Genome-Wide Analyses of Nephrotoxicity in Platinum-Treated Cancer Patients Identify Association with Genetic Variant in RBMS3 and Acute Kidney Injury
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Nephrotoxicity
2.2.1. Acute Kidney Injury
2.2.2. Hypomagnesemia
2.3. Genotyping
2.4. Statistical Analyses
2.4.1. Clinical Characteristics
2.4.2. Genome-Wide Association Analyses
2.4.3. Replication of Candidate Variants
3. Results
3.1. Patient and Treatment Characteristics
3.2. Nephrotoxicity
3.3. Genotyping
3.4. Creatinine-Based Analyses
3.4.1. GWASs
3.4.2. Replication of Previously Reported Associations
3.5. Magnesium-Based GWASs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lebwohl, D.; Canetta, R. Clinical development of platinum complexes in cancer therapy: An historical perspective and an update. Eur. J. Cancer 1998, 34, 1522–1534. [Google Scholar] [CrossRef]
- Arquilla, M.; Thompson, L.M.; Pearlman, L.F.; Simpkins, H. Effect of platinum antitumor agents on DNA and RNA investigated by terbium fluorescence. Cancer Res. 1983, 43, 1211–1216. [Google Scholar] [PubMed]
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005, 4, 307–320. [Google Scholar] [CrossRef]
- Go, R.S.; Adjei, A.A. Review of the Comparative Pharmacology and Clinical Activity of Cisplatin and Carboplatin. J. Clin. Oncol. 1999, 17, 409. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [Green Version]
- Oun, R.; Moussa, Y.E.; Wheate, N.J. The side effects of platinum-based chemotherapy drugs: A review for chemists. Dalton Trans. 2018, 47, 6645–6653. [Google Scholar] [CrossRef]
- Peres, L.A.B.; Júnior, A.D.D.C. Acute nephrotoxicity of cisplatin: Molecular mechanisms. J. Bras. de Nefrol. 2013, 35, 332–340. [Google Scholar] [CrossRef]
- Yao, X.; Panichpisal, K.; Kurtzman, N.; Nugent, K. Cisplatin nephrotoxicity: A review. Am. J. Med. Sci. 2007, 334, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Miller, R.P.; Tadagavadi, R.K.; Ramesh, G.; Reeves, W.B. Mechanisms of Cisplatin Nephrotoxicity. Toxins 2010, 2, 2490–2518. [Google Scholar] [CrossRef] [Green Version]
- Townsend, D.M.; Deng, M.; Zhang, L.; Lapus, M.G.; Hanigan, M.H. Metabolism of Cisplatin to a Nephrotoxin in Proximal Tubule Cells. J. Am. Soc. Nephrol. 2003, 14, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ciarimboli, G.; Ludwig, T.; Lang, D.; Pavenstädt, H.; Koepsell, H.; Piechota, H.-J.; Haier, J.; Jaehde, U.; Zisowsky, J.; Schlatter, E. Cisplatin Nephrotoxicity Is Critically Mediated via the Human Organic Cation Transporter 2. Am. J. Pathol. 2005, 167, 1477–1484. [Google Scholar] [CrossRef] [Green Version]
- Pabla, N.; Murphy, R.F.; Liu, K.; Dong, Z. The copper transporter Ctr1 contributes to cisplatin uptake by renal tubular cells during cisplatin nephrotoxicity. Am. J. Physiol. Physiol. 2009, 296, F505–F511. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, G.; Reeves, W.B. TNF-alpha mediates chemokine and cytokine expression and renal injury in cisplatin nephrotoxicity. J. Clin. Investig. 2002, 110, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Winston, J.A.; Safirstein, R. Reduced renal blood flow in early cisplatin-induced acute renal failure in the rat. Am. J. Physiol. Physiol. 1985, 249, F490–F496. [Google Scholar] [CrossRef] [PubMed]
- Kooijmans, E.C.; Bökenkamp, A.; Tjahjadi, N.S.; Tettero, J.M.; van Dulmen-den, E.B.; Van Der Pal, H.J.; Veening, M.A. Early and late adverse renal effects after potentially nephrotoxic treatment for childhood cancer. Cochrane Database Syst. Rev. 2019, 3, CD008944. [Google Scholar] [CrossRef]
- Stöhr, W.; Paulides, M.; Bielack, S.; Jürgens, H.; Koscielniak, E.; Rossi, R.; Langer, T.; Beck, J. Nephrotoxicity of cisplatin and carboplatin in sarcoma patients: A report from the late effects surveillance system. Pediatr. Blood Cancer 2007, 48, 140–147. [Google Scholar] [CrossRef]
- English, M.W.; Skinner, R.; Pearson, A.D.J.; Price, L.; Wyllie, R.; Craft, A.W. Dose-related nephrotoxicity of carboplatin in children. Br. J. Cancer 1999, 81, 336–341. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, A.R.D.S.; de Brito, G.A.; Baptista, A.L.; Andrade, L.A.S.; Imanishe, M.H.; Pereira, B.J. The influence of acute kidney disease on the clinical outcomes of patients who received cisplatin, carboplatin, and oxaliplatin. Health Sci. Rep. 2022, 5, e479. [Google Scholar] [CrossRef]
- McMahon, K.R.; Rassekh, S.R.; Schultz, K.R.; Blydt-Hansen, T.; Cuvelier, G.D.E.; Mammen, C.; Pinsk, M.; Carleton, B.C.; Tsuyuki, R.T.; Ross, C.J.D.; et al. Epidemiologic Characteristics of Acute Kidney Injury During Cisplatin Infusions in Children Treated for Cancer. JAMA Netw. Open 2020, 3, e203639. [Google Scholar] [CrossRef]
- Duan, Z.-Y.; Liu, J.-Q.; Yin, P.; Li, J.-J.; Cai, G.-Y.; Chen, X.-M. Impact of aging on the risk of platinum-related renal toxicity: A systematic review and meta-analysis. Cancer Treat. Rev. 2018, 69, 243–253. [Google Scholar] [CrossRef]
- Perazella, M.A.; Moeckel, G.W. Nephrotoxicity from Chemotherapeutic Agents: Clinical Manifestations, Pathobiology, and Prevention/Therapy. Semin. Nephrol. 2010, 30, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Zazuli, Z.; Vijverberg, S.; Slob, E.; Liu, G.; Carleton, B.; Veltman, J.; Baas, P.; Masereeuw, R.; Der Zee, A.-H.M.-V. Genetic Variations and Cisplatin Nephrotoxicity: A Systematic Review. Front. Pharmacol. 2018, 9, 1111. [Google Scholar] [CrossRef] [PubMed]
- Filipski, K.K.; Mathijssen, R.H.; Mikkelsen, T.S.; Schinkel, A.H.; Sparreboom, A. Contribution of Organic Cation Transporter 2 (OCT2) to Cisplatin-Induced Nephrotoxicity. Clin. Pharmacol. Ther. 2009, 86, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Goekkurt, E.; Al-Batran, S.-E.; Hartmann, J.T.; Mogck, U.; Schuch, G.; Kramer, M.; Jaeger, E.; Bokemeyer, C.; Ehninger, G.; Stoehlmacher, J. Pharmacogenetic Analyses of a Phase III Trial in Metastatic Gastroesophageal Adenocarcinoma with Fluorouracil and Leucovorin Plus Either Oxaliplatin or Cisplatin: A Study of the Arbeitsgemeinschaft Internistische Onkologie. J. Clin. Oncol. 2009, 27, 2863–2873. [Google Scholar] [CrossRef] [PubMed]
- Khrunin, A.V.; Moisseev, A.; Gorbunova, V.; Limborska, S. Genetic polymorphisms and the efficacy and toxicity of cisplatin-based chemotherapy in ovarian cancer patients. Pharmacogenomics J. 2009, 10, 54–61. [Google Scholar] [CrossRef]
- Chen, S.; Huo, X.; Lin, Y.; Ban, H.; Lin, Y.; Li, W.; Zhang, B.; Au, W.W.; Xu, X. Association of MDR1 and ERCC1 polymorphisms with response and toxicity to cisplatin-based chemotherapy in non-small-cell lung cancer patients. Int. J. Hyg. Environ. Health 2010, 213, 140–145. [Google Scholar] [CrossRef]
- Tzvetkov, M.V.; Behrens, G.; O’Brien, V.P.; Hohloch, K.; Brockmöller, J.; Benöhr, P. Pharmacogenetic analyses of cisplatin-induced nephrotoxicity indicate a renoprotective effect of ERCC1 polymorphisms. Pharmacogenomics 2011, 12, 1417–1427. [Google Scholar] [CrossRef]
- KimCurran, V.; Zhou, C.; Schmid-Bindert, G.; Shengxiang, R.; Zhou, S.; Zhang, L.; Zhang, J. Lack of correlation between ERCC1 (C8092A) Single Nucleotide Polymorphism and efficacy/toxicity of platinum based chemotherapy in Chinese patients with advanced Non-Small Cell Lung Cancer. Adv. Med. Sci. 2011, 56, 30–38. [Google Scholar] [CrossRef]
- Erčulj, N.; Kovač, V.; Hmeljak, J.; Dolžan, V. The influence of platinum pathway polymorphisms on the outcome in patients with malignant mesothelioma. Ann. Oncol. 2011, 23, 961–967. [Google Scholar] [CrossRef]
- Kim, S.-H.; Lee, G.-W.; Lee, M.J.; Cho, Y.J.; Jeong, Y.Y.; Kim, H.-C.; Lee, J.D.; Hwang, Y.S.; Kim, I.-S.; Lee, S.; et al. Clinical significance of ERCC2 haplotype-tagging single nucleotide polymorphisms in patients with unresectable non-small cell lung cancer treated with first-line platinum-based chemotherapy. Lung Cancer 2012, 77, 578–584. [Google Scholar] [CrossRef]
- Windsor, R.E.; Strauss, S.J.; Kallis, C.; Wood, N.E.; Whelan, J.S. Germline genetic polymorphisms may influence chemotherapy response and disease outcome in osteosarcoma: A pilot study. Cancer 2012, 118, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Iwata, K.; Aizawa, K.; Kamitsu, S.; Jingami, S.; Fukunaga, E.; Yoshida, M.; Yoshimura, M.; Hamada, A.; Saito, H. Effects of genetic variants in SLC22A2 organic cation transporter 2 and SLC47A1 multidrug and toxin extrusion 1 transporter on cisplatin-induced adverse events. Clin. Exp. Nephrol. 2012, 16, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Gao, G.; Li, X.; Ren, S.; Li, A.; Xu, J.; Zhang, J.; Zhou, C. Association between Single Nucleotide Polymorphisms (SNPs) and Toxicity of Advanced Non-Small-Cell Lung Cancer Patients Treated with Chemotherapy. PLoS ONE 2012, 7, e48350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Zhou, W. Ameliorative effects of SLC22A2 gene polymorphism 808 G/T and cimetidine on cisplatin-induced nephrotoxicity in Chinese cancer patients. Food Chem. Toxicol. 2012, 50, 2289–2293. [Google Scholar] [CrossRef]
- Khrunin, A.; Ivanova, F.; Moisseev, A.; Khokhrin, D.; Sleptsova, Y.; Gorbunova, V.; Limborska, S. Pharmacogenomics of cisplatin-based chemotherapy in ovarian cancer patients of different ethnic origins. Pharmacogenomics 2012, 13, 171–178. [Google Scholar] [CrossRef]
- Hinai, Y.; Motoyama, S.; Niioka, T.; Miura, M. Absence of effect of SLC22A2 genotype on cisplatin-induced nephrotoxicity in oesophageal cancer patients receiving cisplatin and 5-fluorouracil: Report of results discordant with those of earlier studies. J. Clin. Pharm. Ther. 2013, 38, 498–503. [Google Scholar] [CrossRef]
- Liu, H.E.; Bai, K.-J.; Hsieh, Y.-C.; Yu, M.-C.; Lee, C.-N.; Chang, J.-H.; Hsu, H.-L.; Lu, P.-C.; Chen, S.H.-Y. Multiple Analytical Approaches Demonstrate a Complex Relationship of Genetic and Nongenetic Factors with Cisplatin- and Carboplatin-Induced Nephrotoxicity in Lung Cancer Patients. BioMed Res. Int. 2014, 2014, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Powrózek, T.; Mlak, R.; Krawczyk, P.; Homa, I.; Ciesielka, M.; Koziol, P.; Prendecka, M.; Milanowski, J.; Malecka-Massalska, T. The relationship between polymorphisms of genes regulating DNA repair or cell division and the toxicity of platinum and vinorelbine chemotherapy in advanced NSCLC patients. Clin. Transl. Oncol. 2015, 18, 125–131. [Google Scholar] [CrossRef]
- Chang, C.; Hu, Y.; Hogan, S.L.; Mercke, N.; Gomez, M.; O'Bryant, C.; Bowles, D.W.; George, B.; Wen, X.; Aleksunes, L.M.; et al. Pharmacogenomic Variants May Influence the Urinary Excretion of Novel Kidney Injury Biomarkers in Patients Receiving Cisplatin. Int. J. Mol. Sci. 2017, 18, 1333. [Google Scholar] [CrossRef] [Green Version]
- Zazuli, Z.; Otten, L.S.; Drögemöller, B.I.; Medeiros, M.; Monzon, J.G.; Wright, G.E.B.; Kollmannsberger, C.K.; Bedard, P.L.; Chen, Z.; Gelmon, K.A.; et al. Outcome Definition Influences the Relationship between Genetic Polymorphisms of ERCC1, ERCC2, SLC22A2 and Cisplatin Nephrotoxicity in Adult Testicular Cancer Patients. Genes 2019, 10, 364. [Google Scholar] [CrossRef] [Green Version]
- Zazuli, Z.; de Jong, C.; Xu, W.; Vijverberg, S.J.H.; Masereeuw, R.; Patel, D.; Mirshams, M.; Khan, K.; Cheng, D.; Ordonez-Perez, B.; et al. Association between Genetic Variants and Cisplatin-Induced Nephrotoxicity: A Genome-Wide Approach and Validation Study. J. Pers. Med. 2021, 11, 1233. [Google Scholar] [CrossRef] [PubMed]
- Driessen, C.M.; Ham, J.C.; Loo, M.T.; van Meerten, E.; van Lamoen, M.; Hakobjan, M.H.; Takes, R.P.; van der Graaf, W.T.; Kaanders, J.H.; Coenen, M.J.; et al. Genetic Variants as Predictive Markers for Ototoxicity and Nephrotoxicity in Patients with Locally Advanced Head and Neck Cancer Treated with Cisplatin-Containing Chemoradiotherapy (The PRONE Study). Cancers 2019, 11, 551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levey, A.S.; Stevens, L.A. Estimating GFR Using the CKD Epidemiology Collaboration (CKD-EPI) Creatinine Equation: More Accurate GFR Estimates, Lower CKD Prevalence Estimates, and Better Risk Predictions. Am. J. Kidney Dis. 2010, 55, 622–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, G.J.; Haycock, G.B.; Edelmann, C.M.; Spitzer, A. A simple estimate of glomerular filtration rate in children derived from body length and plasma creatinine. Pediatrics 1976, 58, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Loh, P.-R.; Danecek, P.; Palamara, P.F.; Fuchsberger, C.; Reshef, Y.A.; Finucane, H.K.; Schoenherr, S.; Forer, L.; McCarthy, S.; Abecasis, C.F.G.R.; et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016, 48, 1443–1448. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef] [Green Version]
- Lam, M.; Awasthi, S.; Watson, H.J.; Goldstein, J.; Panagiotaropoulou, G.; Trubetskoy, V.; Karlsson, R.; Frei, O.; Fan, C.-C.; De Witte, W.; et al. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics 2019, 36, 930–933. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Negishi, Y.; Iguchi-Ariga, S.M.; Ariga, H. Protein complexes bearing myc-like antigenicity recognize two distinct DNA sequences. Oncogene 1992, 7, 543–548. [Google Scholar]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [Green Version]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar]
- Fröhlich, H.; Kollmeyer, M.L.; Linz, V.C.; Stuhlinger, M.; Groneberg, D.; Reigl, A.; Zizer, E.; Friebe, A.; Niesler, B.; Rappold, G. Gastrointestinal dysfunction in autism displayed by altered motility and achalasia in Foxp1 (+/-) mice. Proc. Natl. Acad. Sci. USA 2019, 116, 22237–22245. [Google Scholar] [CrossRef] [PubMed]
- Leblond, C.S.; Cliquet, F.; Carton, C.; Huguet, G.; Mathieu, A.; Kergrohen, T.; Buratti, J.; Lemière, N.; Cuisset, L.; Bienvenu, T.; et al. Both rare and common genetic variants contribute to autism in the Faroe Islands. NPJ Genom. Med. 2019, 4, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Chen, P.L.; Nie, C.-J.; Zeng, T.-T.; Liu, H.; Mao, X.; Qin, Y.; Zhu, Y.-H.; Fu, L.; Guan, X.-Y. Downregulation of RBMS3 Is Associated with Poor Prognosis in Esophageal Squamous Cell Carcinoma. Cancer Res. 2011, 71, 6106–6115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Fu, L.; Zhang, L.-Y.; Kwong, D.L.; Yan, L.; Guan, X.-Y. Tumor suppressor genes on frequently deleted chromosome 3p in nasopharyngeal carcinoma. Chin. J. Cancer 2012, 31, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wu, Y.; Fang, Z.; Yan, Q.; Zhang, S.; Sun, R.; Khaliq, J.; Li, Y. Low expression of RBMS3 and SFRP1 are associated with poor prognosis in patients with gastric cancer. Am. J. Cancer Res. 2016, 6, 2679–2689. [Google Scholar]
- Wu, G.; Cao, L.; Zhu, J.; Tan, Z.; Tang, M.; Li, Z.; Hu, Y.; Yu, R.; Zhang, S.; Song, L.; et al. Loss of RBMS3 Confers Platinum Resistance in Epithelial Ovarian Cancer via Activation of miR-126-5p/beta-catenin/CBP signaling. Clin. Cancer Res. 2019, 25, 1022–1035. [Google Scholar] [CrossRef] [Green Version]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT missense predictions for genomes. Nat. Protoc. 2015, 11, 1–9. [Google Scholar] [CrossRef]
- Morita, Y.; Shibutani, T.; Nakanishi, N.; Nishikura, K.; Iwai, S.; Kuraoka, I. Human endonuclease V is a ribonuclease specific for inosine-containing RNA. Nat. Commun. 2013, 4, 2273. [Google Scholar] [CrossRef] [Green Version]
- Vik, E.S.; Nawaz, M.S.; Andersen, P.S.; Fladeby, C.; Bjørås, M.; Dalhus, B.; Alseth, I. Endonuclease V cleaves at inosines in RNA. Nat. Commun. 2013, 4, 2271. [Google Scholar] [CrossRef] [Green Version]
- Hartwig, A. Role of magnesium in genomic stability. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2001, 475, 113–121. [Google Scholar] [CrossRef]
- Pabla, N.; Dong, Z. Cisplatin nephrotoxicity: Mechanisms and renoprotective strategies. Kidney Int. 2008, 73, 994–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai, Y.; Nakao, T.; Kunimura, N.; Kohda, Y.; Gemba, M. Relationship of Intracellular Calcium and Oxygen Radicals to Cisplatin-Related Renal Cell Injury. J. Pharmacol. Sci. 2006, 100, 65–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faubel, S.; Ljubanovic, D.; Reznikov, L.; Somerset, H.; Dinarello, C.A.; Edelstein, C.L. Caspase-1–deficient mice are protected against cisplatin-induced apoptosis and acute tubular necrosis. Kidney Int. 2004, 66, 2202–2213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skinner, R.; Parry, A.; Price, L.; Cole, M.; Craft, A.W.; Pearson, A.D. Persistent nephrotoxicity during 10-year follow-up after cisplatin or carboplatin treatment in childhood: Relevance of age and dose as risk factors. Eur. J. Cancer 2009, 45, 3213–3219. [Google Scholar] [CrossRef] [PubMed]
- Reece, P.A.; Stafford, I.; Russell, J.; Khan, M.; Gill, P.G. Creatinine clearance as a predictor of ultrafilterable platinum disposition in cancer patients treated with cisplatin: Relationship between peak ultrafilterable platinum plasma levels and nephrotoxicity. J. Clin. Oncol. 1987, 5, 304–309. [Google Scholar] [CrossRef]
- van der Vorst, M.J.; Neefjes, E.C.; Toffoli, E.C.; Oosterling-Jansen, J.E.; Vergeer, M.R.; Leemans, C.R.; Kooistra, M.P.; Voortman, J.; Verheul, H.M. Incidence and risk factors for acute kidney injury in head and neck cancer patients treated with concurrent chemoradiation with high-dose cisplatin. BMC Cancer 2019, 19, 1066. [Google Scholar] [CrossRef]
- Tong, G.M.; Rude, R.K. Magnesium Deficiency in Critical Illness. J. Intensiv. Care Med. 2005, 20, 3–17. [Google Scholar] [CrossRef]

| Creatinine (n = 195) | Magnesium (n = 163) | ||||||
|---|---|---|---|---|---|---|---|
| na | n (%), or Median (Range) b | pb | na | n (%), or Median (Range) b | pb | ||
| Demographics | |||||||
| Male sex | 195 | 120 (61.5%) | 0.728 | 163 | 107 (65.6%) | 0.439 | |
| Age at diagnosis (in years) | 195 | 21 (1–72) | 0.030 | 163 | 48.0 (0–72) | 0.451 | |
| <18 years old | 195 | 90 (46.2%) | 0.002 | 163 | 69 (42.3%) | 0.284 | |
| ≥18 years old | 105 (53.8%) | 94 (57.7%) | |||||
| Self-reported Caucasian ethnicity | 195 | 195 (100.0%) | - | 163 | 163 (100.0%) | - | |
| Diabetes mellitus | 195 | 1 (0.5%) | 0.400 | 163 | 1 (0.6%) | 0.768 | |
| Disease and treatment | |||||||
| Diagnosis | |||||||
| Medulloblastoma | 195 | 72 (36.9%) | 0.012 | 163 | 41 (25.2%) | 0.004 | |
| Low-grade glioma | 34 (17.4%) | 36 (22.1%) | |||||
| Head–neck tumor | 89 (45.6%) | 86 (52.8%) | |||||
| Received radiotherapy | 195 | 161 (82.6%) | 0.085 | 163 | 129 (79.1%) | 0.247 | |
| Primary platinum agent | |||||||
| Cisplatin | 195 | 149 (76.4%) | 0.236 | 163 | 132 (81.0%) | 0.170 | |
| Carboplatin | 46 (23.6%) | 31 (19.0%) | |||||
| Cisplatin treatment | |||||||
| Cisplatin cumulative dose (mg/m²) | 150 | 240 (80–900) | 0.010 | 133 | 240 (80–900) | 0.998 | |
| Cisplatin dose per cycle (mg/m²) | 150 | 70 (30–100) | 9 × 10−10 | 133 | 70 (30–100) | 0.495 | |
| Carboplatin treatment | |||||||
| Carboplatin cumulative dose (mg/m²) | 63 | 800 (200–16,047) | 0.170 | 39 | 1600 (200–16,047) | 0.0001 | |
| Carboplatin dose per cycle (mg/m²) | 62 | 550 (35–800) | 0.755 | 38 | 550 (35–800) | 0.131 | |
| Hydration per protocol (in L/m²/cycle) | 191 | 2.5 (0.5–5.3) | 0.137 | 161 | 2.5 (0.8–5.3) | 0.189 | |
| Use of diuretics | |||||||
| Furosemide | 179 | 42 (23.5%) | 0.277 | 111 | 41 (36.9%) | 0.903 | |
| Use of potentially nephrotoxic drugs | |||||||
| Use of one or more nephrotoxic drugs | 195 | 106 (54.4%) | 0.003 | 163 | 77 (47.2%) | 0.422 | |
| Cyclophosphamide | 195 | 47 (24.1%) | 163 | 29 (17.8%) | |||
| Etoposide | 195 | 55 (28.2%) | 163 | 40 (24.5%) | |||
| Vincristine | 195 | 84 (43.1%) | 163 | 56 (34.4%) | |||
| Methotrexate | 195 | 30 (15.4%) | 163 | 15 (9.2%) | |||
| Aminoglycosides | 193 | 17 (8.8%) | 161 | 7 (4.3%) | |||
| Creatinine-Based Analyses | |||||
|---|---|---|---|---|---|
| Total | Cisplatin-Treated | Carboplatin-Treated | |||
| n = 195 | n = 149 | n = 46 | |||
| Continuous | |||||
| Worst eGFR, in mL/min/1.73 m² (median, range) | 89.2 (31.6–179.3) | 89.1 (31.6–138.6) | 90.1 (56.6–179.3) | ||
| Ratio eGFR, worst/baseline (median, range) | 0.9 (0.3–1.3) | 0.9 (0.3–1.3) | 0.9 (0.4–1.2) | ||
| Binary | |||||
| CTCAE v4.03 ‘Acute kidney injury’ | |||||
| Controls: grade 0 (n, %) | 169 (86.7%) | 129 (86.6%) | 40 (87.0%) | ||
| Cases: grade 1 or higher (n, %) | 26 (13.3%) | 20 (13.4%) | 6 (13.0%) | ||
| Grade 1 (n) | 19 | 14 | 5 | ||
| Grade 2 (n) | 6 | 5 | 1 | ||
| Grade 3 (n) | 1 | 1 | 0 | ||
| Grade 4 (n) | 0 | 0 | 0 | ||
| Grade 5 (n) | 0 | 0 | 0 | ||
| Magnesium-based analyses | |||||
| Total | Cisplatin-treated | Carboplatin-treated | |||
| n = 163 | n = 132 | n = 31 | |||
| Continuous | |||||
| Lowest magnesium plasma level, in mmol/L(median, range) | 0.76 (0.19–0.91) | 0.77 (0.48–0.91) | 0.74 (0.19–0.87) | ||
| Binary | |||||
| Controls: CTCAE grade 0, AND no therapeutic magnesium supplementation (n, %) | 125 (76.7%) | 107 (81.1%) | 18 (58.1%) | ||
| Cases: CTCAE grade 1 or higher, OR therapeutic magnesium supplementation (n, %) | 38 (23.3%) | 25 (18.9%) | 13 (41.9%) | ||
| CTCAE v4.03 ‘Hypomagnesemia’ | |||||
| Grade 0 (n) | 132 | 113 | 19 | ||
| Grade 1 (n) | 24 | 18 | 6 | ||
| Grade 2 (n) | 4 | 1 | 3 | ||
| Grade 3 (n) | 1 | 0 | 1 | ||
| Grade 4 (n) | 2 | 0 | 2 | ||
| Grade 5 (n) | 0 | 0 | 0 | ||
| Received therapeutic magnesium supplementation (n, %) | 22 (13.5%) | 10 (9.3%) | 12 (38.7%) | ||
| eGFR (Continuous) | CTCAE-AKI (Binary) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant | Position a | Chr | Gene | Variant Type | Effect Allele | Non-Effect Allele | MAF | Coef. b | 95% CI | p | OR c | 95% CI | p |
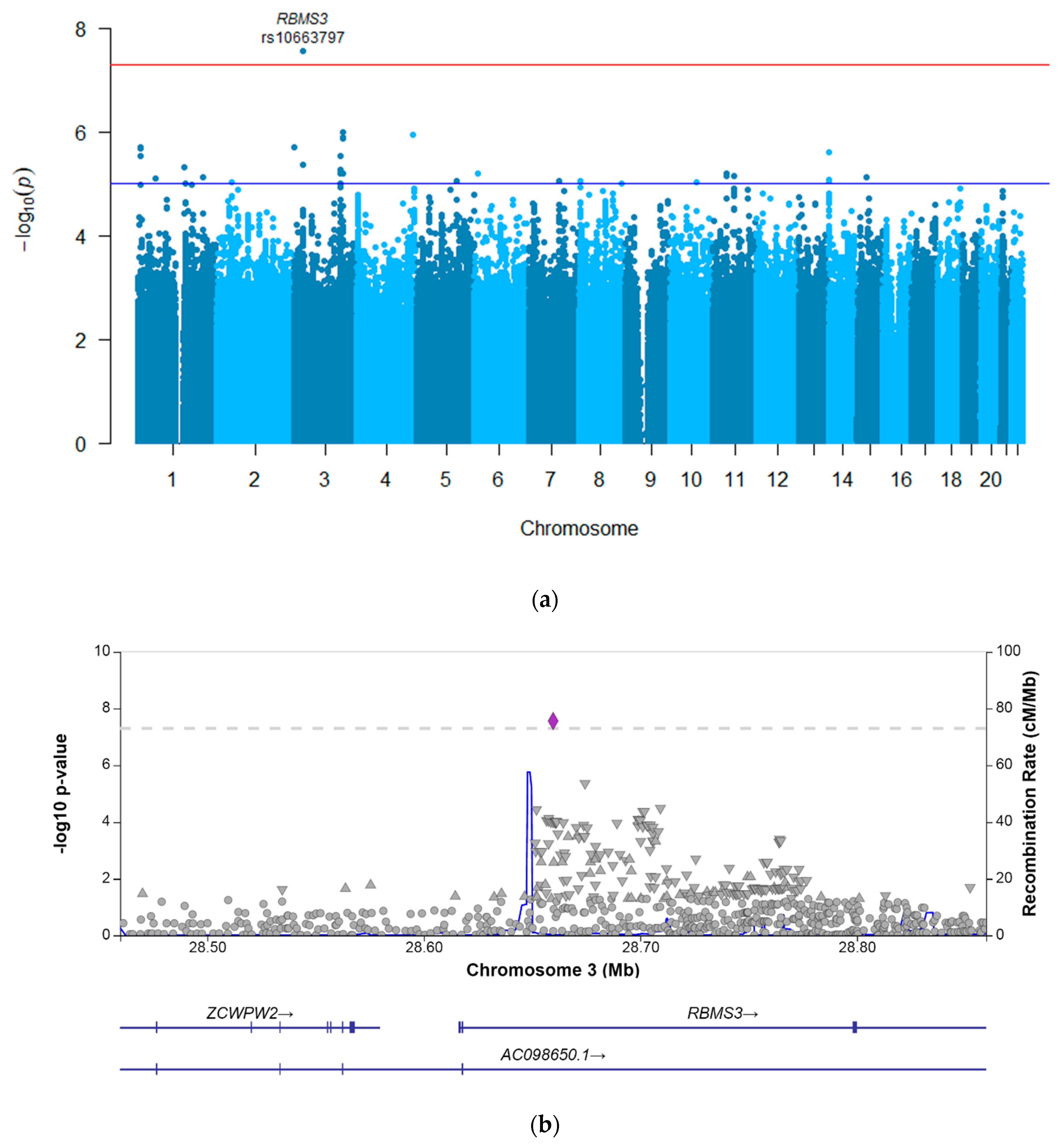
| rs10663797 | 28,659,744 | 3 | RBMS3 | intronic | delAC | insAC | 0.27 | −0.10 | −0.13–−0.06 | 2.72 × 10−8 | 5.69 | 2.54–12.74 | 2.33 × 10−5 |
| Gene | Variant | Data from Discovery Study Zazuli et al. (2021) [41] | Results Current Study (GWAS with eGFR Decline Phenotype) | Comparison | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect Allele | Beta | 95% CI | p | Effect Allele | Coef | 95% CI | p | Effect Allele | Non-Effect Allele | Direction Zazuli et al. a | Direction Current Study a | ||||
| TMEM225B | rs17161766 | A | −28.91 | −38.80 | −19.10 | 7.823 × 10−9 | NA | NA | NA | NA | NA | + | G | + | NA |
| - | chr7:98951080 | CTTAT | −27.19 | −36.50 | −17.90 | 9.485 × 10−9 | NA | NA | NA | NA | NA | + | C | + | NA |
| ARPC1A | rs199659233 | T | 28.65 | 18.70 | 38.60 | 1.473 × 10−8 | C | 0.008 | −0.115 | 0.130 | 0.899 | - | C | - | - |
| ARPC1A | rs556958738 | T | 28.65 | 18.70 | 38.60 | 1.473 × 10−8 | C | 0.008 | −0.115 | 0.131 | 0.899 | - | C | - | - |
| BACH2 | rs4388268 | A | −8.37 | −11.40 | −5.40 | 3.845 × 10−8 | A | 0.013 | −0.020 | 0.045 | 0.443 | + | G | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klumpers, M.J.; Witte, W.D.; Gattuso, G.; Schiavello, E.; Terenziani, M.; Massimino, M.; Gidding, C.E.M.; Vermeulen, S.H.; Driessen, C.M.; Van Herpen, C.M.; et al. Genome-Wide Analyses of Nephrotoxicity in Platinum-Treated Cancer Patients Identify Association with Genetic Variant in RBMS3 and Acute Kidney Injury. J. Pers. Med. 2022, 12, 892. https://doi.org/10.3390/jpm12060892
Klumpers MJ, Witte WD, Gattuso G, Schiavello E, Terenziani M, Massimino M, Gidding CEM, Vermeulen SH, Driessen CM, Van Herpen CM, et al. Genome-Wide Analyses of Nephrotoxicity in Platinum-Treated Cancer Patients Identify Association with Genetic Variant in RBMS3 and Acute Kidney Injury. Journal of Personalized Medicine. 2022; 12(6):892. https://doi.org/10.3390/jpm12060892
Chicago/Turabian StyleKlumpers, Marije J., Ward De Witte, Giovanna Gattuso, Elisabetta Schiavello, Monica Terenziani, Maura Massimino, Corrie E. M. Gidding, Sita H. Vermeulen, Chantal M. Driessen, Carla M. Van Herpen, and et al. 2022. "Genome-Wide Analyses of Nephrotoxicity in Platinum-Treated Cancer Patients Identify Association with Genetic Variant in RBMS3 and Acute Kidney Injury" Journal of Personalized Medicine 12, no. 6: 892. https://doi.org/10.3390/jpm12060892








