Association of Elevated Expression Levels of COL4A1 in Stromal Cells with an Immunosuppressive Tumor Microenvironment in Low-Grade Glioma, Pancreatic Adenocarcinoma, Skin Cutaneous Melanoma, and Stomach Adenocarcinoma
Abstract
:1. Introduction
2. Material and Methods
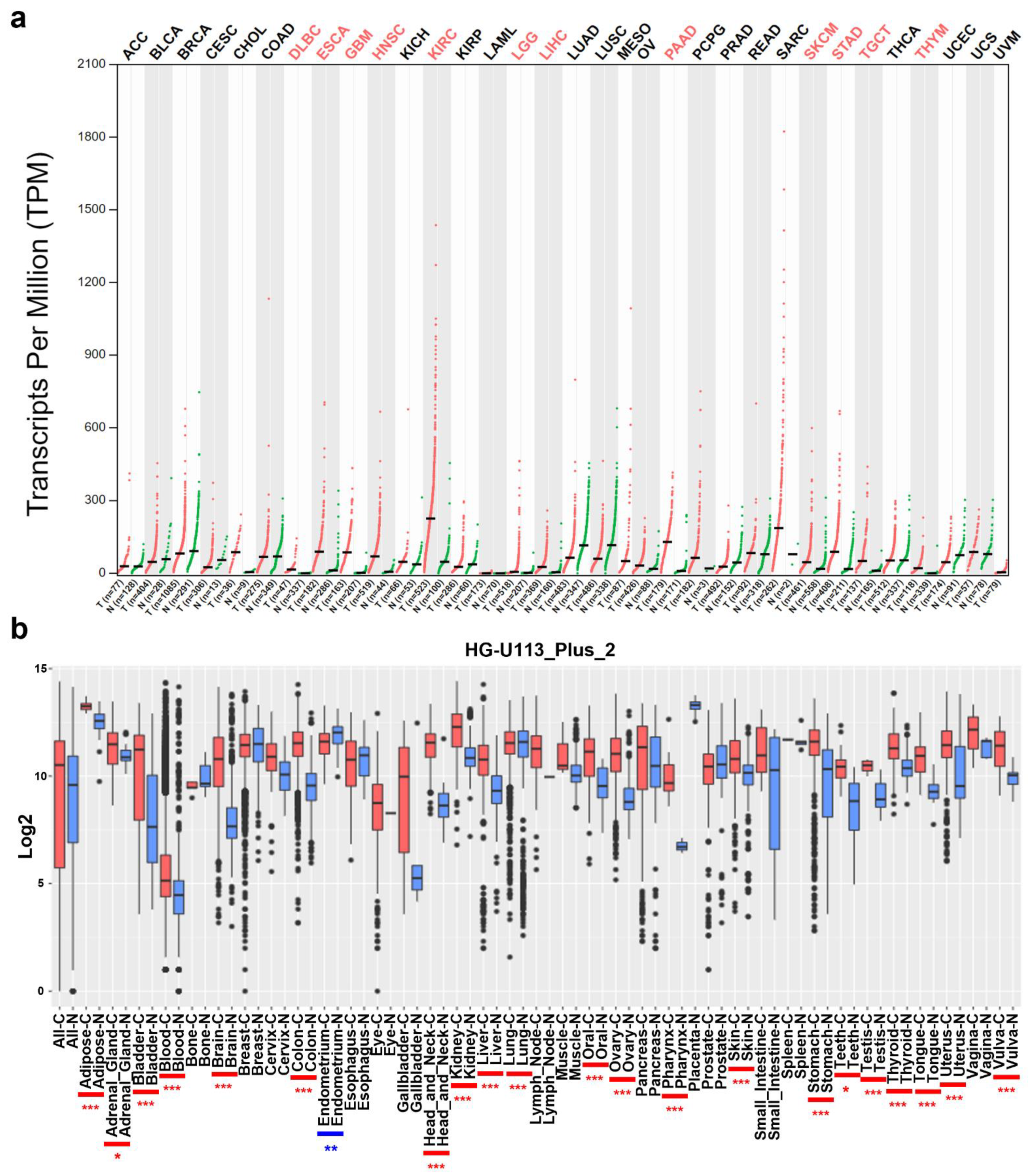
2.1. Analysis of COL4A1 mRNA Expression Levels between Cancer and Normal Tissues
2.2. Survival Analyses of Cancer Patient Groups with High and Low Expression Levels of COL4A1
2.3. Analysis of Heterogenic Expression of COL4A1 and Its Association with Infiltrated CAFs and Endothelial Cells
2.4. Analysis of Correlation between Tumor Infiltration and COL4A1 Expression
2.5. Analyses of Correlations among COL4A1 Expression Levels and Immune Cell Marker Gene and Cytokine Expression Levels
3. Results
3.1. mRNA Levels of COL4A1 in Various Types of Tumors
3.2. Analysis of Correlation between COL4A1 Expression and Patient Survival
3.3. High Expression Levels of COL4A1 in Infiltrated CAFs and TECs among Heterogeneous TME Cells
3.4. Correlation between COL4A1 Expression and Immune Cell Infiltration
3.5. Correlation between the Expression Levels of COL4A1 and Marker Genes Specific to Immune-Suppressive Subtypes
3.6. Correlation between the Expression Levels of COL4A1 and Immunosuppressive Cytokines
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alamowitch, S.; Plaisier, E.; Favrole, P.; Prost, C.; Chen, Z.; Van Agtmael, T.; Marro, B.; Ronco, P. Cerebrovascular disease related to COL4A1 mutations in HANAC syndrome. Neurology 2009, 73, 1873–1882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meuwissen, M.E.; Halley, D.J.; Smit, L.S.; Lequin, M.H.; Cobben, J.M.; de Coo, R.; van Harssel, J.; Sallevelt, S.; Woldringh, G.; van der Knaap, M.S.; et al. The expanding phenotype of COL4A1 and COL4A2 mutations: Clinical data on 13 newly identified families and a review of the literature. Genet. Med. 2015, 17, 843–853. [Google Scholar] [CrossRef] [Green Version]
- Kitzler, T.M.; Schneider, R.; Kohl, S.; Kolvenbach, C.M.; Connaughton, D.M.; Dai, R.; Mann, N.; Nakayama, M.; Majmundar, A.J.; Wu, C.W.; et al. COL4A1 mutations as a potential novel cause of autosomal dominant CAKUT in humans. Hum. Genet. 2019, 138, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Khoshnoodi, J.; Pedchenko, V.; Hudson, B.G. Mammalian collagen IV. Microsc. Res. Tech. 2008, 71, 357–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhang, J.; Chen, Y.; Sohel, H.; Ke, X.; Chen, J.; Li, Y.X. The correlation and role analysis of COL4A1 and COL4A2 in hepatocarcinogenesis. Aging (Albany NY) 2020, 12, 204–223. [Google Scholar] [CrossRef]
- Wang, T.; Jin, H.; Hu, J.; Li, X.; Ruan, H.; Xu, H.; Wei, L.; Dong, W.; Teng, F.; Gu, J.; et al. COL4A1 promotes the growth and metastasis of hepatocellular carcinoma cells by activating FAK-Src signaling. J. Exp. Clin. Cancer Res. 2020, 39, 148. [Google Scholar] [CrossRef]
- Jin, R.; Shen, J.; Zhang, T.; Liu, Q.; Liao, C.; Ma, H.; Li, S.; Yu, Z. The highly expressed COL4A1 genes contributes to the proliferation and migration of the invasive ductal carcinomas. Oncotarget 2017, 8, 58172–58183. [Google Scholar] [CrossRef] [Green Version]
- Miyake, M.; Hori, S.; Morizawa, Y.; Tatsumi, Y.; Toritsuka, M.; Ohnishi, S.; Shimada, K.; Furuya, H.; Khadka, V.S.; Deng, Y.; et al. Collagen type IV alpha 1 (COL4A1) and collagen type XIII alpha 1 (COL13A1) produced in cancer cells promote tumor budding at the invasion front in human urothelial carcinoma of the bladder. Oncotarget 2017, 8, 36099–36114. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Teng, X.; Liu, Z.; Zhang, L.; Liu, Z. Gene expression profile analyze the molecular mechanism of CXCR7 regulating papillary thyroid carcinoma growth and metastasis. J. Exp. Clin. Cancer Res. 2015, 34, 16. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.N.; Zhu, H.L.; Xia, M.T.; Liao, J.; Huang, X.T.; Xiao, J.W.; Yuan, C. A panel of collagen genes are associated with prognosis of patients with gastric cancer and regulated by microRNA-29c-3p: An integrated bioinformatics analysis and experimental validation. Cancer Manag. Res. 2019, 11, 4757–4772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Wang, N.N.; Chang, X.; Wang, S.L.; Wang, L.S.; Yao, J.; Li, Z.S.; Bai, Y. Bioinformatics analysis suggests that COL4A1 may play an important role in gastric carcinoma recurrence. J. Dig. Dis. 2019, 20, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Gu, W.; Sun, B.; Gao, L. Identification of COL4A1 as a potential gene conferring trastuzumab resistance in gastric cancer based on bioinformatics analysis. Mol. Med. Rep. 2018, 17, 6387–6396. [Google Scholar] [CrossRef]
- Yin, W.; Zhu, H.; Tan, J.; Xin, Z.; Zhou, Q.; Cao, Y.; Wu, Z.; Wang, L.; Zhao, M.; Jiang, X.; et al. Identification of collagen genes related to immune infiltration and epithelial-mesenchymal transition in glioma. Cancer Cell Int. 2021, 21, 276. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, N.A.; Sanchez-Salas, R.; Peske, J.D.; Vano, Y.; Becht, E.; Petitprez, F.; Validire, P.; Ingels, A.; Cathelineau, X.; Fridman, W.H.; et al. The clinical role of the TME in solid cancer. Br. J. Cancer 2019, 120, 45–53. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, J.; Zhang, L.; Wei, F.; Lian, Y.; Wu, Y.; Gong, Z.; Zhang, S.; Zhou, J.; Cao, K.; et al. Role of tumor microenvironment in tumorigenesis. J. Cancer 2017, 8, 761–773. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Jahanban-Esfahlan, R.; Seidi, K.; Banimohamad-Shotorbani, B.; Jahanban-Esfahlan, A.; Yousefi, B. Combination of nanotechnology with vascular targeting agents for effective cancer therapy. J. Cell Physiol. 2018, 233, 2982–2992. [Google Scholar] [CrossRef]
- Jahanban-Esfahlan, R.; Seidi, K.; Zarghami, N. Tumor vascular infarction: Prospects and challenges. Int. J. Hematol. 2017, 105, 244–256. [Google Scholar] [CrossRef]
- Sica, A.; Larghi, P.; Mancino, A.; Rubino, L.; Porrta, C.; Totaro, M.G.; Rimoldi, M.; Biswas, S.K.; Allavena, P.; Mantovani, A. Macrophage polarization in tumour progression. Semin. Cancer Biol. 2008, 18, 349–355. [Google Scholar] [CrossRef]
- Mantovani, A.; Marchesi, F.; Malesci, A.; Laghi, L.; Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017, 14, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sica, A.; Locati, M. Macrophage polarization comes of age. Immunity 2005, 23, 344–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T.R.; Cherwinski, H.; Bond, M.W.; Giedlin, M.A.; Coffman, R.L. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986, 136, 2348–2357. [Google Scholar]
- Gordon, S.; Taylor, P.R. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 2005, 5, 953–964. [Google Scholar] [CrossRef]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef] [Green Version]
- Maimela, N.R.; Liu, S.; Zhang, Y. Fates of CD8+ T cells in Tumor Microenvironment. Comput. Struct. Biotechnol. J. 2019, 17, 1–13. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef] [Green Version]
- Chang, L.Y.; Lin, Y.C.; Mahalingam, J.; Huang, C.T.; Chen, T.W.; Kang, C.W.; Peng, H.M.; Chu, Y.Y.; Chiang, J.M.; Dutta, A.; et al. Tumor-derived chemokine CCL5 enhances TGF-beta-mediated killing of CD8(+) T cells in colon cancer by T-regulatory cells. Cancer Res. 2012, 72, 1092–1102. [Google Scholar] [CrossRef] [Green Version]
- Roland, C.L.; Lynn, K.D.; Toombs, J.E.; Dineen, S.P.; Udugamasooriya, D.G.; Brekken, R.A. Cytokine levels correlate with immune cell infiltration after anti-VEGF therapy in preclinical mouse models of breast cancer. PLoS ONE 2009, 4, e7669. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.J.; Dunn, G.P.; Schreiber, R.D. Cancer immunosurveillance and immunoediting: The roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Adv. Immunol. 2006, 90, 1–50. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Han, C.; Wang, S.; Fang, P.; Ma, Z.; Xu, L.; Yin, R. Cancer-associated fibroblasts: An emerging target of anti-cancer immunotherapy. J. Hematol. Oncol. 2019, 12, 86. [Google Scholar] [CrossRef] [PubMed]
- Goveia, J.; Rohlenova, K.; Taverna, F.; Treps, L.; Conradi, L.C.; Pircher, A.; Geldhof, V.; de Rooij, L.; Kalucka, J.; Sokol, L.; et al. An Integrated Gene Expression Landscape Profiling Approach to Identify Lung Tumor Endothelial Cell Heterogeneity and Angiogenic Candidates. Cancer Cell 2020, 37, 21–36.e13. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Shin, H.J.; Lee, K.J.; Gil, M. Multiomic Analysis of Cereblon Expression and Its Prognostic Value in Kidney Renal Clear Cell Carcinoma, Lung Adenocarcinoma, and Skin Cutaneous Melanoma. J. Pers. Med. 2021, 11, 263. [Google Scholar] [CrossRef]
- Park, S.J.; Yoon, B.H.; Kim, S.K.; Kim, S.Y. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med. Genomics 2019, 12, 101. [Google Scholar] [CrossRef]
- Pak, K.; Oh, S.O.; Goh, T.S.; Heo, H.J.; Han, M.E.; Jeong, D.C.; Lee, C.S.; Sun, H.; Kang, J.; Choi, S.; et al. A User-Friendly, Web-Based Integrative Tool (ESurv) for Survival Analysis: Development and Validation Study. J. Med. Internet Res. 2020, 22, e16084. [Google Scholar] [CrossRef]
- Koster, J.; Volckmann, R.; Zwijnenburg, D.; Molenaar, P.; Versteeg, R. Abstract 2490: R2: Genomics analysis and visualization platform. Cancer Res. 2019, 79, 2490. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., 2nd; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Levitin, H.M.; Frattini, V.; Bush, E.C.; Boyett, D.M.; Samanamud, J.; Ceccarelli, M.; Dovas, A.; Zanazzi, G.; Canoll, P.; et al. Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med. 2018, 10, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, J.; Sun, B.F.; Chen, C.Y.; Zhou, J.Y.; Chen, Y.S.; Chen, H.; Liu, L.; Huang, D.; Jiang, J.; Cui, G.S.; et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019, 29, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Yang, M.; Zhang, Y.; Xiao, S.; Lai, X.; Tan, A.; Du, S.; Li, S. Dissecting the Single-Cell Transcriptome Network Underlying Gastric Premalignant Lesions and Early Gastric Cancer. Cell Rep. 2019, 27, 1934–1947.e1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Severson, E.; Pignon, J.C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.Y.; Gil, M.; Kim, K.E. Neuropilin1 Expression Acts as a Prognostic Marker in Stomach Adenocarcinoma by Predicting the Infiltration of Treg Cells and M2 Macrophages. J. Clin. Med. 2020, 9, 1430. [Google Scholar] [CrossRef]
- Pan, J.H.; Zhou, H.; Cooper, L.; Huang, J.L.; Zhu, S.B.; Zhao, X.X.; Ding, H.; Pan, Y.L.; Rong, L. LAYN Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Gastric and Colon Cancers. Front. Immunol. 2019, 10, 6. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; You, W.H.; Li, X.; Wang, P.; Sha, B.; Liang, Y.; Qiu, J.; Zhou, J.; Hu, H.; Lu, L. Single-cell RNA-seq reveals transcriptional landscape and intratumor heterogenicity in gallbladder cancer liver metastasis microenvironment. Ann. Transl. Med. 2021, 9, 889. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, J.; Zhao, Z.; Yang, M.; Chen, M.; Liu, C.; Ji, J.; Zhu, D. Single-cell transcriptome analysis reveals tumor immune microenvironment heterogenicity and granulocytes enrichment in colorectal cancer liver metastases. Cancer Lett. 2020, 470, 84–94. [Google Scholar] [CrossRef]
- Ling, B.; Huang, Z.; Huang, S.; Qian, L.; Li, G.; Tang, Q. Microenvironment Analysis of Prognosis and Molecular Signature of Immune-Related Genes in Lung Adenocarcinoma. Oncol Res. 2021, 28, 561–578. [Google Scholar] [CrossRef] [PubMed]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; He, W.; Wu, C.; Tan, Y.; He, Y.; Xu, B.; Chen, L.; Li, Q.; Jiang, J. Scoring System for Tumor-Infiltrating Lymphocytes and Its Prognostic Value for Gastric Cancer. Front. Immunol. 2019, 10, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively Activated Macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef] [PubMed]
- Furusato, E.; Shen, D.; Cao, X.; Furusato, B.; Nussenblatt, R.B.; Rushing, E.J.; Chan, C.C. Inflammatory cytokine and chemokine expression in sympathetic ophthalmia: A pilot study. Histol. Histopathol. 2011, 26, 1145–1151. [Google Scholar] [CrossRef]
- Yu, T.; Gan, S.; Zhu, Q.; Dai, D.; Li, N.; Wang, H.; Chen, X.; Hou, D.; Wang, Y.; Pan, Q.; et al. Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24. Nat. Commun. 2019, 10, 4353. [Google Scholar] [CrossRef] [Green Version]
- Mauer, J.; Chaurasia, B.; Goldau, J.; Vogt, M.C.; Ruud, J.; Nguyen, K.D.; Theurich, S.; Hausen, A.C.; Schmitz, J.; Bronneke, H.S.; et al. Signaling by IL-6 promotes alternative activation of macrophages to limit endotoxemia and obesity-associated resistance to insulin. Nat. Immunol. 2014, 15, 423–430. [Google Scholar] [CrossRef]
- Zhao, Z.; Cheng, X.; Wang, Y.; Han, R.; Li, L.; Xiang, T.; He, L.; Long, H.; Zhu, B.; He, Y. Metformin inhibits the IL-6-induced epithelial-mesenchymal transition and lung adenocarcinoma growth and metastasis. PLoS ONE 2014, 9, e95884. [Google Scholar] [CrossRef] [Green Version]
- Curl, D.D.; Antovich, T.J. Addressing the issue of cataloging and making chiropractic literature accessible: Part I. Defining the problem. J. Manip. Physiol. Ther. 1991, 14, 79–85. [Google Scholar]
- Sheikhpour, E.; Noorbakhsh, P.; Foroughi, E.; Farahnak, S.; Nasiri, R.; Neamatzadeh, H. A Survey on the Role of Interleukin-10 in Breast Cancer: A Narrative. Rep. Biochem. Mol. Biol. 2018, 7, 30–37. [Google Scholar]
- Sica, A.; Saccani, A.; Bottazzi, B.; Polentarutti, N.; Vecchi, A.; van Damme, J.; Mantovani, A. Autocrine production of IL-10 mediates defective IL-12 production and NF-kappa B activation in tumor-associated macrophages. J. Immunol. 2000, 164, 762–767. [Google Scholar] [CrossRef] [PubMed]
- Massague, J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef] [PubMed]
- Collison, L.W.; Chaturvedi, V.; Henderson, A.L.; Giacomin, P.R.; Guy, C.; Bankoti, J.; Finkelstein, D.; Forbes, K.; Workman, C.J.; Brown, S.A.; et al. IL-35-mediated induction of a potent regulatory T cell population. Nat. Immunol. 2010, 11, 1093–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collison, L.W.; Workman, C.J.; Kuo, T.T.; Boyd, K.; Wang, Y.; Vignali, K.M.; Cross, R.; Sehy, D.; Blumberg, R.S.; Vignali, D.A. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature 2007, 450, 566–569. [Google Scholar] [CrossRef]
- Niedbala, W.; Wei, X.Q.; Cai, B.; Hueber, A.J.; Leung, B.P.; McInnes, I.B.; Liew, F.Y. IL-35 is a novel cytokine with therapeutic effects against collagen-induced arthritis through the expansion of regulatory T cells and suppression of Th17 cells. Eur. J. Immunol. 2007, 37, 3021–3029. [Google Scholar] [CrossRef]
- Pyonteck, S.M.; Akkari, L.; Schuhmacher, A.J.; Bowman, R.L.; Sevenich, L.; Quail, D.F.; Olson, O.C.; Quick, M.L.; Huse, J.T.; Teijeiro, V.; et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 2013, 19, 1264–1272. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Liu, X. Upregulated EBI3 Correlates with Poor Outcome and Tumor Progression in Breast Cancer. Oncol. Res. Treat. 2018, 41, 111–115. [Google Scholar] [CrossRef]
- Peng, L.; Yuan, X.Q.; Zhang, C.Y.; Ye, F.; Zhou, H.F.; Li, W.L.; Liu, Z.Y.; Zhang, Y.Q.; Pan, X.; Li, G.C. High TGF-beta1 expression predicts poor disease prognosis in hepatocellular carcinoma patients. Oncotarget 2017, 8, 34387–34397. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Zhang, H.; Shen, Z.; Lin, C.; Wang, X.; Qin, J.; Qin, X.; Xu, J.; Sun, Y. Increased Expression of CSF-1 Associates With Poor Prognosis of Patients With Gastric Cancer Undergoing Gastrectomy. Medicine 2016, 95, e2675. [Google Scholar] [CrossRef]
- Li, Y.; Gao, P.; Yang, J.; Yu, H.; Zhu, Y.; Si, W. Relationship between IL-10 expression and prognosis in patients with primary breast cancer. Tumour Biol. 2014, 35, 11533–11540. [Google Scholar] [CrossRef]
- Serrano-Gomez, S.J.; Maziveyi, M.; Alahari, S.K. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol. Cancer 2016, 15, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Global Burden of Disease Cancer Collaboration; Fitzmaurice, C.; Abate, D.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdel-Rahman, O.; Abdelalim, A.; Abdoli, A.; Abdollahpour, I.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, H.; Cheng, L.; Yu, J.; Zhang, Z.; Luo, Z.; Chen, D. Identifying the mRNAs associated with Bladder cancer recurrence. Cancer Biomark. 2020, 28, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.W.; Bandopadhayay, P.; Jabado, N. The Power of Human Cancer Genetics as Revealed by Low-Grade Gliomas. Annu. Rev. Genet. 2019, 53, 483–503. [Google Scholar] [CrossRef] [PubMed]
- Kumthekar, P.; Raizer, J.; Singh, S. Low-grade glioma. Cancer Treat. Res. 2015, 163, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Vareedayah, A.A.; Alkaade, S.; Taylor, J.R. Pancreatic Adenocarcinoma. Mo. Med. 2018, 115, 230–235. [Google Scholar]
- Ryan, D.P.; Hong, T.S.; Bardeesy, N. Pancreatic adenocarcinoma. N. Engl. J. Med. 2014, 371, 1039–1049. [Google Scholar] [CrossRef]
- Lodde, G.; Zimmer, L.; Livingstone, E.; Schadendorf, D.; Ugurel, S. [Malignant melanoma]. Hautarzt 2020, 71, 63–77. [Google Scholar] [CrossRef]
- Whiteman, D.C.; Green, A.C.; Olsen, C.M. The Growing Burden of Invasive Melanoma: Projections of Incidence Rates and Numbers of New Cases in Six Susceptible Populations through 2031. J. Investig. Dermatol. 2016, 136, 1161–1171. [Google Scholar] [CrossRef] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Claus, E.B.; Walsh, K.M.; Wiencke, J.K.; Molinaro, A.M.; Wiemels, J.L.; Schildkraut, J.M.; Bondy, M.L.; Berger, M.; Jenkins, R.; Wrensch, M. Survival and low-grade glioma: The emergence of genetic information. Neurosurg. Focus 2015, 38, E6. [Google Scholar] [CrossRef] [PubMed]
- Elyada, E.; Bolisetty, M.; Laise, P.; Flynn, W.F.; Courtois, E.T.; Burkhart, R.A.; Teinor, J.A.; Belleau, P.; Biffi, G.; Lucito, M.S.; et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019, 9, 1102–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, R.L.; Pure, E. Cancer-associated fibroblasts and their influence on tumor immunity and immunotherapy. Elife 2020, 9, e57243. [Google Scholar] [CrossRef]
- Truffi, M.; Mazzucchelli, S.; Bonizzi, A.; Sorrentino, L.; Allevi, R.; Vanna, R.; Morasso, C.; Corsi, F. Nano-Strategies to Target Breast Cancer-Associated Fibroblasts: Rearranging the Tumor Microenvironment to Achieve Antitumor Efficacy. Int. J. Mol. Sci. 2019, 20, 1263. [Google Scholar] [CrossRef] [Green Version]
- Ziani, L.; Chouaib, S.; Thiery, J. Alteration of the Antitumor Immune Response by Cancer-Associated Fibroblasts. Front. Immunol. 2018, 9, 414. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Sakakura, K.; Kudo, T.; Toyoda, M.; Kaira, K.; Oyama, T.; Chikamatsu, K. Cancer-associated fibroblasts promote an immunosuppressive microenvironment through the induction and accumulation of protumoral macrophages. Oncotarget 2017, 8, 8633–8647. [Google Scholar] [CrossRef] [Green Version]
- Kato, T.; Noma, K.; Ohara, T.; Kashima, H.; Katsura, Y.; Sato, H.; Komoto, S.; Katsube, R.; Ninomiya, T.; Tazawa, H.; et al. Cancer-Associated Fibroblasts Affect Intratumoral CD8(+) and FoxP3(+) T Cells Via IL6 in the Tumor Microenvironment. Clin. Cancer Res. 2018, 24, 4820–4833. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, T.; Ishii, G.; Hiraoka, N.; Hirayama, S.; Yamauchi, C.; Aokage, K.; Hishida, T.; Yoshida, J.; Nagai, K.; Ochiai, A. Forkhead box P3 regulatory T cells coexisting with cancer associated fibroblasts are correlated with a poor outcome in lung adenocarcinoma. Cancer Sci. 2013, 104, 409–415. [Google Scholar] [CrossRef]
- Bassani, B.; Baci, D.; Gallazzi, M.; Poggi, A.; Bruno, A.; Mortara, L. Natural Killer Cells as Key Players of Tumor Progression and Angiogenesis: Old and Novel Tools to Divert Their Pro-Tumor Activities into Potent Anti-Tumor Effects. Cancers 2019, 11, 461. [Google Scholar] [CrossRef] [Green Version]
- Sobierajska, K.; Ciszewski, W.M.; Sacewicz-Hofman, I.; Niewiarowska, J. Endothelial Cells in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1234, 71–86. [Google Scholar] [CrossRef]
- Maishi, N.; Hida, K. Tumor endothelial cells accelerate tumor metastasis. Cancer Sci. 2017, 108, 1921–1926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folkman, J. Tumor angiogenesis: Therapeutic implications. N. Engl. J. Med. 1971, 285, 1182–1186. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, M.B.; Garg, A.D.; Agostinis, P. Defining the role of the tumor vasculature in antitumor immunity and immunotherapy. Cell Death Dis. 2018, 9, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hudson, B.G.; Tryggvason, K.; Sundaramoorthy, M.; Neilson, E.G. Alport's syndrome, Goodpasture's syndrome, and type IV collagen. N. Engl. J. Med. 2003, 348, 2543–2556. [Google Scholar] [CrossRef]
- Craft, P.S.; Harris, A.L. Clinical prognostic significance of tumour angiogenesis. Ann. Oncol. 1994, 5, 305–311. [Google Scholar] [CrossRef]
- Solimando, A.G.; Summa, S.; Vacca, A.; Ribatti, D. Cancer-Associated Angiogenesis: The Endothelial Cell as a Checkpoint for Immunological Patrolling. Cancers 2020, 12, 3380. [Google Scholar] [CrossRef] [PubMed]
- Ohue, Y.; Nishikawa, H. Regulatory T (Treg) cells in cancer: Can Treg cells be a new therapeutic target? Cancer Sci. 2019, 110, 2080–2089. [Google Scholar] [CrossRef]
- Najafi, M.; Hashemi Goradel, N.; Farhood, B.; Salehi, E.; Nashtaei, M.S.; Khanlarkhani, N.; Khezri, Z.; Majidpoor, J.; Abouzaripour, M.; Habibi, M.; et al. Macroph.hage polarity in cancer: A review. J. Cell Biochem. 2019, 120, 2756–2765. [Google Scholar] [CrossRef]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Tong, L.; LiuAnwei Liu, F.; Liu, A.; Zeng, S.; Xiong, Q.; Yang, Z.; He, X.; Sun, Y.; Xu, C. Tumorinfiltrating M2 macrophages driven by specific genomic alterations are associated with prognosis in bladder cancer. Oncol. Rep. 2019, 42, 581–594. [Google Scholar] [CrossRef]
- Zhao, X.; Qu, J.; Sun, Y.; Wang, J.; Liu, X.; Wang, F.; Zhang, H.; Wang, W.; Ma, X.; Gao, X.; et al. Prognostic significance of tumor-associated macrophages in breast cancer: A meta-analysis of the literature. Oncotarget 2017, 8, 30576–30586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ino, Y.; Yamazaki-Itoh, R.; Shimada, K.; Iwasaki, M.; Kosuge, T.; Kanai, Y.; Hiraoka, N. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br. J. Cancer 2013, 108, 914–923. [Google Scholar] [CrossRef] [PubMed]
- Kuo, D.S.; Labelle-Dumais, C.; Gould, D.B. COL4A1 and COL4A2 mutations and disease: Insights into pathogenic mechanisms and potential therapeutic targets. Hum. Mol. Genet. 2012, 21, R97–R110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudhakar, A.; Nyberg, P.; Keshamouni, V.G.; Mannam, A.P.; Li, J.; Sugimoto, H.; Cosgrove, D.; Kalluri, R. Human alpha1 type IV collagen NC1 domain exhibits distinct antiangiogenic activity mediated by alpha1beta1 integrin. J. Clin. Investig. 2005, 115, 2801–2810. [Google Scholar] [CrossRef] [PubMed]





| Description | Gene Markers | LGG | PAAD | SKCM | STAD | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| None | Purity | None | Purity | None | Purity | None | Purity | ||||||||||
| Cor | p | Cor | p | Cor | p | Cor | p | Cor | p | Cor | p | Cor | p | Cor | p | ||
| CD8+ T cells | CD8A | 0.149 | *** | 0.203 | *** | 0.367 | *** | 0.303 | *** | −0.001 | 0.984 | −0.088 | 0.059 | 0.133 | ** | 0.109 | * |
| CD8B | 0.069 | 0.117 | 0.11 | * | 0.297 | *** | 0.227 | ** | −0.025 | 0.583 | −0.126 | ** | 0.044 | 0.37 | 0.031 | 0.55 | |
| TAMs | CCL2 | 0.282 | *** | 0.3 | *** | 0.344 | *** | 0.303 | *** | 0.302 | *** | 0.278 | *** | 0.385 | *** | 0.366 | *** |
| CD68 | 0.35 | *** | 0.382 | *** | 0.533 | *** | 0.491 | *** | 0.179 | *** | 0.14 | ** | 0.171 | *** | 0.142 | ** | |
| IL10 | 0.28 | *** | 0.287 | *** | 0.442 | *** | 0.4 | *** | 0.283 | *** | 0.265 | *** | 0.349 | *** | 0.341 | *** | |
| M1 macrophages | NOS2 | −0.039 | 0.377 | −0.006 | 0.894 | 0.315 | *** | 0.338 | *** | 0.326 | *** | 0.318 | *** | 0.105 | * | 0.117 | * |
| IRF5 | 0.235 | *** | 0.29 | *** | 0.2 | ** | 0.177 | * | 0.129 | ** | 0.083 | 0.077 | 0.107 | * | 0.096 | 0.063 | |
| PTGS2 | 0.079 | 0.073 | 0.088 | 0.054 | 0.354 | *** | 0.373 | *** | 0.3 | *** | 0.284 | *** | 0.369 | *** | 0.357 | *** | |
| M2 macrophages | CD163 | 0.486 | *** | 0.473 | *** | 0.631 | *** | 0.577 | *** | 0.332 | *** | 0.322 | *** | 0.4 | *** | 0.378 | *** |
| VSIG4 | 0.232 | *** | 0.253 | *** | 0.595 | *** | 0.538 | *** | 0.263 | *** | 0.244 | *** | 0.341 | *** | 0.337 | *** | |
| MS4A4A | 0.393 | *** | 0.4 | *** | 0.614 | *** | 0.562 | *** | 0.28 | *** | 0.262 | *** | 0.319 | *** | 0.305 | *** | |
| Neutrophils | CEACAM8 | −0.003 | 0.944 | −0.022 | 0.628 | 0.171 | * | 0.115 | 0.133 | 0.075 | 0.102 | 0.086 | 0.066 | 0.064 | 0.191 | 0.084 | 0.103 |
| ITGAM | 0.181 | *** | 0.226 | *** | 0.513 | *** | 0.444 | *** | 0.263 | *** | 0.237 | *** | 0.362 | *** | 0.35 | *** | |
| CCR7 | 0.368 | *** | 0.395 | *** | 0.274 | *** | 0.219 | ** | 0.045 | 0.327 | −0.04 | 0.398 | 0.27 | *** | 0.261 | *** | |
| Th1 | TBX21 | 0.439 | *** | 0.434 | *** | 0.237 | ** | 0.183 | * | 0.034 | 0.465 | −0.054 | 0.247 | 0.187 | *** | 0.188 | *** |
| STAT4 | −0.076 | 0.086 | −0.036 | 0.43 | 0.274 | *** | 0.27 | *** | 0.122 | ** | 0.071 | 0.13 | 0.246 | *** | 0.233 | *** | |
| STAT1 | 0.507 | *** | 0.515 | *** | 0.518 | *** | 0.472 | *** | 0.062 | 0.177 | 0.017 | 0.721 | 0.129 | ** | 0.112 | * | |
| IFNG | 0.206 | *** | 0.224 | *** | 0.264 | *** | 0.216 | ** | −0.025 | 0.585 | −0.114 | * | −0.01 | 0.837 | −0.014 | 0.781 | |
| TNF | −0.031 | 0.486 | −0.032 | 0.487 | 0.263 | *** | 0.233 | ** | 0.046 | 0.323 | −0.028 | 0.555 | 0.163 | *** | 0.146 | ** | |
| Treg | FOXP3 | 0.004 | 0.927 | 0.027 | 0.556 | 0.495 | *** | 0.448 | *** | 0.066 | 0.152 | −0.01 | 0.839 | 0.256 | *** | 0.244 | *** |
| CCR8 | 0.22 | *** | 0.236 | *** | 0.578 | *** | 0.539 | *** | 0.112 | * | 0.059 | 0.209 | 0.324 | *** | 0.312 | *** | |
| STAT5B | 0.166 | *** | 0.137 | ** | 0.443 | *** | 0.499 | *** | 0.13 | ** | 0.136 | ** | 0.469 | *** | 0.458 | *** | |
| TGFB1 | 0.292 | *** | 0.316 | *** | 0.441 | *** | 0.419 | *** | 0.454 | *** | 0.451 | *** | 0.465 | *** | 0.442 | *** | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, H.-J.; Gil, M.; Lee, I.-S. Association of Elevated Expression Levels of COL4A1 in Stromal Cells with an Immunosuppressive Tumor Microenvironment in Low-Grade Glioma, Pancreatic Adenocarcinoma, Skin Cutaneous Melanoma, and Stomach Adenocarcinoma. J. Pers. Med. 2022, 12, 534. https://doi.org/10.3390/jpm12040534
Shin H-J, Gil M, Lee I-S. Association of Elevated Expression Levels of COL4A1 in Stromal Cells with an Immunosuppressive Tumor Microenvironment in Low-Grade Glioma, Pancreatic Adenocarcinoma, Skin Cutaneous Melanoma, and Stomach Adenocarcinoma. Journal of Personalized Medicine. 2022; 12(4):534. https://doi.org/10.3390/jpm12040534
Chicago/Turabian StyleShin, Hyo-Jae, Minchan Gil, and Im-Soon Lee. 2022. "Association of Elevated Expression Levels of COL4A1 in Stromal Cells with an Immunosuppressive Tumor Microenvironment in Low-Grade Glioma, Pancreatic Adenocarcinoma, Skin Cutaneous Melanoma, and Stomach Adenocarcinoma" Journal of Personalized Medicine 12, no. 4: 534. https://doi.org/10.3390/jpm12040534
APA StyleShin, H. -J., Gil, M., & Lee, I. -S. (2022). Association of Elevated Expression Levels of COL4A1 in Stromal Cells with an Immunosuppressive Tumor Microenvironment in Low-Grade Glioma, Pancreatic Adenocarcinoma, Skin Cutaneous Melanoma, and Stomach Adenocarcinoma. Journal of Personalized Medicine, 12(4), 534. https://doi.org/10.3390/jpm12040534






