental Risk Factors Significantly Modify the Association between Ischemic Stroke and the Hero Chaperone C19orf53. Life. 2024; 14(9):1158. https://doi.org/10.3390/life14091158
Chicago/Turabian StyleShilenok, Irina, Ksenia Kobzeva, Alexey Deykin, Vladimir Pokrovsky, Evgeny Patrakhanov, and Olga Bushueva. 2024. "Obesity and Environmental Risk Factors Significantly Modify the Association between Ischemic Stroke and the Hero Chaperone C19orf53" Life 14, no. 9: 1158. https://doi.org/10.3390/life14091158







