Molecular Mechanisms of Drug Resistance in Leishmania spp.
Abstract
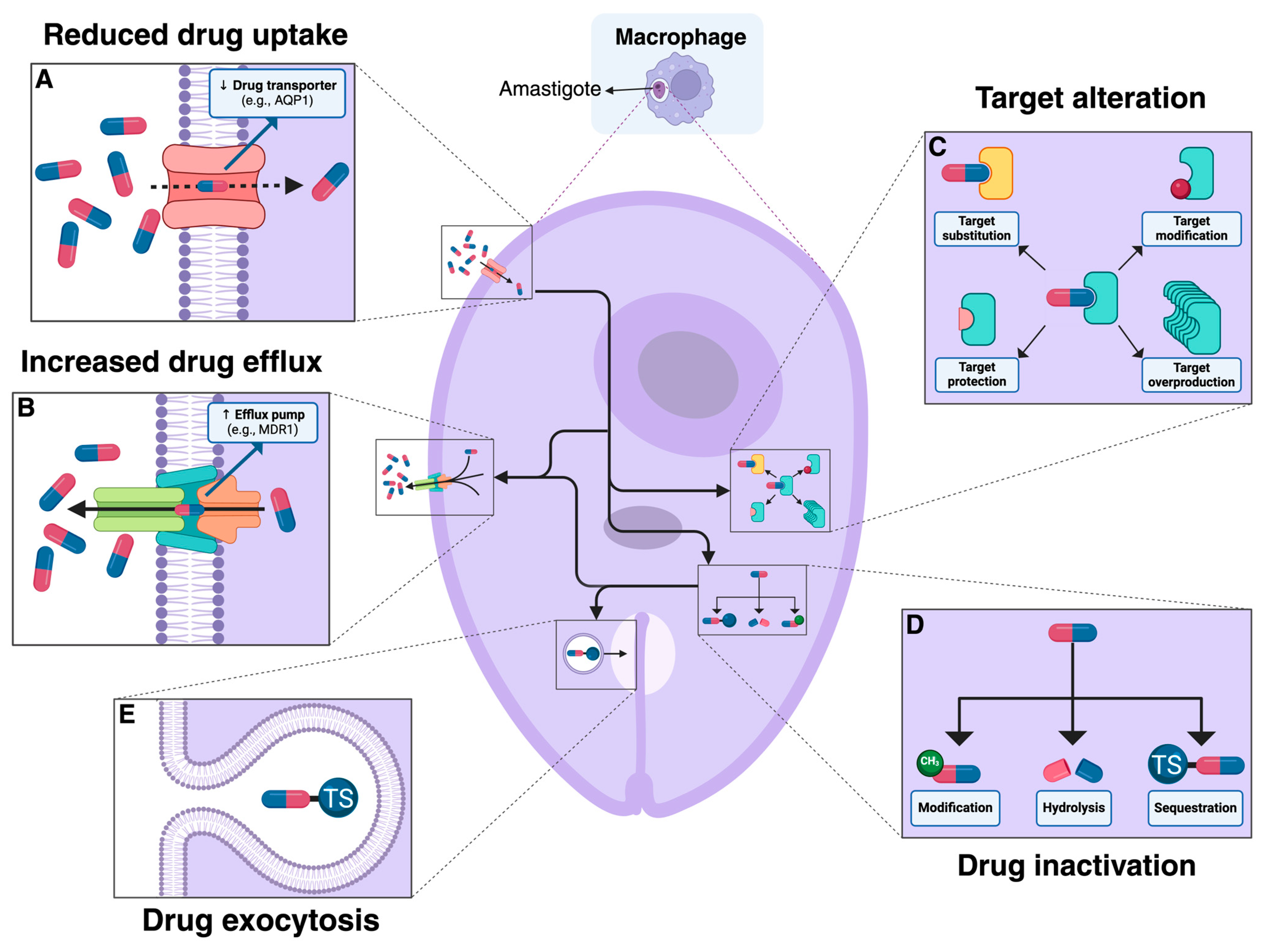
:1. Introduction
2. Available Treatments against Leishmaniasis
3. Genomic Changes and Drug Resistance
4. Changes in Transcriptomes Associated with Drug Resistance
5. Translational Control as a Major Driver of Drug Resistance
6. Changes in Metabolomes and Lipidomes Associated with Drug Resistance
7. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| aa-tRNA | Aminoacyl transfer ribonucleic acid |
| ABC | ATP binding cassette |
| AmB | Amphotericin B |
| APX | Ascorbate Peroxidase |
| AQP1 | Aquaglyceroporin 1 |
| ATP | Adenosine triphosphate |
| BCAT | Branched-chain amino acids |
| CDP | Cytidine-5’-diphosphate |
| CNV | Copy number variation |
| CTP | Cytidine-5’-triphosphate |
| CDPK1 | Calcium dependent protein kinase 1 |
| CL | Cutaneous Leishmaniasis |
| CRISPR-Cas9 | Clustered Regularly Interspaced Short Palindromic Repeats-Protein 9 |
| CYP51 | Sterol 14-demethylase |
| D-LDH | D-lactate dehydrogenase-like protein |
| DNA | Deoxyribonucleic Acid |
| DEGs | Differentially expressed genes |
| FDA | Food and Drug administration |
| G6PDH | Glucose-6-Phosphate Dehydrogenase |
| HAPT1 | High affinity pentamidine transporter |
| HIV | Human immunodeficiency virus |
| JAK-STAT | Janus kinase/signal transducers and activators of transcription |
| LMT | Miltefosine Transporter gene |
| MCL | Muco-Cutaneous Leishmaniasis |
| MDR1 | Multi-drug resistance 1 |
| MLT | Miltefosine |
| mRNA | Messenger ribonucleic acid |
| MRPA | Multidrug-resistance protein A |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NTD | Neglected tropical disease |
| PC | Phosphatidylcholines |
| PE | Phosphatidylethanolamines |
| PL | Phospholipids |
| PMM | Paromomycin |
| PRP1 | Pentamidine resistance protein 1 |
| PTM | Pentamidine |
| PTUs | Polycistronic transcription units |
| PDR-1 | Pectin degradation regulator-1 |
| RBP | RNA Binding protein |
| RBPs | RNA-binding proteins |
| RNAP II | RNA polymerase II |
| RPP | Ribosomal protection protein |
| rRNA | Ribosomal ribonucleic acid |
| SbV | Pentavalent antimony |
| SC5D | Sterol C5 -desaturase |
| SMT | 24-sterol methyltransferase |
| SNPs | Single nucleotide polymorphisms |
| VL | Visceral Leishmaniasis |
References
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 481–511. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.W.K.; Millar, B.C.; Moore, J.E. Antimicrobial Resistance (AMR). Br. J. Biomed. Sci. 2023, 80, 11387. [Google Scholar] [CrossRef]
- Boyce, K.J. The Microevolution of Antifungal Drug Resistance in Pathogenic Fungi. Microorganisms 2023, 11, 2757. [Google Scholar] [CrossRef]
- Kanaujia, R.; Singh, S.; Rudramurthy, S.M. Aspergillosis: An Update on Clinical Spectrum, Diagnostic Schemes, and Management. Curr. Fungal Infect. Rep. 2023, 17, 144–155. [Google Scholar] [CrossRef]
- Cohen, N.R.; Lobritz, M.A.; Collins, J.J. Microbial persistence and the road to drug resistance. Cell Host Microbe 2013, 13, 632–642. [Google Scholar] [CrossRef]
- Tarannum, A.; Rodriguez-Almonacid, C.C.; Salazar-Bravo, J.; Karamysheva, Z.N. Molecular Mechanisms of Persistence in Protozoan Parasites. Microorganisms 2023, 11, 2248. [Google Scholar] [CrossRef]
- Peraman, R.; Sure, S.K.; Dusthackeer, V.N.A.; Chilamakuru, N.B.; Yiragamreddy, P.R.; Pokuri, C.; Kutagulla, V.K.; Chinni, S. Insights on recent approaches in drug discovery strategies and untapped drug targets against drug resistance. Future J. Pharm. Sci. 2021, 7, 56. [Google Scholar] [CrossRef]
- Vitiello, A.; Ferrara, F.; Boccellino, M.; Ponzo, A.; Cimmino, C.; Comberiati, E.; Zovi, A.; Clemente, S.; Sabbatucci, M. Antifungal Drug Resistance: An Emergent Health Threat. Biomedicines 2023, 11, 1063. [Google Scholar] [CrossRef]
- Gaurav, A.; Bakht, P.; Saini, M.; Pandey, S.; Pathania, R. Role of bacterial efflux pumps in antibiotic resistance, virulence, and strategies to discover novel efflux pump inhibitors. Microbiology 2023, 169, 001333. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, V.K.; Pathania, R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J. Med. Res. 2019, 149, 129–145. [Google Scholar] [CrossRef]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Alastruey-Izquierdo, A.; Berman, J.; Bicanic, T.; Bignell, E.M.; Bowyer, P.; Bromley, M.; Bruggemann, R.; Garber, G.; Cornely, O.A.; et al. Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Microbiol. 2022, 20, 557–571. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int. J. Mol. Sci. 2020, 21, 3233. [Google Scholar] [CrossRef]
- Iwamoto, H.; Abe, M.; Yang, Y.; Cui, D.; Seki, T.; Nakamura, M.; Hosaka, K.; Lim, S.; Wu, J.; He, X.; et al. Cancer Lipid Metabolism Confers Antiangiogenic Drug Resistance. Cell Metab. 2018, 28, 104–117.e5. [Google Scholar] [CrossRef]
- Khan, S.U.; Fatima, K.; Aisha, S.; Malik, F. Unveiling the mechanisms and challenges of cancer drug resistance. Cell Commun. Signal 2024, 22, 109. [Google Scholar] [CrossRef]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef]
- Kusnadi, E.P.; Trigos, A.S.; Cullinane, C.; Goode, D.L.; Larsson, O.; Devlin, J.R.; Chan, K.T.; De Souza, D.P.; McConville, M.J.; McArthur, G.A.; et al. Reprogrammed mRNA translation drives resistance to therapeutic targeting of ribosome biogenesis. EMBO J. 2020, 39, e105111. [Google Scholar] [CrossRef]
- Munday, J.C.; Settimo, L.; de Koning, H.P. Transport proteins determine drug sensitivity and resistance in a protozoan parasite, Trypanosoma brucei. Front. Pharmacol. 2015, 6, 32. [Google Scholar] [CrossRef]
- Karamysheva, Z.N.; Gutierrez Guarnizo, S.A.; Karamyshev, A.L. Regulation of Translation in the Protozoan Parasite Leishmania. Int. J. Mol. Sci. 2020, 21, 2981. [Google Scholar] [CrossRef] [PubMed]
- Maltezou, H.C. Drug resistance in visceral leishmaniasis. J. Biomed. Biotechnol. 2010, 2010, 617521. [Google Scholar] [CrossRef] [PubMed]
- Marquis, N.; Gourbal, B.; Rosen, B.P.; Mukhopadhyay, R.; Ouellette, M. Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant Leishmania. Mol. Microbiol. 2005, 57, 1690–1699. [Google Scholar] [CrossRef] [PubMed]
- Ponte-Sucre, A.; Gamarro, F.; Dujardin, J.C.; Barrett, M.P.; Lopez-Velez, R.; Garcia-Hernandez, R.; Pountain, A.W.; Mwenechanya, R.; Papadopoulou, B. Drug resistance and treatment failure in leishmaniasis: A 21st century challenge. PLoS Negl. Trop. Dis. 2017, 11, e0006052. [Google Scholar] [CrossRef]
- Légaré, D.; Ouellette, M. Drug Resistance in Leishmania. In Handbook of Antimicrobial Resistance; Springer: New York, NY, USA, 2014. [Google Scholar]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 2023, 21, 280–295. [Google Scholar] [CrossRef]
- Kamran, M.; Bhattacharjee, R.; Das, S.; Mukherjee, S.; Ali, N. The paradigm of intracellular parasite survival and drug resistance in leishmanial parasite through genome plasticity and epigenetics: Perception and future perspective. Front. Cell Infect. Microbiol. 2023, 13, 1001973. [Google Scholar] [CrossRef]
- Gutierrez Guarnizo, S.A.; Tikhonova, E.B.; Karamyshev, A.L.; Muskus, C.E.; Karamysheva, Z.N. Translational reprogramming as a driver of antimony-drug resistance in Leishmania. Nat. Commun. 2023, 14, 2605. [Google Scholar] [CrossRef]
- Shaw, C.D.; Lonchamp, J.; Downing, T.; Imamura, H.; Freeman, T.M.; Cotton, J.A.; Sanders, M.; Blackburn, G.; Dujardin, J.C.; Rijal, S.; et al. In vitro selection of miltefosine resistance in promastigotes of Leishmania donovani from Nepal: Genomic and metabolomic characterization. Mol. Microbiol. 2016, 99, 1134–1148. [Google Scholar] [CrossRef]
- Burza, S.; Croft, S.L.; Boelaert, M. Leishmaniasis. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef]
- Shmueli, M.; Ben-Shimol, S. Review of Leishmaniasis Treatment: Can We See the Forest through the Trees? Pharmacy 2024, 12, 30. [Google Scholar] [CrossRef]
- Aronson, N.E.; Joya, C.A. Cutaneous Leishmaniasis: Updates in Diagnosis and Management. Infect. Dis. Clin. N. Am. 2019, 33, 101–117. [Google Scholar] [CrossRef] [PubMed]
- CDC. Clinical Care of Leishmaniasis. Available online: https://www.cdc.gov/leishmaniasis/hcp/clinical-care/index.html (accessed on 4 September 2024).
- Maarouf, M.; de Kouchkovsky, Y.; Brown, S.; Petit, P.X.; Robert-Gero, M. In vivo interference of paromomycin with mitochondrial activity of Leishmania. Exp. Cell Res. 1997, 232, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Bharadava, K.; Upadhyay, T.K.; Kaushal, R.S.; Ahmad, I.; Alraey, Y.; Siddiqui, S.; Saeed, M. Genomic Insight of Leishmania Parasite: In-Depth Review of Drug Resistance Mechanisms and Genetic Mutations. ACS Omega 2024, 9, 12500–12514. [Google Scholar] [CrossRef] [PubMed]
- Faris, R.M.; Jarallah, J.S.; Khoja, T.A.; al-Yamani, M.J. Intralesional treatment of cutaneous leishmaniasis with sodium stibogluconate antimony. Int. J. Dermatol. 1993, 32, 610–612. [Google Scholar] [CrossRef]
- Solomon, M.; Baum, S.; Barzilai, A.; Pavlotsky, F.; Trau, H.; Schwartz, E. Treatment of cutaneous leishmaniasis with intralesional sodium stibogluconate. J. Eur. Acad. Dermatol. Venereol. 2009, 23, 1189–1192. [Google Scholar] [CrossRef]
- Herwaldt, B.L.; Berman, J.D. Recommendations for treating leishmaniasis with sodium stibogluconate (Pentostam) and review of pertinent clinical studies. Am. J. Trop. Med. Hyg. 1992, 46, 296–306. [Google Scholar] [CrossRef]
- Heleine, M.; Elenga, N.; Njuieyon, F.; Martin, E.; Piat, C.; Pansart, C.; Couppie, P.; Hernandez, M.; Demar, M.; Blaizot, R. Using pentamidine to treat cutaneous leishmaniasis in children: A 10-year study in French Guiana. Clin. Exp. Dermatol. 2023, 48, 913–915. [Google Scholar] [CrossRef]
- Lai, A.F.E.J.; Vrede, M.A.; Soetosenojo, R.M.; Lai, A.F.R.F. Pentamidine, the drug of choice for the treatment of cutaneous leishmaniasis in Surinam. Int. J. Dermatol. 2002, 41, 796–800. [Google Scholar] [CrossRef]
- Piccica, M.; Lagi, F.; Bartoloni, A.; Zammarchi, L. Efficacy and safety of pentamidine isethionate for tegumentary and visceral human leishmaniasis: A systematic review. J. Travel. Med. 2021, 28, taab065. [Google Scholar] [CrossRef]
- Pokharel, P.; Ghimire, R.; Lamichhane, P. Efficacy and Safety of Paromomycin for Visceral Leishmaniasis: A Systematic Review. J. Trop. Med. 2021, 2021, 8629039. [Google Scholar] [CrossRef]
- Soto, J.; Soto, P.; Ajata, A.; Luque, C.; Tintaya, C.; Paz, D.; Rivero, D.; Berman, J. Topical 15% Paromomycin-Aquaphilic for Bolivian Leishmania braziliensis Cutaneous Leishmaniasis: A Randomized, Placebo-controlled Trial. Clin. Infect. Dis. 2019, 68, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Sosa, N.; Pascale, J.M.; Jimenez, A.I.; Norwood, J.A.; Kreishman-Detrick, M.; Weina, P.J.; Lawrence, K.; McCarthy, W.F.; Adams, R.C.; Scott, C.; et al. Topical paromomycin for New World cutaneous leishmaniasis. PLoS Negl. Trop. Dis. 2019, 13, e0007253. [Google Scholar] [CrossRef] [PubMed]
- Soto, J.; Toledo, J.; Gutierrez, P.; Nicholls, R.S.; Padilla, J.; Engel, J.; Fischer, C.; Voss, A.; Berman, J. Treatment of American cutaneous leishmaniasis with miltefosine, an oral agent. Clin. Infect. Dis. 2001, 33, E57–E61. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Report of a WHO Informal Consultation on Liposomal Amphotericin B in the Treatment of Visceral Leishmaniasis; WHO/CDS/NTD/IDM/2007.4; WHO: Geneva, Switzerland, 16 April 2005. [Google Scholar]
- Frezard, F.; Aguiar, M.M.G.; Ferreira, L.A.M.; Ramos, G.S.; Santos, T.T.; Borges, G.S.M.; Vallejos, V.M.R.; De Morais, H.L.O. Liposomal Amphotericin B for Treatment of Leishmaniasis: From the Identification of Critical Physicochemical Attributes to the Design of Effective Topical and Oral Formulations. Pharmaceutics 2022, 15, 99. [Google Scholar] [CrossRef]
- Battista, T.; Colotti, G.; Ilari, A.; Fiorillo, A. Targeting Trypanothione Reductase, a Key Enzyme in the Redox Trypanosomatid Metabolism, to Develop New Drugs against Leishmaniasis and Trypanosomiases. Molecules 2020, 25, 1924. [Google Scholar] [CrossRef]
- Kumari, D.; Perveen, S.; Sharma, R.; Singh, K. Advancement in leishmaniasis diagnosis and therapeutics: An update. Eur. J. Pharmacol. 2021, 910, 174436. [Google Scholar] [CrossRef]
- Pinto-Martinez, A.K.; Rodriguez-Duran, J.; Serrano-Martin, X.; Hernandez-Rodriguez, V.; Benaim, G. Mechanism of Action of Miltefosine on Leishmania donovani Involves the Impairment of Acidocalcisome Function and the Activation of the Sphingosine-Dependent Plasma Membrane Ca2+ Channel. Antimicrob. Agents Chemother. 2018, 62, 1. [Google Scholar] [CrossRef]
- Rakotomanga, M.; Blanc, S.; Gaudin, K.; Chaminade, P.; Loiseau, P.M. Miltefosine affects lipid metabolism in Leishmania donovani promastigotes. Antimicrob. Agents Chemother. 2007, 51, 1425–1430. [Google Scholar] [CrossRef]
- Shirzadi, M.R. Lipsosomal amphotericin B: A review of its properties, function, and use for treatment of cutaneous leishmaniasis. Res. Rep. Trop. Med. 2019, 10, 11–18. [Google Scholar] [CrossRef]
- Stone, N.R.; Bicanic, T.; Salim, R.; Hope, W. Liposomal Amphotericin B (AmBisome((R))): A Review of the Pharmacokinetics, Pharmacodynamics, Clinical Experience and Future Directions. Drugs 2016, 76, 485–500. [Google Scholar] [CrossRef]
- Shalev-Benami, M.; Zhang, Y.; Rozenberg, H.; Nobe, Y.; Taoka, M.; Matzov, D.; Zimmerman, E.; Bashan, A.; Isobe, T.; Jaffe, C.L.; et al. Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin. Nat. Commun. 2017, 8, 1589. [Google Scholar] [CrossRef] [PubMed]
- Capela, R.; Moreira, R.; Lopes, F. An Overview of Drug Resistance in Protozoal Diseases. Int. J. Mol. Sci. 2019, 20, 5748. [Google Scholar] [CrossRef] [PubMed]
- Chawla, B.; Jhingran, A.; Panigrahi, A.; Stuart, K.D.; Madhubala, R. Paromomycin affects translation and vesicle-mediated trafficking as revealed by proteomics of paromomycin -susceptible -resistant Leishmania donovani. PLoS ONE 2011, 6, e26660. [Google Scholar] [CrossRef]
- Nguewa, P.A.; Fuertes, M.A.; Cepeda, V.; Iborra, S.; Carrion, J.; Valladares, B.; Alonso, C.; Perez, J.M. Pentamidine is an antiparasitic and apoptotic drug that selectively modifies ubiquitin. Chem. Biodivers. 2005, 2, 1387–1400. [Google Scholar] [CrossRef]
- Wong, I.L.; Chan, K.F.; Zhao, Y.; Chan, T.H.; Chow, L.M. Quinacrine and a novel apigenin dimer can synergistically increase the pentamidine susceptibility of the protozoan parasite Leishmania. J. Antimicrob. Chemother. 2009, 63, 1179–1190. [Google Scholar] [CrossRef]
- Maheshwari, A.; Seth, A.; Kaur, S.; Aneja, S.; Rath, B.; Basu, S.; Patel, R.; Dutta, A.K. Cumulative cardiac toxicity of Sodium Stibogluconate and Amphotericin B in treatment of Kala-Azar. Pediatr. Infect. Dis. J. 2011, 30, 180–181. [Google Scholar] [CrossRef]
- Garza-Tovar, T.F.; Sacriste-Hernandez, M.I.; Juarez-Duran, E.R.; Arenas, R. An overview of the treatment of cutaneous leishmaniasis. Fac. Rev. 2020, 9, 28. [Google Scholar] [CrossRef]
- Ashutosh; Sundar, S.; Goyal, N. Molecular mechanisms of antimony resistance in Leishmania. J. Med. Microbiol. 2007, 56, 143–153. [Google Scholar] [CrossRef]
- Magalhaes, L.S.; Bomfim, L.G.; Mota, S.G.; Cruz, G.S.; Correa, C.B.; Tanajura, D.M.; Lipscomb, M.W.; Borges, V.M.; Jesus, A.R.; Almeida, R.P.; et al. Increased thiol levels in antimony-resistant Leishmania infantum isolated from treatment-refractory visceral leishmaniasis in Brazil. Mem. Inst. Oswaldo Cruz 2018, 113, 119–125. [Google Scholar] [CrossRef]
- Wyllie, S.; Cunningham, M.L.; Fairlamb, A.H. Dual action of antimonial drugs on thiol redox metabolism in the human pathogen Leishmania donovani. J. Biol. Chem. 2004, 279, 39925–39932. [Google Scholar] [CrossRef]
- Pan American Health Organization. Leishmaniasis in the Americas: Treatment recommendations.; PAHO: Washington, DC, USA, 2018. [Google Scholar]
- Velez, I.; Lopez, L.; Sanchez, X.; Mestra, L.; Rojas, C.; Rodriguez, E. Efficacy of miltefosine for the treatment of American cutaneous leishmaniasis. Am. J. Trop. Med. Hyg. 2010, 83, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Dorlo, T.P.; Balasegaram, M.; Beijnen, J.H.; de Vries, P.J. Miltefosine: A review of its pharmacology and therapeutic efficacy in the treatment of leishmaniasis. J. Antimicrob. Chemother. 2012, 67, 2576–2597. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Prada, C.; Vincent, I.M.; Brotherton, M.C.; Roberts, M.; Roy, G.; Rivas, L.; Leprohon, P.; Smith, T.K.; Ouellette, M. Different Mutations in a P-type ATPase Transporter in Leishmania Parasites are Associated with Cross-resistance to Two Leading Drugs by Distinct Mechanisms. PLoS Negl. Trop. Dis. 2016, 10, e0005171. [Google Scholar] [CrossRef]
- Younis, B.M.; Mudawi Musa, A.; Monnerat, S.; Abdelrahim Saeed, M.; Awad Gasim Khalil, E.; Elbashir Ahmed, A.; Ahmed Ali, M.; Noureldin, A.; Muthoni Ouattara, G.; Nyakaya, G.M.; et al. Safety and efficacy of paromomycin/miltefosine/liposomal amphotericin B combinations for the treatment of post-kala-azar dermal leishmaniasis in Sudan: A phase II, open label, randomized, parallel arm study. PLoS Negl. Trop. Dis. 2023, 17, e0011780. [Google Scholar] [CrossRef]
- Hendrickx, S.; Van den Kerkhof, M.; Mabille, D.; Cos, P.; Delputte, P.; Maes, L.; Caljon, G. Combined treatment of miltefosine and paromomycin delays the onset of experimental drug resistance in Leishmania infantum. PLoS Negl. Trop. Dis. 2017, 11, e0005620. [Google Scholar] [CrossRef]
- Bussotti, G.; Piel, L.; Pescher, P.; Domagalska, M.A.; Rajan, K.S.; Cohen-Chalamish, S.; Doniger, T.; Hiregange, D.G.; Myler, P.J.; Unger, R.; et al. Genome instability drives epistatic adaptation in the human pathogen Leishmania. Proc. Natl. Acad. Sci. USA 2021, 118, e2113744118. [Google Scholar] [CrossRef]
- Leprohon, P.; Fernandez-Prada, C.; Gazanion, E.; Monte-Neto, R.; Ouellette, M. Drug resistance analysis by next generation sequencing in Leishmania. Int. J. Parasitol. Drugs Drug Resist. 2015, 5, 26–35. [Google Scholar] [CrossRef]
- Santi, A.M.M.; Murta, S.M.F. Impact of Genetic Diversity and Genome Plasticity of Leishmania spp. in Treatment and the Search for Novel Chemotherapeutic Targets. Front. Cell Infect. Microbiol. 2022, 12, 826287. [Google Scholar] [CrossRef]
- Grunebast, J.; Clos, J. Leishmania: Responding to environmental signals and challenges without regulated transcription. Comput. Struct. Biotechnol. J. 2020, 18, 4016–4023. [Google Scholar] [CrossRef]
- Assis, L.H.C.; de Paiva, S.C.; Cano, M.I.N. Behind Base J: The Roles of JBP1 and JBP2 on Trypanosomatids. Pathogens 2023, 12, 467. [Google Scholar] [CrossRef]
- Ubeda, J.M.; Legare, D.; Raymond, F.; Ouameur, A.A.; Boisvert, S.; Rigault, P.; Corbeil, J.; Tremblay, M.J.; Olivier, M.; Papadopoulou, B.; et al. Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 2008, 9, R115. [Google Scholar] [CrossRef] [PubMed]
- Leprohon, P.; Legare, D.; Raymond, F.; Madore, E.; Hardiman, G.; Corbeil, J.; Ouellette, M. Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Res. 2009, 37, 1387–1399. [Google Scholar] [CrossRef] [PubMed]
- Patino, L.H.; Imamura, H.; Cruz-Saavedra, L.; Pavia, P.; Muskus, C.; Mendez, C.; Dujardin, J.C.; Ramirez, J.D. Major changes in chromosomal somy, gene expression and gene dosage driven by Sb(III) in Leishmania braziliensis and Leishmania panamensis. Sci. Rep. 2019, 9, 9485. [Google Scholar] [CrossRef] [PubMed]
- Patino, L.H.; Muskus, C.; Munoz, M.; Ramirez, J.D. Genomic analyses reveal moderate levels of ploidy, high heterozygosity and structural variations in a Colombian isolate of Leishmania (Leishmania) amazonensis. Acta Trop. 2020, 203, 105296. [Google Scholar] [CrossRef]
- Laffitte, M.N.; Leprohon, P.; Papadopoulou, B.; Ouellette, M. Plasticity of the Leishmania genome leading to gene copy number variations and drug resistance. F1000Research 2016, 5, 2350. [Google Scholar] [CrossRef]
- Dumetz, F.; Cuypers, B.; Imamura, H.; Zander, D.; D’Haenens, E.; Maes, I.; Domagalska, M.A.; Clos, J.; Dujardin, J.C.; De Muylder, G. Molecular Preadaptation to Antimony Resistance in Leishmania donovani on the Indian Subcontinent. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Shaw, C.D.; Imamura, H.; Downing, T.; Blackburn, G.; Westrop, G.D.; Cotton, J.A.; Berriman, M.; Sanders, M.; Rijal, S.; Coombs, G.H.; et al. Genomic and Metabolomic Polymorphism among Experimentally Selected Paromomycin-Resistant Leishmania donovani Strains. Antimicrob. Agents Chemother. 2019, 64. [Google Scholar] [CrossRef]
- Gazanion, E.; Fernandez-Prada, C.; Papadopoulou, B.; Leprohon, P.; Ouellette, M. Cos-Seq for high-throughput identification of drug target and resistance mechanisms in the protozoan parasite Leishmania. Proc. Natl. Acad. Sci. USA 2016, 113, E3012–E3021. [Google Scholar] [CrossRef]
- Monte-Neto, R.; Laffitte, M.C.; Leprohon, P.; Reis, P.; Frezard, F.; Ouellette, M. Intrachromosomal amplification, locus deletion and point mutation in the aquaglyceroporin AQP1 gene in antimony resistant Leishmania (Viannia) guyanensis. PLoS Negl. Trop. Dis. 2015, 9, e0003476. [Google Scholar] [CrossRef]
- Mukherjee, A.; Boisvert, S.; Monte-Neto, R.L.; Coelho, A.C.; Raymond, F.; Mukhopadhyay, R.; Corbeil, J.; Ouellette, M. Telomeric gene deletion and intrachromosomal amplification in antimony-resistant Leishmania. Mol. Microbiol. 2013, 88, 189–202. [Google Scholar] [CrossRef]
- Xiang, L.; Laranjeira-Silva, M.F.; Maeda, F.Y.; Hauzel, J.; Andrews, N.W.; Mittra, B. Ascorbate-Dependent Peroxidase (APX) from Leishmania amazonensis Is a Reactive Oxygen Species-Induced Essential Enzyme That Regulates Virulence. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed]
- Abadi, M.F.S.; Moradabadi, A.; Vahidi, R.; Shojaeepour, S.; Rostami, S.; Rad, I.; Dabiri, S. High resolution melting analysis and detection of Leishmania resistance: The role of multi drug resistance 1 gene. Genes Environ. 2021, 43, 36. [Google Scholar] [CrossRef] [PubMed]
- Rastrojo, A.; Garcia-Hernandez, R.; Vargas, P.; Camacho, E.; Corvo, L.; Imamura, H.; Dujardin, J.C.; Castanys, S.; Aguado, B.; Gamarro, F.; et al. Genomic and transcriptomic alterations in Leishmania donovani lines experimentally resistant to antileishmanial drugs. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 246–264. [Google Scholar] [CrossRef] [PubMed]
- Mwenechanya, R.; Kovarova, J.; Dickens, N.J.; Mudaliar, M.; Herzyk, P.; Vincent, I.M.; Weidt, S.K.; Burgess, K.E.; Burchmore, R.J.S.; Pountain, A.W.; et al. Sterol 14alpha-demethylase mutation leads to amphotericin B resistance in Leishmania mexicana. PLoS Negl. Trop. Dis. 2017, 11, e0005649. [Google Scholar] [CrossRef]
- Coelho, A.C.; Boisvert, S.; Mukherjee, A.; Leprohon, P.; Corbeil, J.; Ouellette, M. Multiple mutations in heterogeneous miltefosine-resistant Leishmania major population as determined by whole genome sequencing. PLoS Negl. Trop. Dis. 2012, 6, e1512. [Google Scholar] [CrossRef]
- Morais-Teixeira, E.; Damasceno, Q.S.; Galuppo, M.K.; Romanha, A.J.; Rabello, A. The in vitro leishmanicidal activity of hexadecylphosphocholine (miltefosine) against four medically relevant Leishmania species of Brazil. Mem. Inst. Oswaldo Cruz 2011, 106, 475–478. [Google Scholar] [CrossRef]
- Hendrickx, S.; Reis-Cunha, J.L.; Forrester, S.; Jeffares, D.C.; Caljon, G. Experimental Selection of Paromomycin Resistance in Leishmania donovani Amastigotes Induces Variable Genomic Polymorphisms. Microorganisms 2021, 9, 1546. [Google Scholar] [CrossRef]
- Rugani, J.N.; Gontijo, C.M.F.; Frezard, F.; Soares, R.P.; Monte-Neto, R.L.D. Antimony resistance in Leishmania (Viannia) braziliensis clinical isolates from atypical lesions associates with increased ARM56/ARM58 transcripts and reduced drug uptake. Mem. Inst. Oswaldo Cruz 2019, 114, e190111. [Google Scholar] [CrossRef]
- Cordeiro, A.T.; Thiemann, O.H.; Michels, P.A. Inhibition of Trypanosoma brucei glucose-6-phosphate dehydrogenase by human steroids and their effects on the viability of cultured parasites. Bioorg. Med. Chem. 2009, 17, 2483–2489. [Google Scholar] [CrossRef]
- Pountain, A.W.; Weidt, S.K.; Regnault, C.; Bates, P.A.; Donachie, A.M.; Dickens, N.J.; Barrett, M.P. Genomic instability at the locus of sterol C24-methyltransferase promotes amphotericin B resistance in Leishmania parasites. PLoS Neglected Trop. Dis. 2019, 13, e0007052. [Google Scholar] [CrossRef]
- Singh, A.K.; Papadopoulou, B.; Ouellette, M. Gene amplification in amphotericin B-resistant Leishmania tarentolae. Exp. Parasitol. 2001, 99, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Douanne, N.; Dong, G.; Amin, A.; Bernardo, L.; Blanchette, M.; Langlais, D.; Olivier, M.; Fernandez-Prada, C. Leishmania parasites exchange drug-resistance genes through extracellular vesicles. Cell Rep. 2022, 40, 111121. [Google Scholar] [CrossRef] [PubMed]
- Gommers-Ampt, J.; Lutgerink, J.; Borst, P. A novel DNA nucleotide in<i>Trypanosoma brucei</i>only present in the mammalian phase of the life-cycle. Nucleic Acids Res. 1991, 19, 1745–1751. [Google Scholar] [CrossRef]
- Gommers-Ampt, J.H.; Van Leeuwen, F.; De Beer, A.L.J.; Vliegenthart, J.F.G.; Dizdaroglu, M.; Kowalak, J.A.; Crain, P.F.; Borst, P. β-d-glucosyl-hydroxymethyluracil: A novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell 1993, 75, 1129–1136. [Google Scholar] [CrossRef]
- van Luenen, H.G.; Farris, C.; Jan, S.; Genest, P.-A.; Tripathi, P.; Velds, A.; Ron, M.K.; Nieuwland, M.; Haydock, A.; Ramasamy, G.; et al. Glucosylated Hydroxymethyluracil, DNA Base J, Prevents Transcriptional Readthrough in Leishmania. Cell 2012, 150, 909–921. [Google Scholar] [CrossRef]
- Haile, S.; Papadopoulou, B. Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr. Opin. Microbiol. 2007, 10, 569–577. [Google Scholar] [CrossRef]
- Cortazzo da Silva, L.; Aoki, J.I.; Floeter-Winter, L.M. Finding Correlations Between mRNA and Protein Levels in Leishmania Development: Is There a Discrepancy? Front. Cell Infect. Microbiol. 2022, 12, 852902. [Google Scholar] [CrossRef]
- Andrade, J.M.; Goncalves, L.O.; Liarte, D.B.; Lima, D.A.; Guimaraes, F.G.; de Melo Resende, D.; Santi, A.M.M.; de Oliveira, L.M.; Velloso, J.P.L.; Delfino, R.G.; et al. Comparative transcriptomic analysis of antimony resistant and susceptible Leishmania infantum lines. Parasit. Vectors 2020, 13, 600. [Google Scholar] [CrossRef]
- Patino, L.H.; Muskus, C.; Ramirez, J.D. Transcriptional responses of Leishmania (Leishmania) amazonensis in the presence of trivalent sodium stibogluconate. Parasit. Vectors 2019, 12, 348. [Google Scholar] [CrossRef]
- García-Hernández, R.; Perea-Martínez, A.; Manzano, J.I.; Terrón-Camero, L.C.; Andrés-León, E.; Gamarro, F. Transcriptome Analysis of Intracellular Amastigotes of Clinical Leishmania infantum Lines from Therapeutic Failure Patients after Infection of Human Macrophages. Microorganisms 2022, 10, 1304. [Google Scholar] [CrossRef]
- Medina, J.; Cruz-Saavedra, L.; Patiño, L.H.; Muñoz, M.; Ramírez, J.D. Comparative analysis of the transcriptional responses of five Leishmania species to trivalent antimony. Parasit. Vectors 2021, 14, 419. [Google Scholar] [CrossRef] [PubMed]
- Laffitte, M.-C.N.; Leprohon, P.; Légaré, D.; Ouellette, M. Deep-sequencing revealing mutation dynamics in the miltefosine transporter gene in Leishmania infantum selected for miltefosine resistance. Parasitol. Res. 2016, 115, 3699–3703. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Victoria, F.J.; Gamarro, F.; Ouellette, M.; Castanys, S. Functional Cloning of the Miltefosine Transporter. J. Biol. Chem. 2003, 278, 49965–49971. [Google Scholar] [CrossRef]
- Pérez-Victoria, F.J.; Sánchez-Cañete, M.P.; Castanys, S.; Gamarro, F. Phospholipid Translocation and Miltefosine Potency Require Both L. donovani Miltefosine Transporter and the New Protein LdRos3 in Leishmania Parasites. J. Biol. Chem. 2006, 281, 23766–23775. [Google Scholar] [CrossRef]
- Seifert, K.; Pérez-Victoria, F.J.; Stettler, M.; Sánchez-Cañete, M.P.; Castanys, S.; Gamarro, F.; Croft, S.L. Inactivation of the miltefosine transporter, LdMT, causes miltefosine resistance that is conferred to the amastigote stage of Leishmania donovani and persists in vivo. Int. J. Antimicrob. Agents 2007, 30, 229–235. [Google Scholar] [CrossRef]
- Espada, C.R.; Magalhaes, R.M.; Cruz, M.C.; Machado, P.R.; Schriefer, A.; Carvalho, E.M.; Hornillos, V.; Alves, J.M.; Cruz, A.K.; Coelho, A.C.; et al. Investigation of the pathways related to intrinsic miltefosine tolerance in Leishmania (Viannia) braziliensis clinical isolates reveals differences in drug uptake. Int. J. Parasitol. Drugs Drug Resist. 2019, 11, 139–147. [Google Scholar] [CrossRef]
- Kulshrestha, A.; Sharma, V.; Singh, R.; Salotra, P. Comparative transcript expression analysis of miltefosine-sensitive and miltefosine-resistant Leishmania donovani. Parasitol. Res. 2014, 113, 1171–1184. [Google Scholar] [CrossRef]
- Suman, S.S.; Equbal, A.; Zaidi, A.; Ansari, M.Y.; Singh, K.P.; Singh, K.; Purkait, B.; Sahoo, G.C.; Bimal, S.; Das, P.; et al. Up-regulation of cytosolic tryparedoxin in Amp B resistant isolates of Leishmania donovani and its interaction with cytosolic tryparedoxin peroxidase. Biochimie 2016, 121, 312–325. [Google Scholar] [CrossRef]
- Purkait, B.; Singh, R.; Wasnik, K.; Das, S.; Kumar, A.; Paine, M.; Dikhit, M.; Singh, D.; Sardar, A.H.; Ghosh, A.K.; et al. Up-regulation of silent information regulator 2 (Sir2) is associated with amphotericin B resistance in clinical isolates of Leishmania donovani. J. Antimicrob. Chemother. 2015, 70, 1343–1356. [Google Scholar] [CrossRef]
- Verma, A.; Bhandari, V.; Deep, D.K.; Sundar, S.; Dujardin, J.C.; Singh, R.; Salotra, P. Transcriptome profiling identifies genes/pathways associated with experimental resistance to paromomycin in Leishmania donovani. Int. J. Parasitol. Drugs Drug Resist. 2017, 7, 370–377. [Google Scholar] [CrossRef]
- Chu, J.; Pelletier, J. Therapeutic Opportunities in Eukaryotic Translation. Cold Spring Harb. Perspect. Biol. 2018, 10, a032995. [Google Scholar] [CrossRef] [PubMed]
- Arribere, J.A.; Kuroyanagi, H.; Hundley, H.A. mRNA Editing, Processing and Quality Control in Caenorhabditis elegans. Genetics 2020, 215, 531–568. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Almonacid, C.C.; Kellogg, M.K.; Karamyshev, A.L.; Karamysheva, Z.N. Ribosome Specialization in Protozoa Parasites. Int. J. Mol. Sci. 2023, 24, 7484. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Leprohon, P.; Bigot, S.; Padmanabhan, P.K.; Mukherjee, A.; Roy, G.; Gingras, H.; Mestdagh, A.; Papadopoulou, B.; Ouellette, M. Coupling chemical mutagenesis to next generation sequencing for the identification of drug resistance mutations in Leishmania. Nat. Commun. 2019, 10, 5627. [Google Scholar] [CrossRef]
- Singh, A.K.; Roberts, S.; Ullman, B.; Madhubala, R. A quantitative proteomic screen to identify potential drug resistance mechanism in alpha-difluoromethylornithine (DFMO) resistant Leishmania donovani. J. Proteom. 2014, 102, 44–59. [Google Scholar] [CrossRef]
- Gutierrez Guarnizo, S.A.; Tikhonova, E.B.; Zabet-Moghaddam, M.; Zhang, K.; Muskus, C.; Karamyshev, A.L.; Karamysheva, Z.N. Drug-Induced Lipid Remodeling in Leishmania Parasites. Microorganisms 2021, 9, 790. [Google Scholar] [CrossRef]
- Gutierrez Guarnizo, S.A.; Karamysheva, Z.N.; Galeano, E.; Muskus, C.E. Metabolite Biomarkers of Leishmania Antimony Resistance. Cells 2021, 10, 1063. [Google Scholar] [CrossRef]
- Lee, L.J.; Papadopoli, D.; Jewer, M.; Del Rincon, S.; Topisirovic, I.; Lawrence, M.G.; Postovit, L.M. Cancer Plasticity: The Role of mRNA Translation. Trends Cancer 2021, 7, 134–145. [Google Scholar] [CrossRef]
- T’Kindt, R.; Scheltema, R.A.; Jankevics, A.; Brunker, K.; Rijal, S.; Dujardin, J.C.; Breitling, R.; Watson, D.G.; Coombs, G.H.; Decuypere, S. Metabolomics to unveil and understand phenotypic diversity between pathogen populations. PLoS Negl. Trop. Dis. 2010, 4, e904. [Google Scholar] [CrossRef]
- Scheltema, R.A.; Decuypere, S.; T’Kindt, R.; Dujardin, J.C.; Coombs, G.H.; Breitling, R. The potential of metabolomics for Leishmania research in the post-genomics era. Parasitology 2010, 137, 1291–1302. [Google Scholar] [CrossRef]
- Canuto, G.A.; Castilho-Martins, E.A.; Tavares, M.; Lopez-Gonzalvez, A.; Rivas, L.; Barbas, C. CE-ESI-MS metabolic fingerprinting of Leishmania resistance to antimony treatment. Electrophoresis 2012, 33, 1901–1910. [Google Scholar] [CrossRef] [PubMed]
- Berg, M.; Mannaert, A.; Vanaerschot, M.; Van Der Auwera, G.; Dujardin, J.C. (Post-) Genomic approaches to tackle drug resistance in Leishmania. Parasitology 2013, 140, 1492–1505. [Google Scholar] [CrossRef] [PubMed]
- Berg, M.; Vanaerschot, M.; Jankevics, A.; Cuypers, B.; Maes, I.; Mukherjee, S.; Khanal, B.; Rijal, S.; Roy, S.; Opperdoes, F.; et al. Metabolic adaptations of Leishmania donovani in relation to differentiation, drug resistance, and drug pressure. Mol. Microbiol. 2013, 90, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Vincent, I.M.; Weidt, S.; Rivas, L.; Burgess, K.; Smith, T.K.; Ouellette, M. Untargeted metabolomic analysis of miltefosine action in Leishmania infantum reveals changes to the internal lipid metabolism. Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 20–27. [Google Scholar] [CrossRef]
- Glatz, J.F. Challenges in Fatty Acid and lipid physiology. Front. Physiol. 2011, 2, 45. [Google Scholar] [CrossRef]
- Rakotomanga, M.; Saint-Pierre-Chazalet, M.; Loiseau, P.M. Alteration of fatty acid and sterol metabolism in miltefosine-resistant Leishmania donovani promastigotes and consequences for drug-membrane interactions. Antimicrob. Agents Chemother. 2005, 49, 2677–2686. [Google Scholar] [CrossRef]
- Mbongo, N.; Loiseau, P.M.; Billion, M.A.; Robert-Gero, M. Mechanism of amphotericin B resistance in Leishmania donovani promastigotes. Antimicrob. Agents Chemother. 1998, 42, 352–357. [Google Scholar] [CrossRef]
- de Azevedo, A.F.; Dutra, J.L.; Santos, M.L.; Santos Dde, A.; Alves, P.B.; de Moura, T.R.; de Almeida, R.P.; Fernandes, M.F.; Scher, R.; Fernandes, R.P. Fatty acid profiles in Leishmania spp. isolates with natural resistance to nitric oxide and trivalent antimony. Parasitol. Res. 2014, 113, 19–27. [Google Scholar] [CrossRef]
- Basselin, M.; Robert-Gero, M. Alterations in membrane fluidity, lipid metabolism, mitochondrial activity, and lipophosphoglycan expression in pentamidine-resistant Leishmania. Parasitol. Res. 1998, 84, 78–83. [Google Scholar] [CrossRef]
- Wassef, M.K.; Fioretti, T.B.; Dwyer, D.M. Lipid analyses of isolated surface membranes of Leishmania donovani promastigotes. Lipids 1985, 20, 108–115. [Google Scholar] [CrossRef]
- Zhang, K.; Beverley, S.M. Phospholipid and sphingolipid metabolism in Leishmania. Mol. Biochem. Parasitol. 2010, 170, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Goad, L.J.; Holz, G.G., Jr.; Beach, D.H. Sterols of Leishmania species. Implications for biosynthesis. Mol. Biochem. Parasitol. 1984, 10, 161–170. [Google Scholar] [CrossRef] [PubMed]
- de Souza, W.; Rodrigues, J.C. Sterol Biosynthesis Pathway as Target for Anti-trypanosomatid Drugs. Interdiscip. Perspect. Infect. Dis. 2009, 2009, 642502. [Google Scholar] [CrossRef]
- Mathur, R.; Das, R.P.; Ranjan, A.; Shaha, C. Elevated ergosterol protects Leishmania parasites against antimony-generated stress. FASEB J. 2015, 29, 4201–4213. [Google Scholar] [CrossRef]
- Cauchetier, E.; Loiseau, P.M.; Lehman, J.; Rivollet, D.; Fleury, J.; Astier, A.; Deniau, M.; Paul, M. Characterisation of atovaquone resistance in Leishmania infantum promastigotes. Int. J. Parasitol. 2002, 32, 1043–1051. [Google Scholar] [CrossRef]
- Imbert, L.; Cojean, S.; Libong, D.; Chaminade, P.; Loiseau, P.M. Sitamaquine-resistance in Leishmania donovani affects drug accumulation and lipid metabolism. Biomed. Pharmacother. 2014, 68, 893–897. [Google Scholar] [CrossRef]
- Bringaud, F.; Barrett, M.P.; Zilberstein, D. Multiple roles of proline transport and metabolism in trypanosomatids. Front. Biosci. 2012, 17, 349–374. [Google Scholar] [CrossRef] [PubMed]
- Rojo, D.; Canuto, G.A.; Castilho-Martins, E.A.; Tavares, M.F.; Barbas, C.; Lopez-Gonzalvez, A.; Rivas, L. A Multiplatform Metabolomic Approach to the Basis of Antimonial Action and Resistance in Leishmania infantum. PLoS ONE 2015, 10, e0130675. [Google Scholar] [CrossRef]
- Colotti, G.; Ilari, A. Polyamine metabolism in Leishmania: From arginine to trypanothione. Amino Acids 2011, 40, 269–285. [Google Scholar] [CrossRef]
- Karamysheva, Z.N.; Tikhonova, E.B.; Grozdanov, P.N.; Huffman, J.C.; Baca, K.R.; Karamyshev, A.; Denison, R.B.; MacDonald, C.C.; Zhang, K.; Karamyshev, A.L. Polysome Profiling in Leishmania, Human Cells and Mouse Testis. J. Vis. Exp. 2018, 134, e57600. [Google Scholar] [CrossRef]


| Current Drugs for Leishmaniasis Treatment | Mode of Action and Parasite Targeting | References |
|---|---|---|
| Pentavalent Antimony (SbV) | Inhibits the mitochondrial enzyme trypanothione reductase, increasing the parasite’s susceptibility to oxidative stress generated by the macrophage during infection. It can obstruct major energy-driven pathways such as fatty acid oxidation and glycolysis. | [35,48,49] |
| Miltefosine (MLT) | Inhibits the enzyme cytochrome c oxidase located in the mitochondria, directly affecting energy production in the parasite. Also inhibits phosphatidylcholine synthesis, which affects lipid metabolism through the CDP-choline pathway by acting on CTP-phosphocholine cytidylyltransferase activity. | [50,51] |
| Liposomal amphotericin B (AmB) | Forms transmembrane channels through the cell wall and is known to have a high affinity for ergosterol, causing micropores in the membrane, increasing permeability and ion loss, and resulting in cell death. | [52,53] |
| Paromomycin (PMM) | Inhibits the cytosolic ribosome, affecting protein synthesis through binding to the 16S ribosomal unit and creating an alteration in its structure. | [54,55,56] |
| Pentamidine (PTM) | Inhibits DNA and protein synthesis and causes cell-cycle arrest in the G2/M phase. Inhibits RNA polymerase, leading to apoptosis. Inhibits arginine transport. | [57,58] |
| Drug | Gene Name | Genomic Changes | Effect Associated with Drug Resistance | Reference |
|---|---|---|---|---|
| Antimony | MRPA | Amplification | Increases drug efflux | [80,92] |
| APX | Amplification | Protection from ROS accumulation | [84] | |
| G6PDH | Amplification | Protection from ROS accumulation | [84,93] | |
| AQP1 | Amplification, Deletion | Reduces drug uptake | [83] | |
| MDR1 | Point mutation | Increases drug efflux | [86] | |
| AmB | SMT | Deletion | Reduces drug uptake | [87] |
| SC5D | Point mutation | Alters sterol biosynthesis | [94] | |
| CYP51 | Point mutation | Alters sterol biosynthesis | [88] | |
| LMT | Deletion, Point mutation | Alters sterol biosynthesis | [67] | |
| Miltefosine | LMT | Deletion | Reduces drug uptake | [89] |
| Paromomycin | Gene 18S RNA | Point mutation | Decreases binding of PMM | [81] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moncada-Diaz, M.J.; Rodríguez-Almonacid, C.C.; Quiceno-Giraldo, E.; Khuong, F.T.H.; Muskus, C.; Karamysheva, Z.N. Molecular Mechanisms of Drug Resistance in Leishmania spp. Pathogens 2024, 13, 835. https://doi.org/10.3390/pathogens13100835
Moncada-Diaz MJ, Rodríguez-Almonacid CC, Quiceno-Giraldo E, Khuong FTH, Muskus C, Karamysheva ZN. Molecular Mechanisms of Drug Resistance in Leishmania spp. Pathogens. 2024; 13(10):835. https://doi.org/10.3390/pathogens13100835
Chicago/Turabian StyleMoncada-Diaz, Maria Juliana, Cristian Camilo Rodríguez-Almonacid, Eyson Quiceno-Giraldo, Francis T. H. Khuong, Carlos Muskus, and Zemfira N. Karamysheva. 2024. "Molecular Mechanisms of Drug Resistance in Leishmania spp." Pathogens 13, no. 10: 835. https://doi.org/10.3390/pathogens13100835






