Mitogenome-Based Phylogeny with Divergence Time Estimates Revealed the Presence of Cryptic Species within Heptageniidae (Insecta, Ephemeroptera)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling and Species Identification
2.2. Mitogenome Sequencing and Assembling
2.3. Mitogenome Annotation and Structural Analysis
2.4. Dataset Selection and Phylogenetic Analyses
2.5. Divergence Time Estimation
3. Results
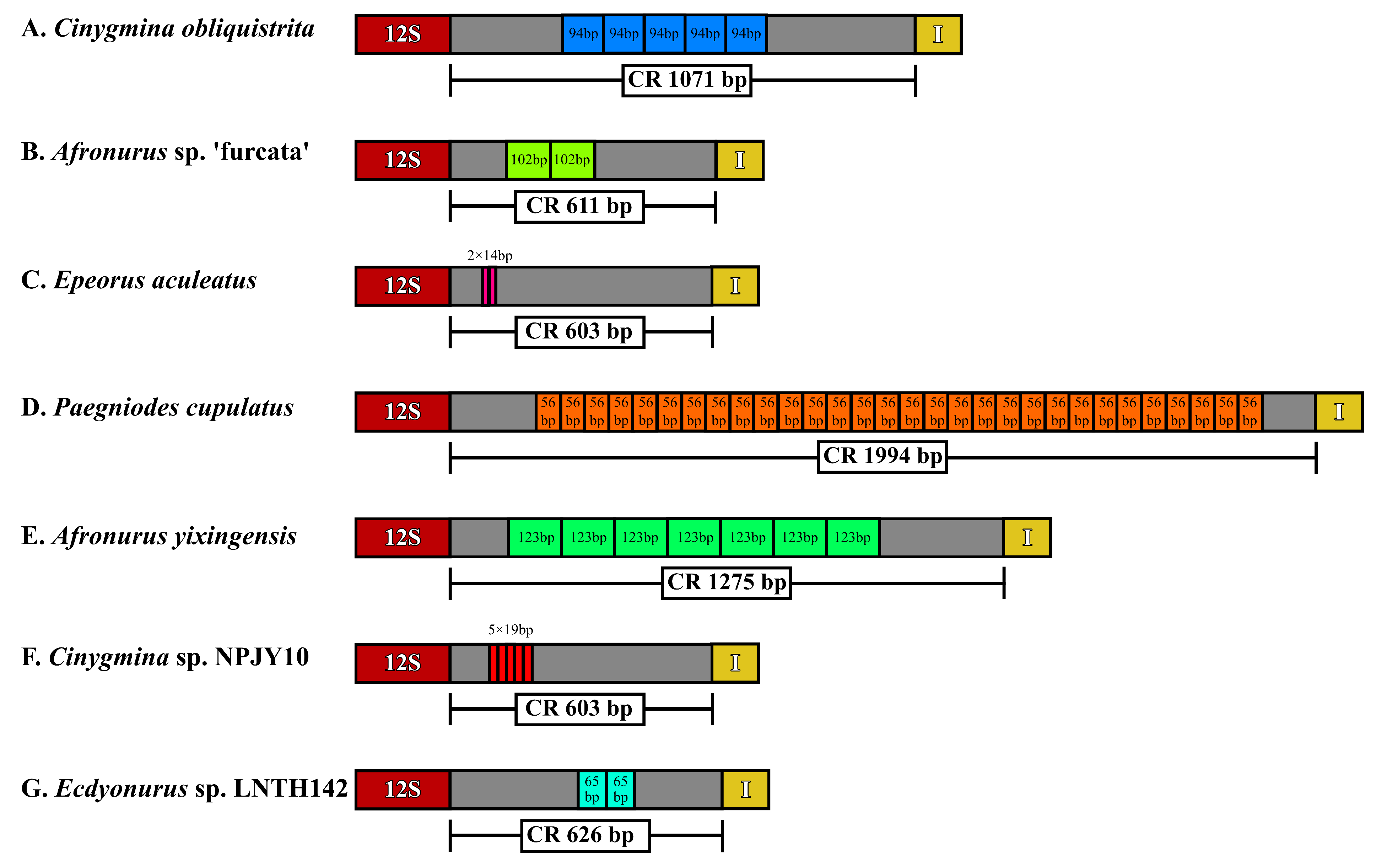
3.1. Characteristics of Mitogenomes
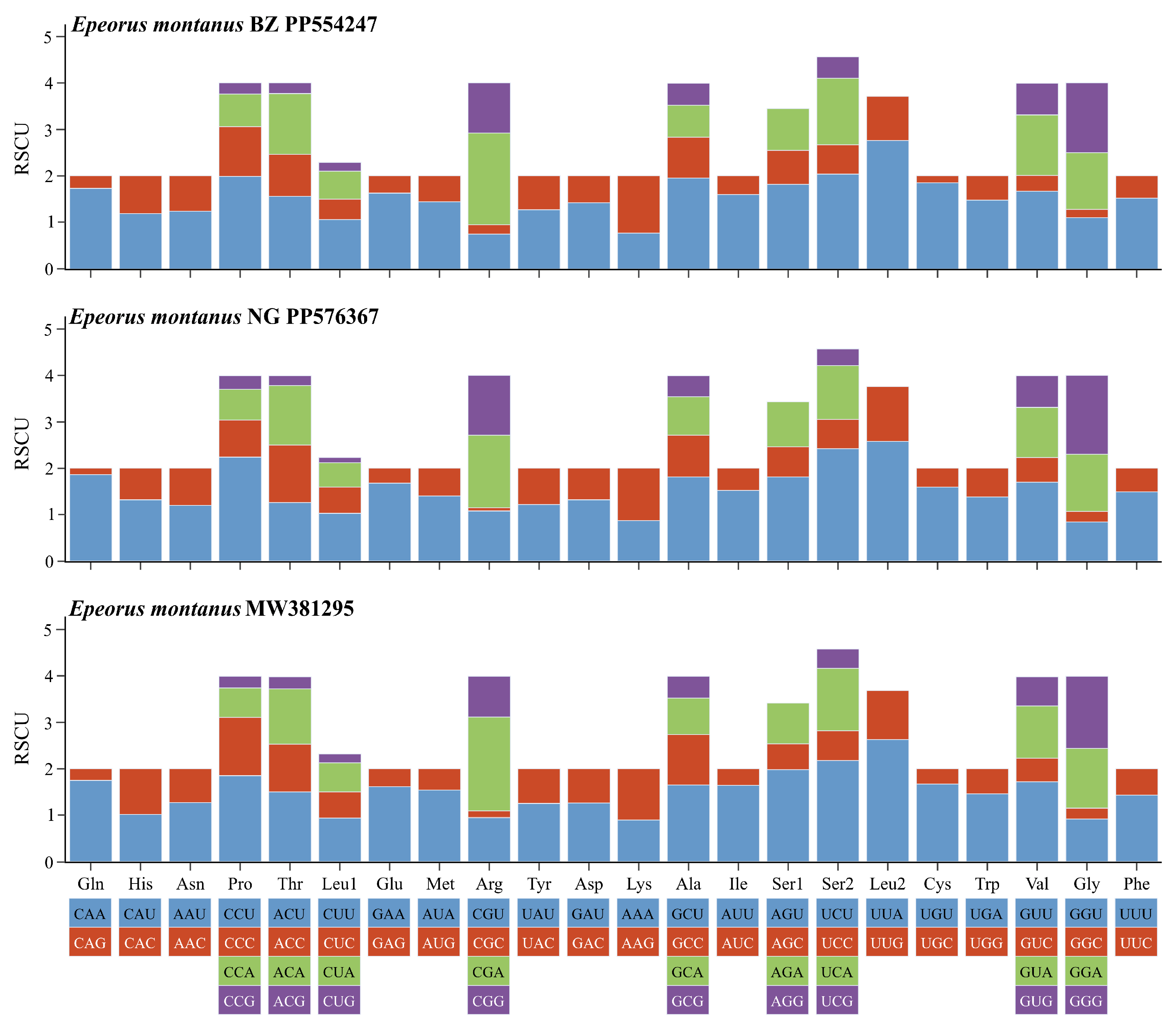
3.2. Characteristics of the Mitogenomes of E. montanus
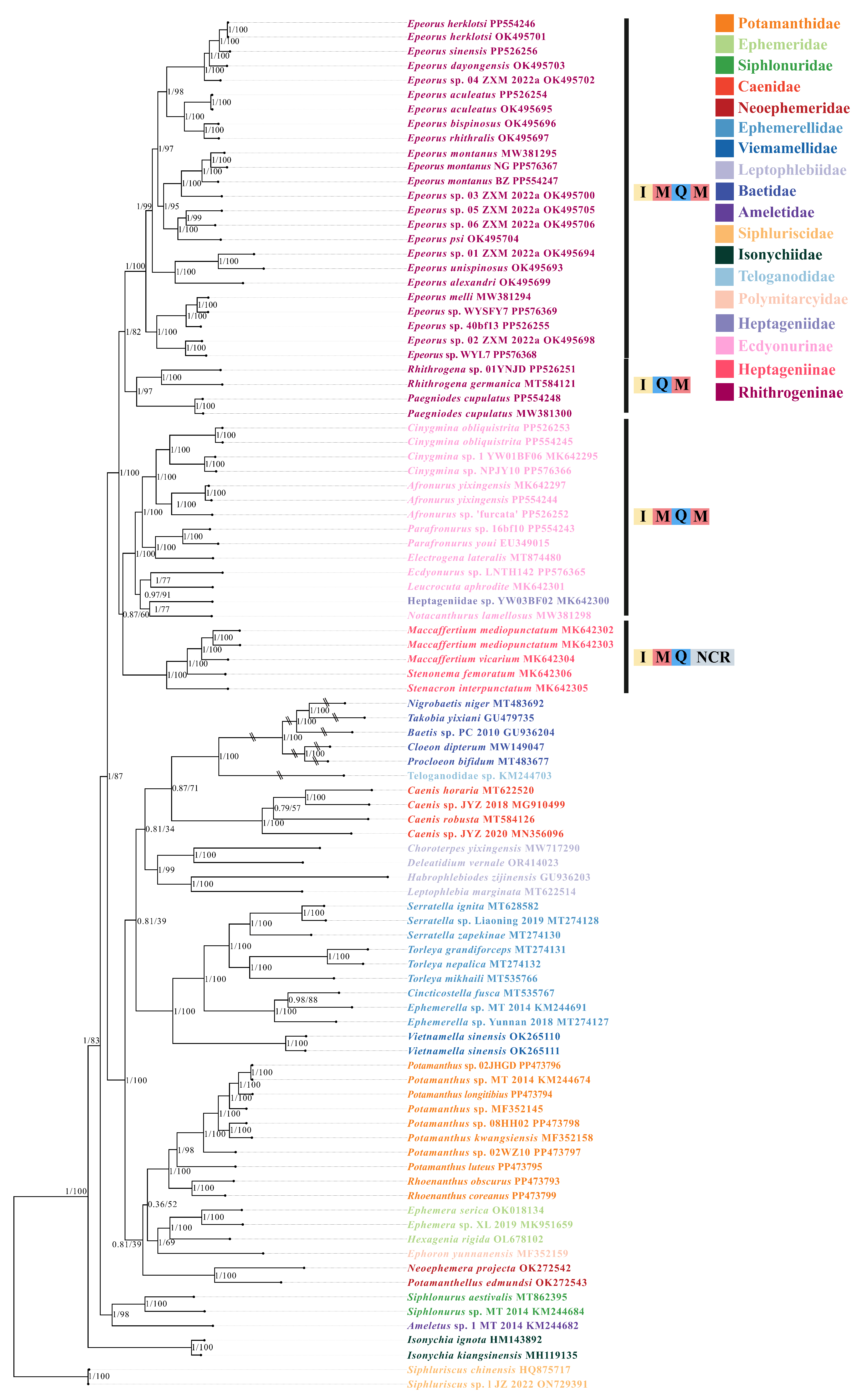
3.3. Phylogenetic Relationships of Heptageniidae
3.4. Estimation of Divergence Time
4. Discussion
4.1. Phylogenetic and Gene Rearrangement Analyses
4.2. The Evolutional Time of Ephemeroptera
4.3. Identification of Cryptic Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brittain, J.E. Biology of mayflies. Annu. Rev. Entomol. 1982, 27, 119–147. [Google Scholar] [CrossRef]
- Ogden, T.H.; Gattolliat, J.L.; Sartori, M.; Staniczek, A.H.; SoldÁN, T.; Whiting, M.F. Towards a new paradigm in mayfly phylogeny (Ephemeroptera): Combined analysis of morphological and molecular data. Syst. Entomol. 2009, 34, 616–634. [Google Scholar] [CrossRef]
- Barber-James, H.M.; Gattolliat, J.L.; Sartori, M.; Hubbard, M.D. Global diversity of mayflies (Ephemeroptera, Insecta) in freshwater. Hydrobiologia 2007, 595, 339–350. [Google Scholar] [CrossRef]
- Webb, J.M. Heptageniidae of the world. Part II: Key to the genera. Can. J. Arthropod Ident. 2008, 7, 1–55. [Google Scholar]
- Jacobus, L.M.; Macadam, C.R.; Sartori, M. Mayflies (Ephemeroptera) and their contributions to ecosystem services. Insects 2019, 10, 6. [Google Scholar] [CrossRef]
- McCafferty, W.P. The Cladistics, Classification, and Evolution of the Heptagenioidea (Ephemeroptera); The American Entomological Institute: Gainesville, FL, USA, 2004; pp. 649–664. [Google Scholar]
- Flowers, R.W.; Hilsenhoff, W.L. Heptageniidae (Ephemeroptera) of Wisconsin. Great Lakes Entomol. 2017, 8, 5. [Google Scholar] [CrossRef]
- Bauernfeind, E.; Soldán, T. The Mayflies of Europe; Apollo Books: Ollerup, Denmark, 2012; p. 781. [Google Scholar]
- Kluge, N.J. The Phylogenetic System of Ephemeroptera; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2004; p. 442. [Google Scholar]
- Ball, S.L.; Hebert, P.D.N.; Burian, S.K.; Webb, J.M. Biological identifications of mayflies (Ephemeroptera) using DNA barcodes. J. N. Am. Benthol. Soc. 2005, 24, 508. [Google Scholar] [CrossRef]
- Ogden, T.H.; Whiting, M.F. Phylogeny of Ephemeroptera (mayflies) based on molecular evidence. Mol. Phylogenet. 2005, 37, 625–643. [Google Scholar] [CrossRef]
- Sun, L.; Sabo, A.; Meyer, M.D.; Randolph, R.P.; Jacobus, L.M.; McCafferty, W.P.; Ferris, V.R. Tests of Current Hypotheses of Mayfly (Ephemeroptera) Phylogeny Using Molecular (18s rDNA) Data. Ann. Entomol. Soc. Am. 2006, 99, 241–252. [Google Scholar] [CrossRef]
- Cai, Y.Y.; Gao, Y.J.; Zhang, L.P.; Yu, D.N.; Storey, K.B.; Zhang, J.Y. The mitochondrial genome of Caenis sp. (Ephemeroptera: Caenidae) and the phylogeny of Ephemeroptera in Pterygota. Mitochondrial DNA Part B 2018, 3, 577–579. [Google Scholar] [CrossRef]
- Cao, S.S.; Xu, X.D.; Jia, Y.Y.; Guan, J.Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. The complete mitochondrial genome of Choroterpides apiculata (Ephemeroptera: Leptophlebiidae) and its phylogenetic relationships. Mitochondrial DNA Part B 2020, 5, 1159–1160. [Google Scholar] [CrossRef]
- Gao, X.Y.; Zhang, S.S.; Zhang, L.P.; Yu, D.N.; Zhang, J.Y.; Cheng, H.Y. The complete mitochondrial genome of Epeorus herklotsi (Ephemeroptera: Heptageniidae) and its phylogeny. Mitochondrial DNA Part B 2018, 3, 303–304. [Google Scholar] [CrossRef]
- Guan, J.Y.; Zhang, Z.Y.; Cao, Y.R.; Xu, X.D.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. The complete mitochondrial genome of Choroterpes (Euthralus) yixingensis (Ephemeroptera: Leptophlebiidae) and its mitochondrial protein-coding gene expression under imidacloprid stress. Gene 2021, 800, 145833. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, W.; Ma, Z.; Zhou, C. Novel gene rearrangement pattern in the mitochondrial genomes of Torleya mikhaili and Cincticostella fusca (Ephemeroptera: Ephemerellidae). Int. J. Biol. Macromol. 2020, 165, 3106–3114. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Shen, C.Y.; Zhao, Y.Y.; Lin, Y.J.; Wu, L.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. The genetic diversity and the divergence time in extant primitive mayfly, Siphluriscus chinensis Ulmer, 1920 using the mitochondrial genome. Genes 2022, 13, 1780. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Wu, L.; Ayivi, S.P.G.; Storey, K.B.; Ma, Y.; Yu, D.N.; Zhang, J.Y. Cryptic species exist in Vietnamella sinensis Hsu, 1936 (Insecta: Ephemeroptera) from studies of complete mitochondrial genomes. Insects 2022, 13, 412. [Google Scholar] [CrossRef]
- Tong, Y.; Wu, L.; Lin, Y.J.; Ayivi, S.P.G.; Storey, K.B.; Zhang, J.Y.; Yu, D.N. The first complete mitochondrial genome of Hexagenia rigida Mc Dunnough, 1924 (Ephemeroptera: Ephemeridae) and its phylogeny. Mitochondrial DNA Part B 2022, 7, 1093–1095. [Google Scholar] [CrossRef]
- Xu, X.D.; Guan, J.Y.; Zhang, Z.Y.; Cao, Y.R.; Cai, Y.Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Insight into the phylogenetic relationships among three subfamilies within Heptageniidae (Insecta: Ephemeroptera) along with low-temperature selection pressure analyses using mitogenomes. Insects 2021, 12, 656. [Google Scholar] [CrossRef]
- Xu, X.D.; Jia, Y.Y.; Cao, S.S.; Zhang, Z.Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Six complete mitochondrial genomes of mayflies from three genera of Ephemerellidae (Insecta: Ephemeroptera) with inversion and translocation of trnI rearrangement and their phylogenetic relationships. PeerJ 2020, 8, e9740. [Google Scholar] [CrossRef]
- Xu, X.D.; Jia, Y.Y.; Dai, X.Y.; Ma, J.L.; Storey, K.B.; Zhang, J.Y.; Yu, D.N. The mitochondrial genome of Caenis sp. (Ephemeroptera: Caenidae) from Fujian and the phylogeny of Caenidae within Ephemeroptera. Mitochondrial DNA Part B 2019, 5, 192–193. [Google Scholar] [CrossRef]
- Ye, Q.M.; Zhang, S.S.; Cai, Y.Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. The complete mitochondrial genome of Isonychia kiangsinensis (Ephemeroptera: Isonychiidae). Mitochondrial DNA Part B 2018, 3, 541–542. [Google Scholar] [CrossRef]
- Yu, D.N.; Yu, P.P.; Zhang, L.P.; Storey, K.B.; Gao, X.Y.; Zhang, J.Y. Increasing 28 mitogenomes of Ephemeroptera, Odonata and Plecoptera support the Chiastomyaria hypothesis with three different outgroup combinations. PeerJ 2021, 9, e11402. [Google Scholar] [CrossRef]
- Ogden, T.H.; Breinholt, J.W.; Bybee, S.M.; Miller, D.B.; Sartori, M.; Shiozawa, D.; Whiting, M.F. Mayfly phylogenomics: Initial evaluation of anchored hybrid enrichment data for the order Ephemeroptera. Zoosymposia 2019, 16, 167–181. [Google Scholar]
- Wang, L.; Li, B.; Jiang, J.; Tong, X. The complete mitochondrial genome of Ephemera serica (Ephemeroptera: Ephemeridae) and phylogenetic analysis. Mitochondrial DNA Part B 2022, 7, 461–463. [Google Scholar] [CrossRef] [PubMed]
- Zurwerra, A.; Metzler, M.; Tomka, I. Biochemical systematics and evolution of the European Heptageniidae (Ephemeroptera). Arch. Hydrobiol. 1987, 109, 481–510. [Google Scholar] [CrossRef]
- Ma, Z.; Li, R.; Zhu, B.; Zheng, X.; Zhou, C. Comparative mitogenome analyses of subgenera and species groups in Epeorus (Ephemeroptera: Heptageniidae). Insects 2022, 13, 599. [Google Scholar] [CrossRef]
- García-Girón, J.; Múrria, C.; Arnedo, M.A.; Bonada, N.; Cañedo-Argüelles, M.; Derka, T.; Fernández-Calero, J.M.; Li, Z.; Tierno de Figueroa, J.M.; Xie, Z.; et al. A time-calibrated ‘Tree of Life’ of aquatic insects for knitting historical patterns of evolution and measuring extant phylogenetic biodiversity across the world. Earth Sci. Rev. 2024, 252, 104767. [Google Scholar] [CrossRef]
- Webb, J.M.; Sun, L.; McCafferty, W.P.; Ferris, V.R. A new species and new synonym in Heptagenia Walsh (Ephemeroptera: Heptageniidae: Heptageniinae) based on molecular and morphological evidence. J. Insect Sci. 2007, 7, 63. [Google Scholar] [CrossRef]
- Wang, T.Q.; McCafferty, W.P. Heptageniidae (Ephemeroptera) of the world. Part I: Phylogenetic higher classification. T. Am. Entomol. Soc. 2004, 130, 11–45. [Google Scholar]
- Monjardim, M.; Paresque, R.; Salles, F.F. Phylogeny and classification of Leptophlebiidae (Ephemeroptera) with an emphasis on Neotropical fauna. Syst. Entomol. 2020, 45, 415–429. [Google Scholar] [CrossRef]
- Sartori, M.; Brittain, J.E. Order Ephemeroptera. In Ecology and General Biology, Vol I: Thorp and Covich’s Freshwater Invertebrates, 4th ed.; Thorp, J.H., Rogers, D.C., Eds.; Academic Press: New York, NY, USA, 2015; pp. 873–891. [Google Scholar]
- Sroka, P.; Godunko, R.J.; Prokop, J. Fluctuation in the diversity of mayflies (Insecta, Ephemerida) as documented in the fossil record. Sci. Rep. 2023, 13, 16052. [Google Scholar] [CrossRef]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef]
- Kohli, M.L.; Letsch, H.; Greve, C.; Béthoux, O.; Deregnaucourt, I.; Liu, S.; Zhou, X.; Donath, A.; Mayer, C.; Podsiadlowski, L.; et al. Evolutionary history and divergence times of Odonata (dragonflies and damselflies) revealed through transcriptomics. iScience 2021, 14, 103324. [Google Scholar] [CrossRef]
- Vuataz, L.; Rutschmann, S.; Monaghan, M.T.; Sartori, M. Molecular phylogeny and timing of diversification in Alpine Rhithrogena (Ephemeroptera: Heptageniidae). BMC Evol. Biol. 2016, 16, 194. [Google Scholar] [CrossRef]
- Winker, K. Sibling species were first recognized by William Derham (1718). Auk 2005, 122, 706–707. [Google Scholar] [CrossRef]
- Struck, T.H.; Cerca, J. Cryptic species and their evolutionary significance. Encycl. Life Sci. 2019, 2019, 1–9. [Google Scholar]
- Stauffer-Olsen, N.J.; O’Grady, P.M.; Resh, V.H. Cytochrome oxidase I sequences from northern and southern California suggest cryptic Baetis (Ephemeroptera: Baetidae) species. West. N. Am. Nat. 2019, 79, 204–218. [Google Scholar] [CrossRef]
- Bisconti, R.; Canestrelli, D.; Tenchini, R.; Belfiore, C.; Buffagni, A.; Nascetti, G. Cryptic diversity and multiple origins of the widespread mayfly species group Baetis rhodani (Ephemeroptera: Baetidae) on northwestern Mediterranean islands. Ecol. Evol. 2016, 6, 7901–7910. [Google Scholar] [CrossRef]
- Rutschmann, S.; Gattolliat, J.L.; Hughes, S.J.; Báez, M.; Sartori, M.; Monaghan, M.T. Evolution and island endemism of morphologically cryptic Baetis and Cloeon species (Ephemeroptera, Baetidae) on the Canary Islands and Madeira. Freshwater Biol. 2014, 59, 2516–2527. [Google Scholar] [CrossRef]
- Williams, H.C.; Ormerod, S.J.; Bruford, M.W. Molecular systematics and phylogeography of the cryptic species complex Baetis rhodani (Ephemeroptera, Baetidae). Mol. Phylogenetics Evol. 2006, 40, 370–382. [Google Scholar] [CrossRef]
- Lalueza-Fox, C.; Vuataz, L.; Sartori, M.; Wagner, A.; Monaghan, M.T. Toward a DNA taxonomy of Alpine Rhithrogena (Ephemeroptera: Heptageniidae) using a mixed yule-coalescent analysis of mitochondrial and nuclear DNA. PLoS ONE 2011, 6, e19728. [Google Scholar] [CrossRef]
- Yang, X.J.; Zhao, Z.S.; Zhang, Y.M.; Ying, J.P.; Wang, S.H.; Yuan, M.L.; Zhang, Q.L. A method for isolating highly purified and active mitochondria from insects. J. Insect Physiol. 2022, 140, 104402. [Google Scholar] [CrossRef]
- Goodsell, D.S. Mitochondrion. Biochem. Mol. Biol. Educ. 2010, 38, 134–140. [Google Scholar] [CrossRef]
- Nakao, M.; McManus, D.P.; Schantz, P.M.; Craig, P.S.; Ito, A. A molecular phylogeny of the genus Echinococcus inferred from complete mitochondrial genomes. Parasitology 2006, 134, 713–722. [Google Scholar] [CrossRef]
- Yong, H.S.; Song, S.L.; Suana, I.W.; Eamsobhana, P.; Lim, P.E. Complete mitochondrial genome of Orthetrum dragonflies and molecular phylogeny of Odonata. Biochem. Syst. Ecol. 2016, 69, 124–131. [Google Scholar] [CrossRef]
- Liu, Y.; Li, H.; Song, F.; Zhao, Y.; Wilson, J.J.; Cai, W. Higher-level phylogeny and evolutionary history of Pentatomomorpha (Hemiptera: Heteroptera) inferred from mitochondrial genome sequences. Syst. Entomol. 2019, 44, 810–819. [Google Scholar] [CrossRef]
- Li, H.; Shao, R.; Song, N.; Song, F.; Jiang, P.; Li, Z.; Cai, W. Higher-level phylogeny of paraneopteran insects inferred from mitochondrial genome sequences. Sci. Rep. 2015, 5, 8527. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, Y.; Liu, Q.; Tian, L.; Li, H.; Song, F.; Cai, W. Exploring the mitogenomes of Mantodea: New insights from structural diversity and higher-level phylogenomic analyses. Int. J. Mol. Sci. 2023, 24, 10570. [Google Scholar] [CrossRef]
- Xu, H.; Liu, X.; Wang, P.; Li, H.; Wu, S. Phylogenetic implications of mitogenomic sequences and gene rearrangements of scale insects (Hemiptera, Coccoidea). Insects 2023, 14, 257. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef]
- Logan, D.C. The mitochondrial compartment. J. Exp. Bot. 2006, 57, 1225–1243. [Google Scholar] [CrossRef]
- Cameron, S.L. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst. Entomol. 2014, 39, 400–411. [Google Scholar] [CrossRef]
- Li, D.; Qin, J.C.; Zhou, C.F. The phylogeny of Ephemeroptera in Pterygota revealed by the mitochondrial genome of Siphluriscus chinensis (Hexapoda: Insecta). Gene 2014, 545, 132–140. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Zhang, J.; Zhou, C.; Gai, Y.; Song, D.; Zhou, K. The complete mitochondrial genome of Parafronurus youi (Insecta: Ephemeroptera) and phylogenetic position of the Ephemeroptera. Gene 2008, 424, 18–24. [Google Scholar] [CrossRef]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. In Bioinformatics Methods and Protocols; Humana Press: Totowa, NJ, USA, 2000; Volume 132, pp. 71–91. [Google Scholar]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Allio, R.; Schomaker-Bastos, A.; Romiguier, J.; Prosdocimi, F.; Nabholz, B.; Delsuc, F. MitoFinder: Efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol. Ecol. Resour. 2020, 20, 892–905. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogen. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S.; Battistuzzi, F.U. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Serif (Europe) Ltd. Affinity Designer 2. 2014. Available online: https://affinity.serif.com/zh-cn/ (accessed on 12 April 2024).
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Kück, P.; Meid, S.A.; Groß, C.; Wägele, J.W.; Misof, B. AliGROOVE–visualization of heterogeneous sequence divergence within multiple sequence alignments and detection of inflated branch support. BMC Bioinf. 2014, 15, 294. [Google Scholar] [CrossRef]
- Tang, M.; Tan, M.; Meng, G.; Yang, S.; Su, X.; Liu, S.; Song, W.; Li, Y.; Wu, Q.; Zhang, A.; et al. Multiplex sequencing of pooled mitochondrial genomes—A crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 2014, 42, e166. [Google Scholar] [CrossRef]
- Rutschmann, S.; Chen, P.; Zhou, C.; Monaghan, M.T. Mitochondrial genomes infer phylogenetic relationships among the oldest extant winged insects (Palaeoptera). BioRxiv 2017, 164459. [Google Scholar] [CrossRef]
- Rutschmann, S.; Chen, P.; Zhou, C.; Monaghan, M.T. Three mitochondrial genomes of early-winged insects (Ephemeroptera: Baetidae and Leptophlebiidae). Mitochondrial DNA Part B 2021, 6, 2969–2971. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Lei, Z.; Li, W.; Zhang, W.; Zhou, C. Comparative mitogenomic analysis of Heptageniid mayflies (Insecta: Ephemeroptera): Conserved intergenic spacer and tRNA gene duplication. Insects 2021, 12, 170. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.Q.; Gao, Y.J.; Chen, Y.X.; Zhan, L.M.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Comparative mitogenome of phylogenetic relationships and divergence time analysis within Potamanthidae (Insecta: Ephemeroptera). Insects 2024, 15, 357. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Ho, S.Y.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef]
- Liegeois, M.; Sartori, M.; Schwander, T.; Orive, M. Extremely widespread parthenogenesis and a trade-off between alternative forms of reproduction in mayflies (Ephemeroptera). J. Hered. 2021, 112, 45–57. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A. Figtree Version 1.4.0. 2012. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 12 April 2024).
- Huang, J.D.; Ren, D.; Sinitshenkova, N.D.; Shih, C.K. New fossil mayflies (Insecta: Ephemeroptera) from the Middle Jurassic of Daohugou, Inner Mongolia, China. Insect Sci. 2008, 15, 193–198. [Google Scholar] [CrossRef]
- Godunko, R.J.; Martynov, A.V.; Staniczek, A.H. First fossil record of the mayfly family Vietnamellidae (Insecta, Ephemeroptera) from Burmese amber confirms its Oriental origin and gives new insights into its evolution. ZooKeys 2021, 1036, 99. [Google Scholar] [CrossRef]
- Staniczek, A.H.; Godunko, R.J.; Krzeminski, W. A new fossil mayfly species of the genus Borinquena Traver, 1938 (Insecta: Ephemeroptera: Leptophlebiidae: Atalophlebiinae) from Miocene Dominican amber. Annal Zool. 2017, 67, 113–119. [Google Scholar] [CrossRef]
- Staniczek, A.H.; Godunko, R.J.; Kluge, N.J. Fossil record of the mayfly family Ephemerellidae (Insecta: Ephemeroptera), with description of new species and first report of Ephemerellinae from Baltic amber. J. Syst. Palaeontol. 2018, 16, 1319–1335. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.Y.; Cao, L.J.; Tang, P.; van Achterberg, K.; Hoffmann, A.A.; Chen, H.Y.; Chen, X.X.; Wei, S.J. Gene arrangement and sequence of mitochondrial genomes yield insights into the phylogeny and evolution of bees and sphecid wasps (Hymenoptera: Apoidea). Mol. Phylogenet. 2018, 124, 1–9. [Google Scholar] [CrossRef]
- Tyagi, K.; Chakraborty, R.; Cameron, S.L.; Sweet, A.D.; Chandra, K.; Kumar, V. Rearrangement and evolution of mitochondrial genomes in Thysanoptera (Insecta). Sci. Rep. 2020, 10, 695. [Google Scholar] [CrossRef]
- Sinitshenkova, N.D. Main ecological events in aquatic insects history. Act. Zool. Cracov. 2003, 46, 381–392. [Google Scholar]
- Katz, M.E.; Finkel, Z.V.; Grzebyk, D.; Knoll, A.H.; Falkowski, P.G. Evolutionary trajectories and biogeochemical impacts of marine eukaryotic phytoplankton. Annu. Rev. Ecol. Evol. Syst. 2004, 35, 523–556. [Google Scholar] [CrossRef]
- Föllmi, K.B. Early Cretaceous life, climate and anoxia. Cretaceous Res. 2012, 35, 230–257. [Google Scholar] [CrossRef]
- Corrêa, E.C.; Roque, F.d.O.; Utz, R.M.; Correa, J.d.S.; de Souza, F.L.; Covich, A.P. Effects of macroconsumers on benthic communities: Rapid increases in dry-season accrual of calcium in a tropical karst stream. PLoS ONE 2018, 13, e0209102. [Google Scholar] [CrossRef]
- Abrams, P.A. The evolution of predator-prey interactions: Theory and evidence. Annu. Rev. Ecol. Evol. S. 2000, 31, 79–105. [Google Scholar] [CrossRef]
- Seehausen, O. African cichlid fish: A model system in adaptive radiation research. Proc. Biol. Sci. 2006, 273, 1987–1998. [Google Scholar] [CrossRef] [PubMed]
- Morii, Y.; Prozorova, L.; Chiba, S. Parallel evolution of passive and active defence in land snails. Sci. Rep. 2016, 6, 35600. [Google Scholar] [CrossRef] [PubMed]
- Hrivniak, Ľ.; Sroka, P.; Bojková, J.; Godunko, R.J.; Imanpour Namin, J.; Bagheri, S.; Nejat, F.; Abdoli, A.; Staniczek, A.H. Diversity and distribution of Epeorus (Caucasiron) (Ephemeroptera, Heptageniidae) in Iran, with descriptions of three new species. ZooKeys 2020, 947, 71–102. [Google Scholar] [CrossRef] [PubMed]
- Hrivniak, Ľ.; Sroka, P.; Bojková, J.; Godunko, R.J.; Soldán, T.; Staniczek, A.H. The impact of Miocene orogeny for the diversification of Caucasian Epeorus (Caucasiron) mayflies (Ephemeroptera: Heptageniidae). Mol. Phylogenet. Evol. 2020, 146, 106735. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, Z.; Han, F.; Zhang, Q.; Pan, J.; Ma, J.; Yang, B. Stable isotope evidence for a Paleogene high-altitude setting of the Sikeshu drainage basin in the northern Tianshan, western China. Geomorphology 2019, 342, 51–60. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]



| Number | Species | Subfamily | Length | Sampling Localities | Accession No. |
|---|---|---|---|---|---|
| 02WZ07 | Afronurus sp. ‘furcata’ | Ecdyonurinae | 15,436 | Taishun, Zhejiang | PP526252 |
| FZZX10 | Afronurus yixingensis | Ecdyonurinae | 16,120 | Fuzhou, Jiangxi | PP554244 |
| 02JZWC | Cinygmina obliquistrita | Ecdyonurinae | 15,249 | Zhuji, Zhejiang | PP554245 |
| 02WZ03 | Cinygmina obliquistrita | Ecdyonurinae | 15,916 | Taishun, Zhejiang | PP526253 |
| NPJY10 | Cinygmina sp. NPJY10 | Ecdyonurinae | 15,447 | Nanping, Fujian | PP576366 |
| 16bf10 | Parafronurus sp. 16bf10 | Ecdyonurinae | 15,527 | Longquan, Zhejiang | PP554243 |
| LNTH142 | Ecdyonurus sp. LNTH142 | Ecdyonurinae | 15,453 | Tonghua, Liaoning | PP576365 |
| 07BF95 | Epeorus aculeatus | Rhithrogeninae | 15,466 | Tonglu, Zhejiang | PP526254 |
| 02WYNT | Epeorus herklotsi | Rhithrogeninae | 15,498 | Lishui, Zhejiang | PP554246 |
| 40bf13 | Epeorus sp. 40bf13 | Rhithrogeninae | 15,508 | Wuyi, Zhejiang | PP526255 |
| BZD4 | Epeorus montanus BZ | Rhithrogeninae | 15,476 | Akesu, Xinjiang | PP554247 |
| NLTYT6 | Epeorus montanus NG | Rhithrogeninae | 15,472 | Xinyuan, Xinjiang | PP576367 |
| 02WZ01 | Epeorus sinensis | Rhithrogeninae | 15,508 | Taishun, Zhejiang | PP526256 |
| WYL7 | Epeorus sp. WYL7 | Rhithrogeninae | 15,546 | Taishun, Zhejiang | PP576368 |
| WYSFY7 | Epeorus sp. WYSFY7 | Rhithrogeninae | 15,496 | Taishun, Zhejiang | PP576369 |
| 34bf05 | Paegniodes cupulatus | Rhithrogeninae | 16,784 | Kaihua, Zhejiang | PP554248 |
| 01YNJD | Rhithrogena germanica | Rhithrogeninae | 15,398 | Jingdong, Yunnan | PP526251 |
| E. montanus BZ PP554247 | E. montanus NG PP576367 | E. montanus MW381295 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | Strand | Length (bp) | AT% | AT-skew | CG-skew | Length (bp) | AT% | AT-skew | CG-skew | Length (bp) | AT% | AT-skew | CG-skew |
| Full mitogenome | Heavy | 15,476 | 65.3 | −0.023 | −0.197 | 15,472 | 64.8 | −0.025 | −0.21 | 15472 | 64.8 | −0.019 | −0.21 |
| PCG | Heavy | 6882 | 62.9 | −0.195 | −0.167 | 6882 | 61.9 | −0.205 | −0.176 | 6882 | 61.9 | −0.192 | −0.186 |
| Light | 4326 | 66.2 | −0.227 | 0.279 | 4326 | 66.1 | −0.226 | 0.296 | 4326 | 66 | −0.225 | 0.27 | |
| tRNA | Heavy | 987 | 64.8 | −0.037 | 0.026 | 987 | 64.6 | −0.022 | 0.009 | 986 | 64.7 | −0.019 | 0.105 |
| Light | 529 | 65.4 | −0.012 | 0.279 | 529 | 65.8 | −0.011 | 0.282 | 529 | 64.8 | −0.009 | 0.28 | |
| rRNA | Light | 2066 | 67.2 | 0.03 | 0.26 | 2065 | 67.1 | 0.029 | 0.261 | 2065 | 67.3 | 0.026 | 0.262 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Z.-Q.; Shen, C.-Y.; Cheng, H.-Y.; Chen, Y.-X.; Wu, H.-Y.; Storey, K.B.; Yu, D.-N.; Zhang, J.-Y. Mitogenome-Based Phylogeny with Divergence Time Estimates Revealed the Presence of Cryptic Species within Heptageniidae (Insecta, Ephemeroptera). Insects 2024, 15, 745. https://doi.org/10.3390/insects15100745
Guo Z-Q, Shen C-Y, Cheng H-Y, Chen Y-X, Wu H-Y, Storey KB, Yu D-N, Zhang J-Y. Mitogenome-Based Phylogeny with Divergence Time Estimates Revealed the Presence of Cryptic Species within Heptageniidae (Insecta, Ephemeroptera). Insects. 2024; 15(10):745. https://doi.org/10.3390/insects15100745
Chicago/Turabian StyleGuo, Zhi-Qiang, Chen-Yang Shen, Hong-Yi Cheng, Yu-Xin Chen, Hui-Yuan Wu, Kenneth B. Storey, Dan-Na Yu, and Jia-Yong Zhang. 2024. "Mitogenome-Based Phylogeny with Divergence Time Estimates Revealed the Presence of Cryptic Species within Heptageniidae (Insecta, Ephemeroptera)" Insects 15, no. 10: 745. https://doi.org/10.3390/insects15100745







