Oral Microbiome: A Review of Its Impact on Oral and Systemic Health
Abstract
:1. Introduction
- Inclusion criteria:
- Peer-reviewed articles;
- Research focusing on the oral microbiome and related disease conditions;
- Articles in English.
- Exclusion criteria:
- Non-peer-reviewed articles;
- Studies focusing on non-oral microbiomes;
- Articles not available in English.
2. The Composition and Role of the Oral Microbiome
3. Factors Influencing the Oral Microbiome
3.1. Diet
3.2. Smoking
3.3. Alcohol Consumption
3.4. Other Factors That Influence the Oral Microbiome
4. Oral Microbiome Dysbiosis
5. Oral–Gut Axis
6. Oral Microbiome and Oral Diseases
6.1. Dental Caries
6.2. Gingivitis
6.3. Periodontitis
6.4. Halitosis
6.5. Taste Impairment
6.6. Burning Mouth Syndrome
6.7. Oral Thrush
7. Oral Microbiome and Systemic Diseases
7.1. Gastrointestinal Disorder: Inflammatory Bowel Disease
7.2. Cardiovascular Diseases: Atherosclerosis
7.3. Endocrine Disorders: Diabetes Mellitus
7.4. Obesity
7.5. Neurological Disorders: Alzheimer’s Disease
7.6. Parkinson’s Disease
7.7. Autoimmune Conditions: Rheumatoid Arthritis
7.8. Systemic Lupus Erythematosus
7.9. Cancer
8. Oral Health and Dental Care Practices
8.1. Probiotics
8.2. Peptides in Oral Health and Their Role in Oral Care
8.3. Personalized Oral Care Approaches
9. Chemical Reactions
9.1. Reaction 1
9.2. Reaction 2
9.3. Reaction 3
10. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACPAs | Anticitrullinated protein antibodies |
| α-synuclein | Alpha-synuclein |
| ALS | Agglutinin-like sequence |
| AD | Alzheimer’s disease |
| BBB | Blood–brain barrier |
| CaF2 | Calcium fluoride |
| CD | Crohn’s Disease |
| CH3SH | Methyl mercaptan |
| CH3SSCH3 | Dimethyl sulfide |
| CPC | Cetylpyridinium chloride |
| CVD | Cardiovascular disease |
| EPS | Extracellular polymer |
| F− | Fluoride ions |
| FAP | Fluorapatite |
| GPI | Glycosylphosphatidylinositol |
| HA | Hydroxyapatite |
| HF | Hydrogen fluoride |
| HWP1 | Hyphal wall protein |
| HSPs | Heat shock proteins |
| IBD | Inflammatory bowel disease |
| IL | Interleukin |
| LDH | Lactate dehydrogenase |
| LPS | Lipopolysaccharide |
| LtxA | Leukotoxin A |
| MMP9 | Matrix metalloproteinase 9 |
| nAChRs | Nicotine acetylcholine receptors |
| NETs | Neutrophil extracellular traps |
| NF-κB | Nuclear factor kappa-B |
| NGS | Next-Generation Sequencing |
| OH− | Hydroxyl ions |
| PD | Parkinson’s Disease |
| PPAD | Porphyromonas gingivalis peptidylarginine deiminase |
| RA | Rheumatoid arthritis |
| RS. | Thiol radicals |
| RSH | Thiol compounds |
| RSSR | Disulfides |
| RT-PCR | Quantitative Real-Time PCR |
| SAPs | Secreted aspartyl proteinases |
| SLE | Systemic Lupus Erythematosus |
| SspB | Surface protein SspB |
| Th | T helper |
| VSCs | Volatile sulfur compounds |
References
- Deo, P.N.; Deshmukh, R. Oral microbiome: Unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019, 23, 122–128. [Google Scholar] [CrossRef]
- Wade, W.G. The oral microbiome in health and disease. Pharmacol. Res. 2013, 69, 137–143. [Google Scholar] [CrossRef]
- Kilian, M.; Chapple, I.L.C.; Hannig, M.; Marsh, P.D.; Meuric, V.; Pedersen, A.M.L.; Tonetti, M.S.; Wade, W.G.; Zaura, E. The oral microbiome—An update for oral healthcare professionals. Br. Dent. J. 2016, 221, 657–666. [Google Scholar] [CrossRef]
- Forshaw, R.J. Dental health and disease in ancient Egypt. Br. Dent. J. 2009, 206, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Alam, Y.H.; Kim, R.; Jang, C. Metabolism and Health Impacts of Dietary Sugars. J. Lipid Atheroscler. 2022, 11, 20–38. [Google Scholar] [CrossRef]
- Zarco, M.; Vess, T.; Ginsburg, G. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012, 18, 109–120. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Li, Y.; Cao, Y.; Xue, J.; Zhou, X. The oral microbiome diversity and its relation to human diseases. Folia Microbiol. 2015, 60, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Minty, M.; Vinel, A.; Canceill, T.; Loubières, P.; Burcelin, R.; Kaddech, M.; Blasco-Baque, V.; Laurencin-Dalicieux, S. Oral Microbiota: A Major Player in the Diagnosis of Systemic Diseases. Diagnostics 2021, 11, 1376. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-H.; Chung, S.W.; Auh, Q.-S.; Hong, S.-J.; Lee, Y.-A.; Jung, J.; Lee, G.-J.; Park, H.J.; Shin, S.-I.; Hong, J.-Y. Progress in Oral Microbiome Related to Oral and Systemic Diseases: An Update. Diagnostics 2021, 11, 1283. [Google Scholar] [CrossRef]
- Malla, M.A.; Dubey, A.; Kumar, A.; Yadav, S.; Hashem, A.; Abd Allah, E.F. Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment. Front. Immunol. 2019, 9, 2868. [Google Scholar] [CrossRef]
- Pozhitkov, A.E.; Beikler, T.; Flemmig, T.; Noble, P.A. High-throughput methods for analysis of the human oral microbiome. Periodontology 2000 2011, 55, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Alcaraz, L.D.; Belda-Ferre, P.; Cabrera-Rubio, R.; Romero, H.; Simon-Soro, A.; Pignatelli, M.; Mira, A. Identifying a healthy oral microbiome through metagenomics. Clin. Microbiol. Infect. 2012, 18, 54–57. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.R.; Gabaldón, T. The Human Oral Microbiome in Health and Disease: From Sequences to Ecosystems. Microorganisms 2020, 8, 308. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qi, Y.; Lo, E.C.M.; McGrath, C.; Mei, M.L.; Dai, R. Using next-generation sequencing to detect oral microbiome change following periodontal interventions: A systematic review. Oral Dis. 2021, 27, 1073–1089. [Google Scholar] [CrossRef] [PubMed]
- Lazarevic, V.; Whiteson, K.; Huse, S.; Hernandez, D.; Farinelli, L.; Østerås, M.; Schrenzel, J.; François, P. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J. Microbiol. Methods 2009, 79, 266–271. [Google Scholar] [CrossRef]
- Jeong, J.; Mun, S.; Oh, Y.; Cho, C.-S.; Yun, K.; Ahn, Y.; Chung, W.-H.; Lim, M.Y.; Lee, K.E.; Hwang, T.S.; et al. A qRT-PCR Method Capable of Quantifying Specific Microorganisms Compared to NGS-Based Metagenome Profiling Data. Microorganisms 2022, 10, 324. [Google Scholar] [CrossRef]
- Jung, J.Y.; Yoon, H.K.; An, S.; Lee, J.W.; Ahn, E.-R.; Kim, Y.-J.; Park, H.-C.; Lee, K.; Hwang, J.H.; Lim, S.-K. Rapid oral bacteria detection based on real-time PCR for the forensic identification of saliva. Sci. Rep. 2018, 8, 10852. [Google Scholar] [CrossRef]
- Lochman, J.; Zapletalova, M.; Poskerova, H.; Izakovicova Holla, L.; Borilova Linhartova, P. Rapid Multiplex Real-Time PCR Method for the Detection and Quantification of Selected Cariogenic and Periodontal Bacteria. Diagnostics 2019, 10, 8. [Google Scholar] [CrossRef]
- Morillo, J.M.; Lau, L.; Sanz, M.; Herrera, D.; Silva, A. Quantitative real-time PCR based on single copy gene sequence for detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. J. Periodontal Res. 2003, 38, 518–524. [Google Scholar] [CrossRef]
- Fan, X.; Peters, B.A.; Min, D.; Ahn, J. Comparison of the oral microbiome in mouthwash and whole saliva samples. PLoS ONE 2018, 13, e0194729. [Google Scholar] [CrossRef]
- Jo, R.; Nishimoto, Y.; Umezawa, K.; Yama, K.; Aita, Y.; Ichiba, Y.; Murakami, S.; Kakizawa, Y.; Kumagai, T.; Yamada, T.; et al. Comparison of oral microbiome profiles in stimulated and unstimulated saliva, tongue, and mouth-rinsed water. Sci. Rep. 2019, 9, 16124. [Google Scholar] [CrossRef] [PubMed]
- Ames, N.J.; Ranucci, A.; Moriyama, B.; Wallen, G.R. The Human Microbiome and Understanding the 16S rRNA Gene in Translational Nursing Science. Nurs. Res. 2017, 66, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Bhatia, S.; Sodhi, A.S.; Batra, N. Oral microbiome and health. AIMS Microbiol. 2018, 4, 42–66. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.R.; Yu, W.-H.; Lakshmanan, A.; Wade, W.G. The Human Oral Microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef]
- Huang, R.; Li, M.; Gregory, R.L. Bacterial interactions in dental biofilm. Virulence 2011, 2, 435–444. [Google Scholar] [CrossRef]
- Morou-Bermudez, E.; Burne, R.A. Genetic and Physiologic Characterization of Urease of Actinomyces naeslundii. Infect. Immun. 1999, 67, 504–512. [Google Scholar] [CrossRef]
- Zhang, Y.; Ding, Y.; Guo, Q. Probiotic Species in the Management of Periodontal Diseases: An Overview. Front. Cell. Infect. Microbiol. 2022, 12, 806463. [Google Scholar] [CrossRef]
- Marsh, P.D. Role of the Oral Microflora in Health. Microb. Ecol. Health Dis. 2000, 12, 130–137. [Google Scholar] [CrossRef]
- Bradshaw, D.J.; Marsh, P.D.; Watson, G.K.; Allison, C. Role of Fusobacterium nucleatum and Coaggregation in Anaerobe Survival in Planktonic and Biofilm Oral Microbial Communities during Aeration. Infect. Immun. 1998, 66, 4729–4732. [Google Scholar] [CrossRef]
- Könönen, E.; Fteita, D.; Gursoy, U.K.; Gursoy, M. Prevotella species as oral residents and infectious agents with potential impact on systemic conditions. J. Oral Microbiol. 2022, 14, 2079814. [Google Scholar] [CrossRef]
- Baker, J.L.; Bor, B.; Agnello, M.; Shi, W.; He, X. Ecology of the Oral Microbiome: Beyond Bacteria. Trends Microbiol. 2017, 25, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Diaz, P.I.; Dongari-Bagtzoglou, A. Critically Appraising the Significance of the Oral Mycobiome. J. Dent. Res. 2021, 100, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Krom, B.P.; Kidwai, S.; Ten Cate, J.M. Candida and Other Fungal Species: Forgotten Players of Healthy Oral Microbiota. J. Dent. Res. 2014, 93, 445–451. [Google Scholar] [CrossRef]
- Matarazzo, F.; Ribeiro, A.C.; Feres, M.; Faveri, M.; Mayer, M.P. Diversity and quantitative analysis of Archaea in aggressive periodontitis and periodontally healthy subjects. J. Clin. Periodontol. 2011, 38, 621–627. [Google Scholar] [CrossRef]
- Lepp, P.W.; Brinig, M.M.; Ouverney, C.C.; Palm, K.; Armitage, G.C.; Relman, D.A. Methanogenic Archaea and human periodontal disease. Proc. Natl. Acad. Sci. USA 2004, 101, 6176–6181. [Google Scholar] [CrossRef]
- Szafrański, S.P.; Slots, J.; Stiesch, M. The human oral phageome. Periodontology 2000 2021, 86, 79–96. [Google Scholar] [CrossRef]
- Liang, G.; Bushman, F.D. The human virome: Assembly, composition and host interactions. Nat. Rev. Microbiol. 2021, 19, 514–527. [Google Scholar] [CrossRef] [PubMed]
- Ly, M.; Abeles, S.R.; Boehm, T.K.; Robles-Sikisaka, R.; Naidu, M.; Santiago-Rodriguez, T.; Pride, D.T. Altered Oral Viral Ecology in Association with Periodontal Disease. mBio 2014, 5, e01133-14. [Google Scholar] [CrossRef]
- Abbas, A.A.; Taylor, L.J.; Dothard, M.I.; Leiby, J.S.; Fitzgerald, A.S.; Khatib, L.A.; Collman, R.G.; Bushman, F.D. Redondoviridae, a Family of Small, Circular DNA Viruses of the Human Oro-Respiratory Tract Associated with Periodontitis and Critical Illness. Cell Host Microbe 2019, 25, 719–729.e4. [Google Scholar] [CrossRef]
- Spezia, P.G.; Macera, L.; Mazzetti, P.; Curcio, M.; Biagini, C.; Sciandra, I.; Turriziani, O.; Lai, M.; Antonelli, G.; Pistello, M.; et al. Redondovirus DNA in human respiratory samples. J. Clin. Virol. 2020, 131, 104586. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Zeng, L.; Burne, R.A. Fueling the caries process: Carbohydrate metabolism and gene regulation by Streptococcus mutans. J. Oral Microbiol. 2014, 6, 24878. [Google Scholar] [CrossRef] [PubMed]
- Santonocito, S.; Giudice, A.; Polizzi, A.; Troiano, G.; Merlo, E.M.; Sclafani, R.; Grosso, G.; Isola, G. A Cross-Talk between Diet and the Oral Microbiome: Balance of Nutrition on Inflammation and Immune System’s Response during Periodontitis. Nutrients 2022, 14, 2426. [Google Scholar] [CrossRef]
- Schlagenhauf, U. On the Role of Dietary Nitrate in the Maintenance of Systemic and Oral Health. Dent. J. 2022, 10, 84. [Google Scholar] [CrossRef]
- Vanhatalo, A.; Blackwell, J.R.; L’Heureux, J.E.; Williams, D.W.; Smith, A.; Van Der Giezen, M.; Winyard, P.G.; Kelly, J.; Jones, A.M. Nitrate-responsive oral microbiome modulates nitric oxide homeostasis and blood pressure in humans. Free Radic. Biol. Med. 2018, 124, 21–30. [Google Scholar] [CrossRef]
- Pytko-Polończyk, J.; Stawarz-Janeczek, M.; Kryczyk-Poprawa, A.; Muszyńska, B. Antioxidant-Rich Natural Raw Materials in the Prevention and Treatment of Selected Oral Cavity and Periodontal Diseases. Antioxidants 2021, 10, 1848. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.-J.; Liao, Y.; He, Y.-Q.; Zheng, M.-Q.; Tong, X.-T.; Xue, W.-Q.; Zhang, J.-B.; Yuan, L.-L.; Zhang, W.-L.; Jia, W.-H. Association Between Oral Microbiota and Cigarette Smoking in the Chinese Population. Front. Cell. Infect. Microbiol. 2021, 11, 658203. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Peters, B.A.; Dominianni, C.; Zhang, Y.; Pei, Z.; Yang, L.; Ma, Y.; Purdue, M.P.; Jacobs, E.J.; Gapstur, S.M.; et al. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 2016, 10, 2435–2446. [Google Scholar] [CrossRef]
- Wagenknecht, D.; Balhaddad, A.; Gregory, R. Effects of Nicotine on Oral Microorganisms, Human Tissues, and the Interactions between Them. Curr. Oral Health Rep. 2018, 5, 78–87. [Google Scholar] [CrossRef]
- Beghini, F.; Renson, A.; Zolnik, C.P.; Geistlinger, L.; Usyk, M.; Moody, T.U.; Thorpe, L.; Dowd, J.B.; Burk, R.; Segata, N.; et al. Tobacco exposure associated with oral microbiota oxygen utilization in the New York City Health and Nutrition Examination Study. Ann. Epidemiol. 2019, 34, 18–25.e3. [Google Scholar] [CrossRef]
- Yussof, A.; Yoon, P.; Krkljes, C.; Schweinberg, S.; Cottrell, J.; Chu, T.; Chang, S.L. A meta-analysis of the effect of binge drinking on the oral microbiome and its relation to Alzheimer’s disease. Sci. Rep. 2020, 10, 19872. [Google Scholar] [CrossRef]
- Riedel, F.; Goessler, U.R.; Hormann, K. Alcohol-related diseases of the mouth and throat. Dig. Dis. 2005, 23, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Enberg, N.; Alho, H.; Loimaranta, V.; Lenander-Lumikari, M. Saliva flow rate, amylase activity, and protein and electrolyte concentrations in saliva after acute alcohol consumption. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2001, 92, 292–298. [Google Scholar] [CrossRef]
- Maier, H.; Born, I.A.; Mall, G. Effect of chronic ethanol and nicotine consumption on the function and morphology of the salivary glands. Klin. Wochenschr. 1988, 66 (Suppl. 11), 140–150. [Google Scholar]
- Szabo, G. Consequences of Alcohol Consumption on Host Defence. Alcohol Alcohol. 1999, 34, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Szabo, G.; Mandrekar, P.; Girouard, L.; Catalano, D. Regulation of human monocyte functions by acute ethanol treatment: Decreased tumor necrosis factor-alpha, interleukin-1 beta and elevated interleukin-10, and transforming growth factor-beta production. Alcohol. Clin. Exp. Res. 1996, 20, 900–907. [Google Scholar] [CrossRef] [PubMed]
- Pepersack, T.; Fuss, M.; Otero, J.; Bergmann, P.; Valsamis, J.; Corvilain, J. Longitudinal study of bone metabolism after ethanol withdrawal in alcoholic patients. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 1992, 7, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Ogden, G.R.; Wight, A.J.; Rice, P. Effect of alcohol on the oral mucosa assessed by quantitative cytomorphometry. J. Oral Pathol. Med. Off. Publ. Int. Assoc. Oral Pathol. Am. Acad. Oral Pathol. 1999, 28, 216–220. [Google Scholar] [CrossRef]
- Fan, X.; Peters, B.A.; Jacobs, E.J.; Gapstur, S.M.; Purdue, M.P.; Freedman, N.D.; Alekseyenko, A.V.; Wu, J.; Yang, L.; Pei, Z.; et al. Drinking alcohol is associated with variation in the human oral microbiome in a large study of American adults. Microbiome 2018, 6, 59. [Google Scholar] [CrossRef]
- Barb, J.J.; Maki, K.A.; Kazmi, N.; Meeks, B.K.; Krumlauf, M.; Tuason, R.T.; Brooks, A.T.; Ames, N.J.; Goldman, D.; Wallen, G.R. The oral microbiome in alcohol use disorder: A longitudinal analysis during inpatient treatment. J. Oral Microbiol. 2021, 14, 2004790. [Google Scholar] [CrossRef]
- Li, X.; Zhao, K.; Chen, J.; Ni, Z.; Yu, Z.; Hu, L.; Qin, Y.; Zhao, J.; Peng, W.; Lu, L.; et al. Diurnal changes of the oral microbiome in patients with alcohol dependence. Front. Cell Infect. Microbiol. 2022, 12, 1068908. [Google Scholar] [CrossRef]
- Liao, Y.; Tong, X.-T.; Jia, Y.-J.; Liu, Q.-Y.; Wu, Y.-X.; Xue, W.-Q.; He, Y.-Q.; Wang, T.-M.; Zheng, X.-H.; Zheng, M.-Q.; et al. The Effects of Alcohol Drinking on Oral Microbiota in the Chinese Population. Int. J. Environ. Res. Public Health 2022, 19, 5729. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.M.; Gleber-Netto, F.O.; Fernandes, G.R.; Amorim, M.; Barbosa, L.F.; Francisco, A.L.N.; Guerra de Andrade, A.; Setubal, J.C.; Kowalski, L.P.; Nunes, D.N.; et al. Alcohol and tobacco consumption affects bacterial richness in oral cavity mucosa biofilms. BMC Microbiol. 2014, 14, 250. [Google Scholar] [CrossRef] [PubMed]
- Gm, A. Oral Biofilm and Its Impact on Oral Health, Psychological and Social Interaction. Int. J. Oral Dent. Health 2021, 7, 127. [Google Scholar] [CrossRef]
- Peng, X.; Cheng, L.; You, Y.; Tang, C.; Ren, B.; Li, Y.; Xu, X.; Zhou, X. Oral microbiota in human systematic diseases. Int. J. Oral Sci. 2022, 14, 14. [Google Scholar] [CrossRef]
- Patangia, D.V.; Anthony Ryan, C.; Dempsey, E.; Paul Ross, R.; Stanton, C. Impact of antibiotics on the human microbiome and consequences for host health. MicrobiologyOpen 2022, 11, e1260. [Google Scholar] [CrossRef] [PubMed]
- Farah, C.S.; Lynch, N.; McCullough, M.J. Oral fungal infections: An update for the general practitioner. Aust. Dent. J. 2010, 55 (Suppl. 1), 48–54. [Google Scholar] [CrossRef]
- Roberts, A.P.; Kreth, J. The impact of horizontal gene transfer on the adaptive ability of the human oral microbiome. Front. Cell. Infect. Microbiol. 2014, 4, 124. [Google Scholar] [CrossRef]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef]
- Segata, N.; Haake, S.K.; Mannon, P.; Lemon, K.P.; Waldron, L.; Gevers, D.; Huttenhower, C.; Izard, J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012, 13, R42. [Google Scholar] [CrossRef]
- Nagpal, R.; Mainali, R.; Ahmadi, S.; Wang, S.; Singh, R.; Kavanagh, K.; Kitzman, D.W.; Kushugulova, A.; Marotta, F.; Yadav, H. Gut microbiome and aging: Physiological and mechanistic insights. Nutr. Healthy Aging 2018, 4, 267–285. [Google Scholar] [CrossRef]
- Sovran, B.; Hugenholtz, F.; Elderman, M.; Van Beek, A.A.; Graversen, K.; Huijskes, M.; Boekschoten, M.V.; Savelkoul, H.F.J.; De Vos, P.; Dekker, J.; et al. Age-associated Impairment of the Mucus Barrier Function is Associated with Profound Changes in Microbiota and Immunity. Sci. Rep. 2019, 9, 1437. [Google Scholar] [CrossRef] [PubMed]
- Toda, K.; Hisata, K.; Satoh, T.; Katsumata, N.; Odamaki, T.; Mitsuyama, E.; Katayama, T.; Kuhara, T.; Aisaka, K.; Shimizu, T.; et al. Neonatal oral fluid as a transmission route for bifidobacteria to the infant gut immediately after birth. Sci. Rep. 2019, 9, 8692. [Google Scholar] [CrossRef] [PubMed]
- P744. Proton pump inhibitors affect the gut microbiome. J. Crohns Colitis 2016, 10 (Suppl. S1), S487. [Google Scholar] [CrossRef]
- Park, S.-Y.; Hwang, B.-O.; Lim, M.; Ok, S.-H.; Lee, S.-K.; Chun, K.-S.; Park, K.-K.; Hu, Y.; Chung, W.-Y.; Song, N.-Y. Oral-Gut Microbiome Axis in Gastrointestinal Disease and Cancer. Cancers 2021, 13, 2124. [Google Scholar] [CrossRef] [PubMed]
- Khor, B.; Snow, M.; Herrman, E.; Ray, N.; Mansukhani, K.; Patel, K.A.; Said-Al-Naief, N.; Maier, T.; Machida, C.A. Interconnections between the Oral and Gut Microbiomes: Reversal of Microbial Dysbiosis and the Balance between Systemic Health and Disease. Microorganisms 2021, 9, 496. [Google Scholar] [CrossRef]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S.; Kiguchi, Y.; Yasuma, K.; Watanabe, E.; Tanoue, T.; et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef]
- Kitamoto, S.; Nagao-Kitamoto, H.; Hein, R.; Schmidt, T.M.; Kamada, N. The Bacterial Connection between the Oral Cavity and the Gut Diseases. J. Dent. Res. 2020, 99, 1021–1029. [Google Scholar] [CrossRef]
- Walker, M.Y.; Pratap, S.; Southerland, J.H.; Farmer-Dixon, C.M.; Lakshmyya, K.; Gangula, P.R. Role of oral and gut microbiome in nitric oxide-mediated colon motility. Nitric Oxide Biol. Chem. 2018, 73, 81–88. [Google Scholar] [CrossRef]
- Schmidt, T.S.; Hayward, M.R.; Coelho, L.P.; Li, S.S.; Costea, P.I.; Voigt, A.Y.; Wirbel, J.; Maistrenko, O.M.; Alves, R.J.; Bergsten, E.; et al. Extensive transmission of microbes along the gastrointestinal tract. eLife 2019, 8, e42693. [Google Scholar] [CrossRef]
- Bowen, W.H.; Burne, R.A.; Wu, H.; Koo, H. Oral Biofilms: Pathogens, Matrix and Polymicrobial Interactions in Microenvironments. Trends Microbiol. 2018, 26, 229–242. [Google Scholar] [CrossRef]
- Suwannakul, S.; Stafford, G.P.; Whawell, S.A.; Douglas, C.W.I. Identification of bistable populations of Porphyromonas gingivalis that differ in epithelial cell invasion. Microbiology 2010, 156, 3052–3064. [Google Scholar] [CrossRef]
- Tanner, A.C.R.; Kressirer, C.A.; Rothmiller, S.; Johansson, I.; Chalmers, N.I. The Caries Microbiome: Implications for Reversing Dysbiosis. Adv. Dent. Res. 2018, 29, 78–85. [Google Scholar] [CrossRef]
- Selwitz, R.H.; Ismail, A.I.; Pitts, N.B. Dental caries. Lancet 2007, 369, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Nyvad, B. Caries Ecology Revisited: Microbial Dynamics and the Caries Process. Caries Res. 2008, 42, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef]
- Seminario, A.; Broukal, Z.; Ivancaková, R. Mutans streptococci and the development of dental plaque. Prague Med. Rep. 2005, 106, 349–358. [Google Scholar]
- Madléna, M.; Dombi, C.; Gintner, Z.; Bánóczy, J. Effect of amine fluoride/stannous fluoride toothpaste and mouthrinse on dental plaque accumulation and gingival health. Oral Dis. 2004, 10, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Daliri, E.B.-M.; Kim, N.; Kim, J.-R.; Yoo, D.; Oh, D.-H. Microbial Etiology and Prevention of Dental Caries: Exploiting Natural Products to Inhibit Cariogenic Biofilms. Pathogens 2020, 9, 569. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Nijampatnam, B.; Hua, Z.; Nguyen, T.; Zou, J.; Cai, X.; Michalek, S.M.; Velu, S.E.; Wu, H. Structure-Based Discovery of Small Molecule Inhibitors of Cariogenic Virulence. Sci. Rep. 2017, 7, 5974. [Google Scholar] [CrossRef]
- Simón-Soro, A.; Guillen-Navarro, M.; Mira, A. Metatranscriptomics reveals overall active bacterial composition in caries lesions. J. Oral Microbiol. 2014, 6, 25443. [Google Scholar] [CrossRef]
- Simón-Soro, A.; Mira, A. Solving the etiology of dental caries. Trends Microbiol. 2015, 23, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.L.; Beall, C.J.; Kutsch, S.R.; Firestone, N.D.; Leys, E.J.; Griffen, A.L. Beyond Streptococcus mutans: Dental Caries Onset Linked to Multiple Species by 16S rRNA Community Analysis. PLoS ONE 2012, 7, e47722. [Google Scholar] [CrossRef] [PubMed]
- Mantzourani, M.; Fenlon, M.; Beighton, D. Association between Bifidobacteriaceae and the clinical severity of root caries lesions. Oral Microbiol. Immunol. 2009, 24, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Badet, C.; Thebaud, N.B. Ecology of lactobacilli in the oral cavity: A review of literature. Open Microbiol. J. 2008, 2, 38–48. [Google Scholar] [CrossRef]
- Simón-Soro, Á.; Tomás, I.; Cabrera-Rubio, R.; Catalan, M.D.; Nyvad, B.; Mira, A. Microbial Geography of the Oral Cavity. J. Dent. Res. 2013, 92, 616–621. [Google Scholar] [CrossRef]
- Intan Suhana, M.A.; Farha, A.; Hassan, B.M. Inflammation of the Gums. Malays. Fam. Physician Off. J. Acad. Fam. Physicians Malays. 2020, 15, 71–73. [Google Scholar]
- Rathee, M.; Jain, P. Gingivitis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. Available online: http://www.ncbi.nlm.nih.gov/books/NBK557422/ (accessed on 3 May 2024).
- Gao, L.; Xu, T.; Huang, G.; Jiang, S.; Gu, Y.; Chen, F. Oral microbiomes: More and more importance in oral cavity and whole body. Protein Cell 2018, 9, 488–500. [Google Scholar] [CrossRef]
- Lu, M.; Xuan, S.; Wang, Z. Oral microbiota: A new view of body health. Food Sci. Hum. Wellness 2019, 8, 8–15. [Google Scholar] [CrossRef]
- Moore, W.E.; Moore, L.V. The bacteria of periodontal diseases. Periodontology 2000 1994, 5, 66–77. [Google Scholar] [CrossRef]
- Curtis, M.A.; Diaz, P.I.; Van Dyke, T.E. The role of the microbiota in periodontal disease. Periodontology 2000 2020, 83, 14–25. [Google Scholar] [CrossRef]
- Kistler, J.O.; Booth, V.; Bradshaw, D.J.; Wade, W.G. Bacterial community development in experimental gingivitis. PLoS ONE 2013, 8, e71227. [Google Scholar] [CrossRef] [PubMed]
- Abdulkareem, A.A.; Al-Taweel, F.B.; Al-Sharqi, A.J.B.; Gul, S.S.; Sha, A.; Chapple, I.L.C. Current concepts in the pathogenesis of periodontitis: From symbiosis to dysbiosis. J. Oral Microbiol. 2023, 15, 2197779. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Darveau, R.P.; Curtis, M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012, 10, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Hasegawa, M.; Inohara, N. The Role of Oral Pathobionts in Dysbiosis during Periodontitis Development. J. Dent. Res. 2014, 93, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, U.; Sharma, G.; Juneja, M.; Nagpal, A. Halitosis: Current concepts on etiology, diagnosis and management. Eur. J. Dent. 2016, 10, 292–300. [Google Scholar] [CrossRef]
- Cortelli, J.R.; Barbosa, M.D.S.; Westphal, M.A. Halitosis: A review of associated factors and therapeutic approach. Braz. Oral Res. 2008, 22, 44–54. [Google Scholar] [CrossRef]
- Hampelska, K.; Jaworska, M.M.; Babalska, Z.Ł.; Karpiński, T.M. The Role of Oral Microbiota in Intra-Oral Halitosis. J. Clin. Med. 2020, 9, 2484. [Google Scholar] [CrossRef]
- Yang, F.; Huang, S.; He, T.; Catrenich, C.; Teng, F.; Bo, C.; Chen, J.; Liu, J.; Li, J.; Song, Y.; et al. Microbial Basis of Oral Malodor Development in Humans. J. Dent. Res. 2013, 92, 1106–1112. [Google Scholar] [CrossRef]
- Nakano, Y.; Yoshimura, M.; Koga, T. Correlation between oral malodor and periodontal bacteria. Microbes Infect. 2002, 4, 679–683. [Google Scholar] [CrossRef]
- Ratcliff, P.A.; Johnson, P.W. The Relationship Between Oral Malodor, Gingivitis, and Periodontitis. A Review. J. Periodontol. 1999, 70, 485–489. [Google Scholar] [CrossRef]
- Porter, S.R.; Scully, C. Oral malodour (halitosis). BMJ 2006, 333, 632–635. [Google Scholar] [CrossRef]
- Persson, S.; Edlund, M.B.; Claesson, R.; Carlsson, J. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol. Immunol. 1990, 5, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, T.; Suzuki, N.; Nakano, Y.; Yasui, M.; Yoneda, M.; Shimazaki, Y.; Hirofuji, T.; Yamashita, Y. Discrimination of the oral microbiota associated with high hydrogen sulfide and methyl mercaptan production. Sci. Rep. 2012, 2, 215. [Google Scholar] [CrossRef]
- Washio, J.; Sato, T.; Koseki, T.; Takahashi, N. Hydrogen sulfide-producing bacteria in tongue biofilm and their relationship with oral malodour. J. Med. Microbiol. 2005, 54, 889–895. [Google Scholar] [CrossRef]
- Solemdal, K.; Sandvik, L.; Willumsen, T.; Mowe, M.; Hummel, T. The impact of oral health on taste ability in acutely hospitalized elderly. PLoS ONE 2012, 7, e36557. [Google Scholar] [CrossRef]
- Finkelstein, J.A.; Schiffman, S.S. Workshop on taste and smell in the elderly: An overview. Physiol. Behav. 1999, 66, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Lindemann, B. Receptors and transduction in taste. Nature 2001, 413, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Spielman, A.I. Interaction of saliva and taste. J. Dent. Res. 1990, 69, 838–843. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, R. Role of saliva in the maintenance of taste sensitivity. Crit. Rev. Oral Biol. Med. 2000, 11, 216–229. [Google Scholar] [CrossRef]
- Nederfors, T.; Isaksson, R.; Mörnstad, H.; Dahlöf, C. Prevalence of perceived symptoms of dry mouth in an adult Swedish population—Relation to age, sex and pharmacotherapy. Community Dent. Oral Epidemiol. 1997, 25, 211–216. [Google Scholar] [CrossRef]
- Kamel, U.F.; Maddison, P.; Whitaker, R. Impact of primary Sjogren’s syndrome on smell and taste: Effect on quality of life. Rheumatology 2009, 48, 1512–1514. [Google Scholar] [CrossRef] [PubMed]
- Weiffenbach, J.M.; Schwartz, L.K.; Atkinson, J.C.; Fox, P.C. Taste performance in Sjogren’s syndrome. Physiol. Behav. 1995, 57, 89–96. [Google Scholar] [CrossRef]
- Aravindhan, R.; Vidyalakshmi, S.; Kumar, M.S.; Satheesh, C.; Balasubramanium, A.M.; Prasad, V.S. Burning mouth syndrome: A review on its diagnostic and therapeutic approach. J. Pharm. Bioallied Sci. 2014, 6, S21–S25. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-M.; Park, J.W.; Jo, J.H.; Oh, B.; Chung, G. Comparative analysis of the oral microbiome of burning mouth syndrome patients. J. Oral Microbiol. 2022, 14, 2052632. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Verma, R.; Murari, A.; Agrawal, A. Oral candidiasis: An overview. J. Oral Maxillofac. Pathol. JOMFP 2014, 18, S81–S85. [Google Scholar] [CrossRef] [PubMed]
- Rajendra Santosh, A.B.; Muddana, K.; Bakki, S.R. Fungal Infections of Oral Cavity: Diagnosis, Management, and Association with COVID-19. SN Compr. Clin. Med. 2021, 3, 1373–1384. [Google Scholar] [CrossRef]
- Arya, N.R.; Rafiq, N.B. Candidiasis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. Available online: https://www.ncbi.nlm.nih.gov/books/NBK560624/ (accessed on 20 September 2023).
- Oral Thrush: Overview; Institute for Quality and Efficiency in Health Care: Cologne, Germany, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK367586/ (accessed on 20 September 2023).
- Cotter, G.; Kavanagh, K. Adherence mechanisms of Candida albicans. Br. J. Biomed. Sci. 2000, 57, 241–249. [Google Scholar]
- Hoyer, L.L. The ALS gene family of Candida albicans. Trends Microbiol. 2001, 9, 176–180. [Google Scholar] [CrossRef]
- Hoyer, L.L.; Green, C.B.; Oh, S.-H.; Zhao, X. Discovering the Secrets of the Candida albicans Agglutinin-Like Sequence (ALS) Gene Family—A Sticky Pursuit. Med. Mycol. 2008, 46, 1–15. [Google Scholar] [CrossRef]
- Nikou, S.-A.; Kichik, N.; Brown, R.; Ponde, N.O.; Ho, J.; Naglik, J.R.; Richardson, J.P. Candida albicans Interactions with Mucosal Surfaces during Health and Disease. Pathogens 2019, 8, 53. [Google Scholar] [CrossRef]
- Naglik, J.R.; Fostira, F.; Ruprai, J.; Staab, J.F.; Challacombe, S.J.; Sundstrom, P. Candida albicans HWP1 gene expression and host antibody responses in colonization and disease. J. Med. Microbiol. 2006, 55, 1323–1327. [Google Scholar] [CrossRef] [PubMed]
- Sundstrom, P.; Balish, E.; Allen, C.M. Essential role of the Candida albicans transglutaminase substrate, hyphal wall protein 1, in lethal oroesophageal candidiasis in immunodeficient mice. J. Infect. Dis. 2002, 185, 521–530. [Google Scholar] [CrossRef]
- Vila, T.; Sultan, A.S.; Montelongo-Jauregui, D.; Jabra-Rizk, M.A. Oral Candidiasis: A Disease of Opportunity. J. Fungi 2020, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Silverman, R.J.; Nobbs, A.H.; Vickerman, M.M.; Barbour, M.E.; Jenkinson, H.F. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect. Immun. 2010, 78, 4644–4652. [Google Scholar] [CrossRef] [PubMed]
- Desai, J. Candida albicans Hyphae: From Growth Initiation to Invasion. J. Fungi 2018, 4, 10. [Google Scholar] [CrossRef]
- Lewis, M.a.O.; Williams, D.W. Diagnosis and management of oral candidosis. Br. Dent. J. 2017, 223, 675–681. [Google Scholar] [CrossRef]
- Naglik, J.R.; Challacombe, S.J.; Hube, B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 2003, 67, 400–428. [Google Scholar] [CrossRef]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef]
- Höfs, S.; Mogavero, S.; Hube, B. Interaction of Candida albicans with host cells: Virulence factors, host defense, escape strategies, and the microbiota. J. Microbiol. 2016, 54, 149–169. [Google Scholar] [CrossRef]
- Swidergall, M.; Khalaji, M.; Solis, N.V.; Moyes, D.L.; Drummond, R.A.; Hube, B.; Lionakis, M.S.; Murdoch, C.; Filler, S.G.; Naglik, J.R. Candidalysin Is Required for Neutrophil Recruitment and Virulence During Systemic Candida albicans Infection. J. Infect. Dis. 2019, 220, 1477–1488. [Google Scholar] [CrossRef]
- Moyes, D.L.; Wilson, D.; Richardson, J.P.; Mogavero, S.; Tang, S.X.; Wernecke, J.; Höfs, S.; Gratacap, R.L.; Robbins, J.; Runglall, M.; et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 2016, 532, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Phan, Q.T.; Myers, C.L.; Fu, Y.; Sheppard, D.C.; Yeaman, M.R.; Welch, W.H.; Ibrahim, A.S.; Edwards, J.E.; Filler, S.G. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007, 5, e64. [Google Scholar] [CrossRef] [PubMed]
- Seedorf, H.; Griffin, N.W.; Ridaura, V.K.; Reyes, A.; Cheng, J.; Rey, F.E.; Smith, M.I.; Simon, G.M.; Scheffrahn, R.H.; Woebken, D.; et al. Bacteria from Diverse Habitats Colonize and Compete in the Mouse Gut. Cell 2014, 159, 253–266. [Google Scholar] [CrossRef]
- Schmitz, H.; Barmeyer, C.; Fromm, M.; Runkel, N.; Foss, H.D.; Bentzel, C.J.; Riecken, E.O.; Schulzke, J.D. Altered tight junction structure contributes to the impaired epithelial barrier function in ulcerative colitis. Gastroenterology 1999, 116, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Peeters, M.; Geypens, B.; Claus, D.; Nevens, H.; Ghoos, Y.; Verbeke, G.; Baert, F.; Vermeire, S.; Vlietinck, R.; Rutgeerts, P. Clustering of increased small intestinal permeability in families with Crohn’s disease. Gastroenterology 1997, 113, 802–807. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum—Symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef]
- Schincaglia, G.P.; Hong, B.Y.; Rosania, A.; Barasz, J.; Thompson, A.; Sobue, T.; Panagakos, F.; Burleson, J.A.; Dongari-Bagtzoglou, A.; Diaz, P.I. Clinical, Immune, and Microbiome Traits of Gingivitis and Peri-implant Mucositis. J. Dent. Res. 2017, 96, 47–55. [Google Scholar] [CrossRef]
- Sohn, J.; Li, L.; Zhang, L.; Settem, R.P.; Honma, K.; Sharma, A.; Falkner, K.L.; Novak, J.M.; Sun, Y.; Kirkwood, K.L. Porphyromonas gingivalis indirectly elicits intestinal inflammation by altering the gut microbiota and disrupting epithelial barrier function through IL9-producing CD4+ T cells. Mol. Oral Microbiol. 2022, 37, 42–52. [Google Scholar] [CrossRef]
- Abusleme, L.; Dupuy, A.K.; Dutzan, N.; Silva, N.; Burleson, J.A.; Strausbaugh, L.D.; Gamonal, J.; Diaz, P.I. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013, 7, 1016–1025. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, M.; Liu, Y.; Luo, B.; Cui, J.; Huang, L.; Chen, K.; Liu, Y. The oral microbiota and cardiometabolic health: A comprehensive review and emerging insights. Front. Immunol. 2022, 13, 1010368. [Google Scholar] [CrossRef]
- Lucchese, A. Streptococcus mutans antigen I/II and autoimmunity in cardiovascular diseases. Autoimmun. Rev. 2017, 16, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Tang, Q.; Nie, J.; Zhang, C.; Zhou, X.; Yu, S.; Sun, J.; Cheng, X.; Dong, N.; Hu, Y.; et al. BMAL1-Downregulation Aggravates Porphyromonas gingivalis-Induced Atherosclerosis by Encouraging Oxidative Stress. Circ. Res. 2020, 126, e15–e29. [Google Scholar] [CrossRef]
- Stinson, M.W.; Alder, S.; Kumar, S. Invasion and killing of human endothelial cells by viridans group streptococci. Infect. Immun. 2003, 71, 2365–2372. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Wong, L.-Y.; Jia, L.T.; Kuklenyik, Z.; Calafat, A.M. Trends in exposure to polyfluoroalkyl chemicals in the U.S. Population: 1999–2008. Environ. Sci. Technol. 2011, 45, 8037–8045. [Google Scholar] [CrossRef] [PubMed]
- Ramos, H.C.; Rumbo, M.; Sirard, J.-C. Bacterial flagellins: Mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 2004, 12, 509–517. [Google Scholar] [CrossRef]
- Zindel, J.; Kubes, P. DAMPs, PAMPs, and LAMPs in Immunity and Sterile Inflammation. Annu. Rev. Pathol. 2020, 15, 493–518. [Google Scholar] [CrossRef]
- Kaur, M.; Kachlany, S.C. Aggregatibacter actinomycetemcomitans leukotoxin (LtxA, Leukothera) induces cofilin dephosphorylation and actin depolymerization during killing of malignant monocytes. Microbiology 2014, 160, 2443–2452. [Google Scholar] [CrossRef]
- Aarabi, G.; Heydecke, G.; Seedorf, U. Roles of Oral Infections in the Pathomechanism of Atherosclerosis. Int. J. Mol. Sci. 2018, 19, 1978. [Google Scholar] [CrossRef]
- Chhibber-Goel, J.; Singhal, V.; Bhowmik, D.; Vivek, R.; Parakh, N.; Bhargava, B.; Sharma, A. Linkages between oral commensal bacteria and atherosclerotic plaques in coronary artery disease patients. NPJ Biofilms Microbiomes 2016, 2, 7. [Google Scholar] [CrossRef]
- Lourenço, T.G.B.; Heller, D.; Silva-Boghossian, C.M.; Cotton, S.L.; Paster, B.J.; Colombo, A.P.V. Microbial signature profiles of periodontally healthy and diseased patients. J. Clin. Periodontol. 2014, 41, 1027–1036. [Google Scholar] [CrossRef]
- Plachokova, A.S.; Andreu-Sánchez, S.; Noz, M.P.; Fu, J.; Riksen, N.P. Oral Microbiome in Relation to Periodontitis Severity and Systemic Inflammation. Int. J. Mol. Sci. 2021, 22, 5876. [Google Scholar] [CrossRef]
- Morimoto, R.I. Cells in stress: Transcriptional activation of heat shock genes. Science 1993, 259, 1409–1410. [Google Scholar] [CrossRef] [PubMed]
- Goulhen, F.; Grenier, D.; Mayrand, D. Oral microbial heat-shock proteins and their potential contributions to infections. Crit. Rev. Oral Biol. Med. 2003, 14, 399–412. [Google Scholar] [CrossRef]
- Xiao, E.; Mattos, M.; Vieira, G.H.A.; Chen, S.; Corrêa, J.D.; Wu, Y.; Albiero, M.L.; Bittinger, K.; Graves, D.T. Diabetes Enhances IL-17 Expression and Alters the Oral Microbiome to Increase Its Pathogenicity. Cell Host Microbe 2017, 22, 120–128.e4. [Google Scholar] [CrossRef] [PubMed]
- Mashimo, P.A.; Yamamoto, Y.; Slots, J.; Park, B.H.; Genco, R.J. The periodontal microflora of juvenile diabetics. Culture, immunofluorescence, and serum antibody studies. J. Periodontol. 1983, 54, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Da Cruz, G.A.; de Toledo, S.; Sallum, E.A.; Sallum, A.W.; Ambrosano, G.M.B.; de Cássia Orlandi Sardi, J.; da Cruz, S.E.B.; Gonçalves, R.B. Clinical and laboratory evaluations of non-surgical periodontal treatment in subjects with diabetes mellitus. J. Periodontol. 2008, 79, 1150–1157. [Google Scholar] [CrossRef]
- Campus, G.; Salem, A.; Uzzau, S.; Baldoni, E.; Tonolo, G. Diabetes and periodontal disease: A case-control study. J. Periodontol. 2005, 76, 418–425. [Google Scholar] [CrossRef]
- Graves, D.T.; Corrêa, J.D.; Silva, T.A. The Oral Microbiota Is Modified by Systemic Diseases. J. Dent. Res. 2019, 98, 148–156. [Google Scholar] [CrossRef]
- Omori, K.; Ohira, T.; Uchida, Y.; Ayilavarapu, S.; Batista, E.L.; Yagi, M.; Iwata, T.; Liu, H.; Hasturk, H.; Kantarci, A.; et al. Priming of neutrophil oxidative burst in diabetes requires preassembly of the NADPH oxidase. J. Leukoc. Biol. 2008, 84, 292–301. [Google Scholar] [CrossRef]
- Wu, Y.-Y.; Xiao, E.; Graves, D.T. Diabetes mellitus related bone metabolism and periodontal disease. Int. J. Oral Sci. 2015, 7, 63–72. [Google Scholar] [CrossRef]
- Gasmi Benahmed, A.; Gasmi, A.; Doşa, A.; Chirumbolo, S.; Mujawdiya, P.K.; Aaseth, J.; Dadar, M.; Bjørklund, G. Association between the gut and oral microbiome with obesity. Anaerobe 2021, 70, 102248. [Google Scholar] [CrossRef] [PubMed]
- Schamarek, I.; Anders, L.; Chakaroun, R.M.; Kovacs, P.; Rohde-Zimmermann, K. The role of the oral microbiome in obesity and metabolic disease: Potential systemic implications and effects on taste perception. Nutr. J. 2023, 22, 28. [Google Scholar] [CrossRef]
- Scheithauer, T.P.M.; Rampanelli, E.; Nieuwdorp, M.; Vallance, B.A.; Verchere, C.B.; Van Raalte, D.H.; Herrema, H. Gut Microbiota as a Trigger for Metabolic Inflammation in Obesity and Type 2 Diabetes. Front. Immunol. 2020, 11, 571731. [Google Scholar] [CrossRef] [PubMed]
- Goodson, J.M.; Groppo, D.; Halem, S.; Carpino, E. Is obesity an oral bacterial disease? J. Dent. Res. 2009, 88, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic Endotoxemia Initiates Obesity and Insulin Resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef]
- Janem, W.F.; Scannapieco, F.A.; Sabharwal, A.; Tsompana, M.; Berman, H.A.; Haase, E.M.; Miecznikowski, J.C.; Mastrandrea, L.D. Salivary inflammatory markers and microbiome in normoglycemic lean and obese children compared to obese children with type 2 diabetes. PLoS ONE 2017, 12, e0172647. [Google Scholar] [CrossRef]
- Sohail, M.U.; Elrayess, M.A.; Al Thani, A.A.; Al-Asmakh, M.; Yassine, H.M. Profiling the Oral Microbiome and Plasma Biochemistry of Obese Hyperglycemic Subjects in Qatar. Microorganisms 2019, 7, 645. [Google Scholar] [CrossRef]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef]
- Mathur, R.; Barlow, G.M. Obesity and the microbiome. Expert Rev. Gastroenterol. Hepatol. 2015, 9, 1087–1099. [Google Scholar] [CrossRef]
- Endo, Y.; Tomofuji, T.; Ekuni, D.; Irie, K.; Azuma, T.; Tamaki, N.; Yamamoto, T.; Morita, M. Experimental Periodontitis Induces Gene Expression of Proinflammatory Cytokines in Liver and White Adipose Tissues in Obesity. J. Periodontol. 2010, 81, 520–526. [Google Scholar] [CrossRef]
- Maitre, Y.; Mahalli, R.; Micheneau, P.; Delpierre, A.; Amador, G.; Denis, F. Evidence and Therapeutic Perspectives in the Relationship between the Oral Microbiome and Alzheimer’s Disease: A Systematic Review. Int. J. Environ. Res. Public Health 2021, 18, 11157. [Google Scholar] [CrossRef]
- Poole, S.; Singhrao, S.K.; Kesavalu, L.; Curtis, M.A.; Crean, S. Determining the presence of periodontopathic virulence factors in short-term postmortem Alzheimer’s disease brain tissue. J. Alzheimer’s Dis. 2013, 36, 665–677. [Google Scholar] [CrossRef]
- Miklossy, J. Emerging roles of pathogens in Alzheimer disease. Expert Rev. Mol. Med. 2011, 13, e30. [Google Scholar] [CrossRef] [PubMed]
- Miklossy, J. Alzheimer’s disease—A neurospirochetosis. Analysis of the evidence following Koch’s and Hill’s criteria. J. Neuroinflamm. 2011, 8, 90. [Google Scholar] [CrossRef]
- Riviere, G.R.; Riviere, K.H.; Smith, K.S. Molecular and immunological evidence of oral Treponema in the human brain and their association with Alzheimer’s disease. Oral Microbiol. Immunol. 2002, 17, 113–118. [Google Scholar] [CrossRef]
- Sureda, A.; Daglia, M.; Argüelles Castilla, S.; Sanadgol, N.; Fazel Nabavi, S.; Khan, H.; Belwal, T.; Jeandet, P.; Marchese, A.; Pistollato, F.; et al. Oral microbiota and Alzheimer’s disease: Do all roads lead to Rome? Pharmacol. Res. 2020, 151, 104582. [Google Scholar] [CrossRef]
- Bulgart, H.R.; Neczypor, E.W.; Wold, L.E.; Mackos, A.R. Microbial involvement in Alzheimer disease development and progression. Mol. Neurodegener. 2020, 15, 42. [Google Scholar] [CrossRef] [PubMed]
- Weaver, D.F. Amyloid beta is an early responder cytokine and immunopeptide of the innate immune system. Alzheimers Dement. Transl. Res. Clin. Interv. 2020, 6, e12100. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Butler, C.A.; Ayton, S.; Reynolds, E.C.; Dashper, S.G. Porphyromonas gingivalis and the pathogenesis of Alzheimer’s disease. Crit. Rev. Microbiol. 2024, 50, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Soscia, S.J.; Kirby, J.E.; Washicosky, K.J.; Tucker, S.M.; Ingelsson, M.; Hyman, B.; Burton, M.A.; Goldstein, L.E.; Duong, S.; Tanzi, R.E.; et al. The Alzheimer’s disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS ONE 2010, 5, e9505. [Google Scholar] [CrossRef]
- Von Campenhausen, S.; Bornschein, B.; Wick, R.; Bötzel, K.; Sampaio, C.; Poewe, W.; Oertel, W.; Siebert, U.; Berger, K.; Dodel, R. Prevalence and incidence of Parkinson’s disease in Europe. Eur. Neuropsychopharmacol. 2005, 15, 473–490. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, D.; Comella, C.L.; Horn, S. Parkinson’s disease-—Part 1: Pathophysiology, symptoms, burden, diagnosis, and assessment. Am. J. Manag. Care 2008, 14, S40–S48. [Google Scholar]
- Poewe, W.; Seppi, K.; Tanner, C.M.; Halliday, G.M.; Brundin, P.; Volkmann, J.; Schrag, A.E.; Lang, A.E. Parkinson disease. Nat. Rev. Dis. Primers 2017, 3, 17013. [Google Scholar] [CrossRef]
- Adams, B.; Nunes, J.M.; Page, M.J.; Roberts, T.; Carr, J.; Nell, T.A.; Kell, D.B.; Pretorius, E. Parkinson’s Disease: A Systemic Inflammatory Disease Accompanied by Bacterial Inflammagens. Front. Aging Neurosci. 2019, 11, 210. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-K.; Wu, Y.-T.; Chang, Y.-C. Periodontal inflammatory disease is associated with the risk of Parkinson’s disease: A population-based retrospective matched-cohort study. PeerJ 2017, 5, e3647. [Google Scholar] [CrossRef]
- Feng, Y.-K.; Wu, Q.-L.; Peng, Y.-W.; Liang, F.-Y.; You, H.-J.; Feng, Y.-W.; Li, G.; Li, X.-J.; Liu, S.-H.; Li, Y.-C.; et al. Oral P. gingivalis impairs gut permeability and mediates immune responses associated with neurodegeneration in LRRK2 R1441G mice. J. Neuroinflamm. 2020, 17, 347. [Google Scholar] [CrossRef] [PubMed]
- Jeong, E.; Park, J.-B.; Park, Y.-G. Evaluation of the association between periodontitis and risk of Parkinson’s disease: A nationwide retrospective cohort study. Sci. Rep. 2021, 11, 16594. [Google Scholar] [CrossRef]
- Pretorius, E.; Bester, J.; Page, M.J.; Kell, D.B. The Potential of LPS-Binding Protein to Reverse Amyloid Formation in Plasma Fibrin of Individuals with Alzheimer-Type Dementia. Front Aging Neurosci. 2018, 22, 257. [Google Scholar] [CrossRef]
- Huang, X.; Huang, X.; Huang, Y.; Zheng, J.; Lu, Y.; Mai, Z.; Zhao, X.; Cui, L.; Huang, S. The oral microbiome in autoimmune diseases: Friend or foe? J. Transl. Med. 2023, 21, 211. [Google Scholar] [CrossRef]
- Bellando-Randone, S.; Russo, E.; Venerito, V.; Matucci-Cerinic, M.; Iannone, F.; Tangaro, S.; Amedei, A. Exploring the Oral Microbiome in Rheumatic Diseases, State of Art and Future Prospective in Personalized Medicine with an AI Approach. J. Pers. Med. 2021, 11, 625. [Google Scholar] [CrossRef]
- Rosenstein, E.D.; Greenwald, R.A.; Kushner, L.J.; Weissmann, G. Hypothesis: The Humoral Immune Response to Oral Bacteria Provides a Stimulus for the Development of Rheumatoid Arthritis. Inflammation 2004, 28, 311–318. [Google Scholar] [CrossRef]
- Guo, Q.; Wang, Y.; Xu, D.; Nossent, J.; Pavlos, N.J.; Xu, J. Rheumatoid arthritis: Pathological mechanisms and modern pharmacologic therapies. Bone Res. 2018, 6, 15. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Zorba, M.; Melidou, A.; Patsatsi, A.; Ioannou, E.; Kolokotronis, A. The possible role of oral microbiome in autoimmunity. Int. J. Womens Dermatol. 2020, 6, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Marwaha, A.K.; Leung, N.J.; McMurchy, A.N.; Levings, M.K. TH17 Cells in Autoimmunity and Immunodeficiency: Protective or Pathogenic? Front. Immunol. 2012, 3, 129. [Google Scholar] [CrossRef] [PubMed]
- Cascão, R.; Moura, R.A.; Perpétuo, I.; Canhão, H.; Vieira-Sousa, E.; Mourão, A.F.; Rodrigues, A.M.; Polido-Pereira, J.; Queiroz, M.V.; Rosário, H.S.; et al. Identification of a cytokine network sustaining neutrophil and Th17 activation in untreated early rheumatoid arthritis. Arthritis Res. Ther. 2010, 12, R196. [Google Scholar] [CrossRef]
- Białowąs, K.; Radwan-Oczko, M.; Duś-Ilnicka, I.; Korman, L.; Świerkot, J. Periodontal disease and influence of periodontal treatment on disease activity in patients with rheumatoid arthritis and spondyloarthritis. Rheumatol. Int. 2020, 40, 455–463. [Google Scholar] [CrossRef]
- Konig, M.F.; Abusleme, L.; Reinholdt, J.; Palmer, R.J.; Teles, R.P.; Sampson, K.; Rosen, A.; Nigrovic, P.A.; Sokolove, J.; Giles, J.T.; et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci. Transl. Med. 2016, 8, 369ra176. [Google Scholar] [CrossRef]
- Rooney, C.M.; Mankia, K.; Emery, P. The Role of the Microbiome in Driving RA-Related Autoimmunity. Front. Cell Dev. Biol. 2020, 8, 538130. [Google Scholar] [CrossRef]
- Gao, L.; Cheng, Z.; Zhu, F.; Bi, C.; Shi, Q.; Chen, X. The Oral Microbiome and Its Role in Systemic Autoimmune Diseases: A Systematic Review of Big Data Analysis. Front. Big Data 2022, 5, 927520. [Google Scholar] [CrossRef]
- Pessoa, L.; Aleti, G.; Choudhury, S.; Nguyen, D.; Yaskell, T.; Zhang, Y.; Li, W.; Nelson, K.E.; Neto, L.L.S.; Sant’Ana, A.C.P.; et al. Host-Microbial Interactions in Systemic Lupus Erythematosus and Periodontitis. Front. Immunol. 2019, 10, 2602. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Ren, T.; Li, X.; Zhai, Q.; Xu, X.; Zhang, N.; Jiang, P.; Niu, Y.; Lv, L.; Shi, G.; et al. Distinct Microbiomes of Gut and Saliva in Patients with Systemic Lupus Erythematous and Clinical Associations. Front. Immunol. 2021, 12, 626217. [Google Scholar] [CrossRef] [PubMed]
- Clancy, R.M.; Marion, M.C.; Ainsworth, H.C.; Blaser, M.J.; Chang, M.; Howard, T.D.; Izmirly, P.M.; Lacher, C.; Masson, M.; Robins, K.; et al. Salivary dysbiosis and the clinical spectrum in anti-Ro positive mothers of children with neonatal lupus. J. Autoimmun. 2020, 107, 102354. [Google Scholar] [CrossRef]
- Li, D.; Guo, B.; Wu, H.; Tan, L.; Chang, C.; Lu, Q. Interleukin-17 in systemic lupus erythematosus: A comprehensive review. Autoimmunity 2015, 48, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.A.; Suh, J.W.; Sohn, J.H.; Park, J.W.; Lee, H.; Kang, J.L.; Woo, S.Y.; Cho, Y.J. IL-33 induces Th17-mediated airway inflammation via mast cells in ovalbumin-challenged mice. Am. J. Physiol. Lung Cell Mol. Physiol. 2012, 302, L429–L440. [Google Scholar] [CrossRef]
- Naka, T.; Nishimoto, N.; Kishimoto, T. The paradigm of IL-6: From basic science to medicine. Arthritis Res. 2002, 4 (Suppl. 3), S233–S242. [Google Scholar] [CrossRef]
- Yamamura, K.; Izumi, D.; Kandimalla, R.; Sonohara, F.; Baba, Y.; Yoshida, N.; Kodera, Y.; Baba, H.; Goel, A. Intratumoral Fusobacterium Nucleatum Levels Predict Therapeutic Response to Neoadjuvant Chemotherapy in Esophageal Squamous Cell Carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 6170–6179. [Google Scholar] [CrossRef]
- Sun, J.; Tang, Q.; Yu, S.; Xie, M.; Xie, Y.; Chen, G.; Chen, L. Role of the oral microbiota in cancer evolution and progression. Cancer Med. 2020, 9, 6306–6321. [Google Scholar] [CrossRef]
- Sarao, L.K.; Arora, M. Probiotics, prebiotics, and microencapsulation: A review. Crit. Rev. Food Sci. Nutr. 2017, 57, 344–371. [Google Scholar] [CrossRef]
- Jansen, P.M.; Abdelbary, M.M.H.; Conrads, G. A concerted probiotic activity to inhibit periodontitis-associated bacteria. PLoS ONE 2021, 16, e0248308. [Google Scholar] [CrossRef]
- Cosseau, C.; Devine, D.A.; Dullaghan, E.; Gardy, J.L.; Chikatamarla, A.; Gellatly, S.; Yu, L.L.; Pistolic, J.; Falsafi, R.; Tagg, J.; et al. The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect. Immun. 2008, 76, 4163–4175. [Google Scholar] [CrossRef] [PubMed]
- Kaci, G.; Goudercourt, D.; Dennin, V.; Pot, B.; Doré, J.; Ehrlich, S.D.; Renault, P.; Blottière, H.M.; Daniel, C.; Delorme, C. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl. Environ. Microbiol. 2014, 80, 928–934. [Google Scholar] [CrossRef]
- MacDonald, K.W.; Chanyi, R.M.; Macklaim, J.M.; Cadieux, P.A.; Reid, G.; Burton, J.P. Streptococcus salivarius inhibits immune activation by periodontal disease pathogens. BMC Oral Health 2021, 21, 245. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.K.; Bardow, A.; Jensdottir, T.; Lykkeaa, J.; Twetman, S. Effect of chewing gums containing the probiotic bacterium Lactobacillus reuteri on oral malodour. Acta Odontol. Scand. 2012, 70, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.K.; Hasslöf, P.; Stecksén-Blicks, C.; Twetman, S. Co-aggregation and growth inhibition of probiotic lactobacilli and clinical isolates of mutans streptococci: An in vitro study. Acta Odontol. Scand. 2011, 69, 263–268. [Google Scholar] [CrossRef]
- Invernici, M.M.; Salvador, S.L.; Silva, P.H.F.; Soares, M.S.M.; Casarin, R.; Palioto, D.B.; Souza, S.L.S.; Taba, M.; Novaes, A.B.; Furlaneto, F.A.C.; et al. Effects of Bifidobacterium probiotic on the treatment of chronic periodontitis: A randomized clinical trial. J. Clin. Periodontol. 2018, 45, 1198–1210. [Google Scholar] [CrossRef]
- Zeng, Y.; Fadaak, A.; Alomeir, N.; Wu, T.T.; Rustchenko, E.; Qing, S.; Bao, J.; Gilbert, C.; Xiao, J. Lactobacillus plantarum Disrupts S. mutans-C. albicans Cross-Kingdom Biofilms. Front. Cell Infect. Microbiol. 2022, 12, 872012. [Google Scholar] [CrossRef]
- Forbes, J.; Krishnamurthy, K. Biochemistry, Peptide. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. Available online: https://www.ncbi.nlm.nih.gov/books/NBK562260/ (accessed on 3 May 2024).
- Bermúdez, M.; Hoz, L.; Montoya, G.; Nidome, M.; Pérez-Soria, A.; Romo, E.; Soto-Barreras, U.; Garnica-Palazuelos, J.; Aguilar-Medina, M.; Ramos-Payán, R. Bioactive Synthetic Peptides for Oral Tissues Regeneration. Front. Mater. 2021, 8, 655495. [Google Scholar] [CrossRef]
- Scavello, F.; Kharouf, N.; Lavalle, P.; Haikel, Y.; Schneider, F.; Metz-Boutigue, M.H. The antimicrobial peptides secreted by the chromaffin cells of the adrenal medulla link the neuroendocrine and immune systems: From basic to clinical studies. Front. Immunol. 2022, 13, 977175. [Google Scholar] [CrossRef]
- Alkilzy, M.; Santamaria, R.M.; Schmoeckel, J.; Splieth, C.H. Treatment of Carious Lesions Using Self-Assembling Peptides. Adv. Dent. Res. 2018, 29, 42–47. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Zanetti, M. Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 2004, 75, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Khurshid, Z.; Najeeb, S.; Mali, M.; Moin, S.F.; Raza, S.Q.; Zohaib, S.; Sefat, F.; Zafar, M.S. Histatin peptides: Pharmacological functions and their applications in dentistry. Saudi Pharm. J. SPJ 2017, 25, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Oppenheim, F.G.; Xu, T.; McMillian, F.M.; Levitz, S.M.; Diamond, R.D.; Offner, G.D.; Troxler, R.F. Histatins, a novel family of histidine-rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans. J. Biol. Chem. 1988, 263, 7472–7477. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Levitz, S.M.; Diamond, R.D.; Oppenheim, F.G. Anticandidal activity of major human salivary histatins. Infect. Immun. 1991, 59, 2549–2554. [Google Scholar] [CrossRef]
- Wang, Q.-Q.; Wang, S.; Zhao, T.; Li, Y.; Yang, J.; Liu, Y.; Zhang, H.; Miao, L.; Sun, W. Biomimetic oligopeptide formed enamel-like tissue and dentin tubule occlusion via mineralization for dentin hypersensitivity treatment. J. Appl. Biomater. Funct. Mater. 2021, 19, 22808000211005384. [Google Scholar] [CrossRef]
- Kirkham, J.; Brookes, S.J.; Shore, R.C.; Wood, S.R.; Smith, D.A.; Zhang, J.; Chen, H.; Robinson, C. Physico-Chemical Properties of Crystal Surfaces in Matrix–Mineral Interactions during mammalian biomineralisation. Curr. Opin. Colloid Interface Sci. 2002, 7, 124–132. [Google Scholar] [CrossRef]
- Söderling, E.; Pienihäkkinen, K. Effects of xylitol chewing gum and candies on the accumulation of dental plaque: A systematic review. Clin. Oral Investig. 2022, 26, 119–129. [Google Scholar] [CrossRef]
- Nayak, P.A.; Nayak, U.A.; Khandelwal, V. The effect of xylitol on dental caries and oral flora. Clin. Cosmet. Investig. Dent. 2014, 6, 89–94. [Google Scholar] [CrossRef]
- Drake, D.; Villhauer, A.L. An in vitro comparative study determining bactericidal activity of stabilized chlorine dioxide and other oral rinses. J. Clin. Dent. 2011, 22, 1–5. [Google Scholar]
- Shinada, K.; Ueno, M.; Konishi, C.; Takehara, S.; Yokoyama, S.; Zaitsu, T.; Ohnuki, M.; Wright, F.A.C.; Kawaguchi, Y. Effects of a mouthwash with chlorine dioxide on oral malodor and salivary bacteria: A randomized placebo-controlled 7-day trial. Trials 2010, 11, 14. [Google Scholar] [CrossRef]
- Vlachojannis, C.; Chrubasik-Hausmann, S.; Hellwig, E.; Al-Ahmad, A. A Preliminary Investigation on the Antimicrobial Activity of Listerine®, Its Components, and of Mixtures Thereof. Phytother. Res. PTR 2015, 29, 1590–1594. [Google Scholar] [CrossRef]
- Al-Ani, E.; Heaselgrave, W. The Investigation of Thymol Formulations Containing Poloxamer 407 and Hydroxypropyl Methylcellulose to Inhibit Candida Biofilm Formation and Demonstrate Improved Bio-Compatibility. Pharmaceuticals 2022, 15, 71. [Google Scholar] [CrossRef] [PubMed]
- Wińska, K.; Mączka, W.; Łyczko, J.; Grabarczyk, M.; Czubaszek, A.; Szumny, A. Essential Oils as Antimicrobial Agents—Myth or Real Alternative? Molecules 2019, 24, 2130. [Google Scholar] [CrossRef] [PubMed]
- Alshehri, F.A. The use of mouthwash containing essential oils (LISTERINE®) to improve oral health: A systematic review. Saudi Dent. J. 2018, 30, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Rosin-Grget, K.; Lincir, I. Current concept on the anticaries fluoride mechanism of the action. Coll. Antropol. 2001, 25, 703–712. [Google Scholar] [PubMed]
- Vranic, E.; Lacevic, A.; Mehmedagic, A.; Uzunovic, A. Formulation ingredients for toothpastes and mouthwashes. Bosn. J. Basic Med. Sci. 2004, 4, 51–58. [Google Scholar] [CrossRef]
- Zhang, J.; Sardana, D.; Li, K.Y.; Leung, K.C.M.; Lo, E.C.M. Topical Fluoride to Prevent Root Caries: Systematic Review with Network Meta-analysis. J. Dent. Res. 2020, 99, 506–513. [Google Scholar] [CrossRef]
- O’Hagan-Wong, K.; Enax, J.; Meyer, F.; Ganss, B. The use of hydroxyapatite toothpaste to prevent dental caries. Odontology 2022, 110, 223–230. [Google Scholar] [CrossRef]
- Mao, X.; Auer, D.L.; Buchalla, W.; Hiller, K.-A.; Maisch, T.; Hellwig, E.; Al-Ahmad, A.; Cieplik, F. Cetylpyridinium Chloride: Mechanism of Action, Antimicrobial Efficacy in Biofilms, and Potential Risks of Resistance. Antimicrob. Agents Chemother. 2020, 64, e00576-20. [Google Scholar] [CrossRef]
- Dagli, N.; Dagli, R.; Mahmoud, R.S.; Baroudi, K. Essential oils, their therapeutic properties, and implication in dentistry: A review. J. Int. Soc. Prev. Community Dent. 2015, 5, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Shanbhag, V.K.L. Oil pulling for maintaining oral hygiene—A review. J. Tradit. Complement. Med. 2016, 7, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Woolley, J.; Gibbons, T.; Patel, K.; Sacco, R. The effect of oil pulling with coconut oil to improve dental hygiene and oral health: A systematic review. Heliyon 2020, 6, e04789. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Mira Otal, J.; Torrent-Silla, D.; Porcar, M.; Vilanova, C.; Vivancos Cuadras, F. A mouthwash formulated with o-cymen-5-ol and zinc chloride specifically targets potential pathogens without impairing the native oral microbiome in healthy individuals. J. Oral Microbiol. 2023, 15, 2185962. [Google Scholar] [CrossRef]
- Sheng, Q.; Wang, X.; Hou, Z.; Liu, B.; Jiang, M.; Ren, M.; Fu, J.; He, M.; Zhang, J.; Xiang, Y.; et al. Novel functions of o-cymen-5-ol nanoemulsion in reversing colistin resistance in multidrug-resistant Klebsiella pneumoniae infections. Biochem. Pharmacol. 2024, 227, 116384. [Google Scholar] [CrossRef]
- Kakar, A.; Newby, E.E.; Ghosh, S.; Butler, A.; Bosma, M.L. A randomised clinical trial to assess maintenance of gingival health by a novel gel to foam dentifrice containing 0.1%w/w o-cymen-5-ol, 0.6%w/w zinc chloride. Int. Dent. J. 2011, 61, 21–27. [Google Scholar] [CrossRef]
- Pizzey, R.L.; Marquis, R.E.; Bradshaw, D.J. Antimicrobial effects of o-cymen-5-ol and zinc, alone & in combination in simple solutions and toothpaste formulations. Int. Dent. J. 2020, 61, 33–40. [Google Scholar] [CrossRef]
- Aguilera, F.-R.; Viñas, M.; Sierra, J.M.; Vinuesa, T.R.; Fernandez de Henestrosa, A.; Furmanczyk, M.; Trullàs, C.; Jourdan, E.; López-López, J.; Jorba, M. Substantivity of mouth-rinse formulations containing cetylpyridinium chloride and O-cymen-5-ol: A randomized-crossover trial. BMC Oral Health 2022, 22, 646. [Google Scholar] [CrossRef]
- Nogueira, J.-S.-P.; Lins-Filho, P.-C.; Dias, M.-F.; Silva, M.-F.; Guimarães, R.-P. Does comsumption of staining drinks compromise the result of tooth whitening? J. Clin. Exp. Dent. 2019, 11, e1012–e1017. [Google Scholar] [CrossRef]
- Kitsaras, G.; Goodwin, M.; Kelly, M.P.; Pretty, I.A. Bedtime Oral Hygiene Behaviours, Dietary Habits and Children’s Dental Health. Children 2021, 8, 416. [Google Scholar] [CrossRef]
- Attin, T.; Hornecker, E. Tooth brushing and oral health: How frequently and when should tooth brushing be performed? Oral Health Prev. Dent. 2005, 3, 135–140. [Google Scholar] [PubMed]
- Ganss, C.; Schlueter, N.; Preiss, S.; Klimek, J. Tooth brushing habits in uninstructed adults—Frequency, technique, duration and force. Clin. Oral Investig. 2009, 13, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Fleming, E.; Nguyen, D.; Woods, P.D. Prevalence of daily flossing among adults by selected risk factors for periodontal disease—United States, 2009–2014. J. Periodontol. 2018, 89, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhou, Y.; Liu, C.; Zhao, L.; Zhang, L.; Li, H.; Li, Y.; Cheng, X. Effects of water flossing on gingival inflammation and supragingival plaque microbiota: A 12-week randomized controlled trial. Clin. Oral Investig. 2023, 27, 4567. [Google Scholar] [CrossRef]
- Choi, H.-N.; Cho, Y.-S.; Koo, J.-W. The Effect of Mechanical Tongue Cleaning on Oral Malodor and Tongue Coating. Int. J. Environ. Res. Public Health 2021, 19, 108. [Google Scholar] [CrossRef]
- Outhouse, T.L.; Al-Alawi, R.; Fedorowicz, Z.; Keenan, J.V. Tongue scraping for treating halitosis. Cochrane Database Syst. Rev. 2006, 2, CD005519. [Google Scholar] [CrossRef]
- Canga, M.; Malagnino, G.; Malagnino, V.A.; Malagnino, I. Effectiveness of Sealants Treatment in Permanent Molars: A Longitudinal Study. Int. J. Clin. Pediatr. Dent. 2021, 14, 41–45. [Google Scholar] [CrossRef]


| Microbial Component | Predominant Phyla/Genera | Other Notable Genera |
|---|---|---|
| Bacteria | Actinobacteria, Bacteroidetes, Chlamydia, Euryarchaeota, Fusobacteria, Firmicutes, Proteobacteria, Spirochaetes, and Tenericutes |
|
| Fungi | Candida | Cladosporium, Aureobasidium, Saccharomycetales, Aspergillus, Fusarium, and Cryptococcus |
| Archaea | Euryarchaeota | - |
| Eukaryotic virus | Herpesviridae, Papillomaviridae, Anelloviridae, and Redondoviridae | |
| Bacteriophage | Siphoviridae, Myoviridae, and Podoviridae |
| Disease | Commensals | Pathogens | Directionality | Mechanisms | References |
|---|---|---|---|---|---|
| Oral diseases | |||||
| Dental caries | Lactobacillus spp., Veillonella, Propionibacterium, Bifidobacterium, Corynebacterium, and Capnocytophaga | S. mutans | ↑ | Acidogenic bacteria produce acidic by-products, leading to demineralization and cavitation; dysbiosis-driven disorder. | [83] |
| Gingivitis | Streptococcus sp., Actinomyces sp., and Veillonella sp. | P. gingivalis, T. denticola, A. actinomycetemcomitans, Fusobacterium sp., and P. intermedia | ↑ | Caused by the accumulation of microbial plaque on the tooth surface, which penetrates the gingival tissue and leads to inflammation. | [97] |
| Periodontitis | Prevotella melaninogenica | P. gingivalis, T. denticola, T. forsythia, F. nucleatum ss. polymorphum, and P. intermedia | ↑ | Pathogenic bacteria induce inflammation, oxidative stress, immune activation, and tissue damage; potential systemic implications. | [103] |
| Halitosis | Prevotella melaninogenica, Veillonella spp., Peptostreptococcus, Actinomyces spp., Eubacterium, Megasphaera, Selenomonas, Leptotrichia, and Eikenella corrodens | Treponema denticola, P. gingivalis, P. endodontalis, and Tannerella forsythia | ↑ | Anerobic bacteria produce VSCs causing malodor; bacterial degradation of sulfur-containing amino acids. | [107,108] |
| Taste impairment | Lactobacilli | - | ↑ | High levels of acid produced by the bacteria impair taste, affecting taste perception. | [116] |
| Burning mouth syndrome (BMS) | Streptococcus, Rothia, Bergeyella, and Granulicatella | - | ↑ | Alteration in bacterial strains may contribute to the development of BMS influencing pathways involved in inflammation, immune responses, and sensory perception. | [125] |
| Oral thrush | Candida parapsilosis, Candida krusei, and Candida tropicalis | Candida albicans, Candida glabrata, Candida dubliniensis, and Candida guilliermondii | ↑ | Overgrowth of Candida species due to factors like poor oral hygiene, weakened immune system, or underlying medical conditions; adhesion to host surfaces and tissue invasion through various mechanisms; hyphal formation, biofilm production, and secretion of enzymes that degrade host immune factors. | [127] |
| Systemic diseases | |||||
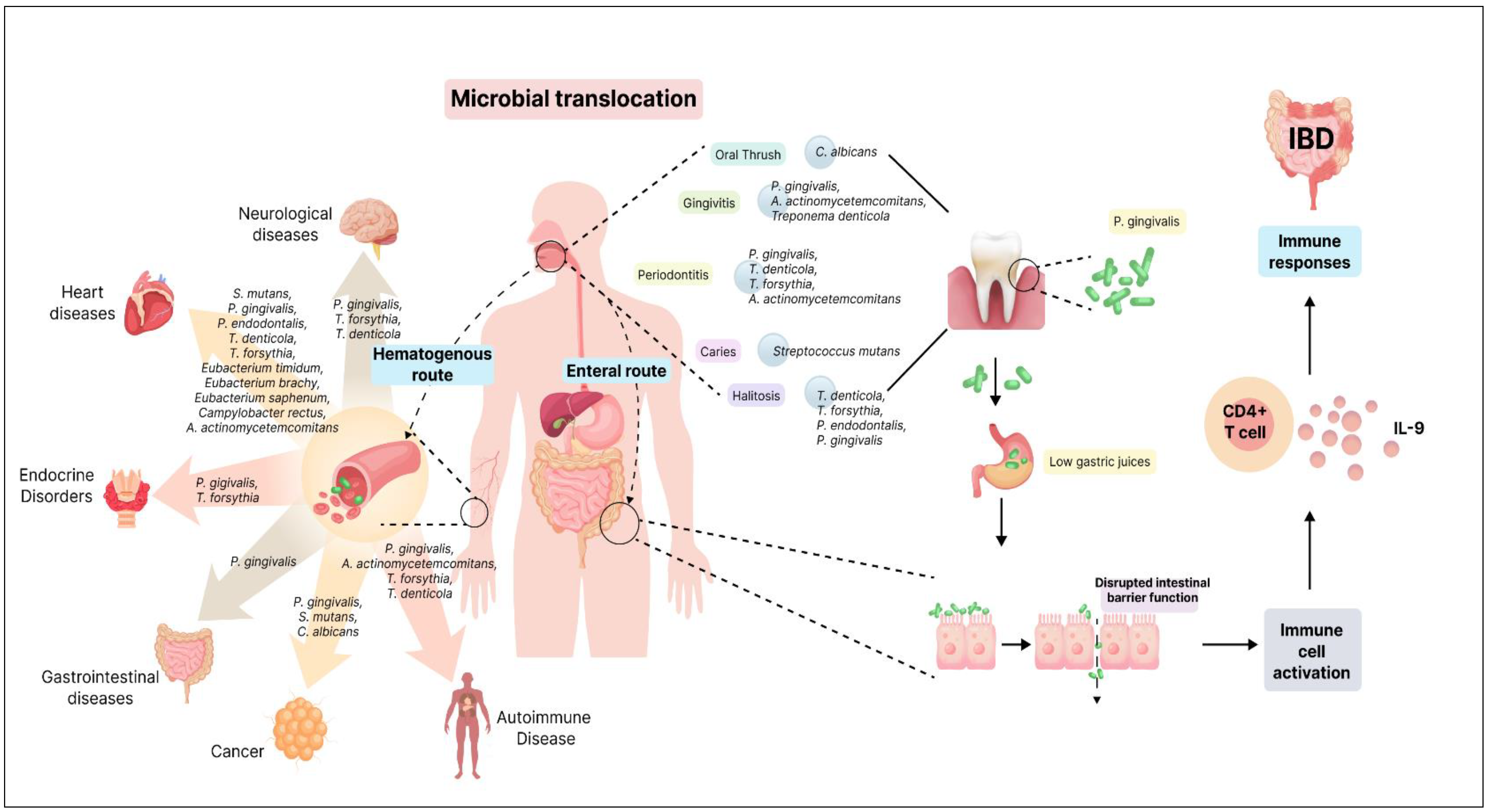
| IBD | - | P. gingivalis and F. nucleatum | ↑ | Oral-resident bacteria may infiltrate the gut microbiome; gut dysbiosis due to altered gut epithelial permeability. | [148] |
| Atherosclerosis | Prevotella nigrescens and Parvimonas micra | S. mutans, P. gingivalis, P. endodontalis, T. denticola, T. forsythia, Prevotella intermedia, Aggregatibacter actinomycetemcomitans, Eubacterium timidum, Eubacterium brachy, Eubacterium saphenum, and Campylobacter rectus | ↑ | Oral dysbiosis triggers local inflammation, systemic inflammatory responses, oxidative stress, immune activation, and platelet aggregation. | [155,156] |
| Diabetes | Capnocytophaga | P. gingivalis and T. forsythia | ↑ | Hyperglycemia and oxidative stress create a conducive environment for microbial dysbiosis; elevated inflammatory reactions. | [172] |
| Obesity | Proteobacteria, Chloroflexi, and Firmicutes | - | ↑ | Potential migration of oral bacteria to the gut; alterations in bacterial populations impact metabolic homeostasis. | [179,180] |
| AD | - | P. gingivalis, T. forsythia, and T. denticola | ↑ | Oral microbiota’s production of inflammatory agents potentially contributes to neuroinflammation and AD progression. | [186,187] |
| PD | - | P. gingivalis | ↑ | P. gingivalis infection is correlated with PD, with studies demonstrating the presence of gingipain R1 (RgpA) in the bloodstream, indicating systemic dissemination. P. gingivalis may contribute to PD pathogenesis by inducing systemic inflammation, promoting hypercoagulability, and exacerbating neurodegeneration. | [197] |
| RA | Prevotella, Veillonella, and Lactobacillus salivarius | P. gingivalis and A. actinomycetemcomitans | ↑ | P. gingivalis implicated in RA onset via citrullination and ACPA production; dysbiosis of oral microbiota exacerbates joint inflammation. | [205] |
| SLE | Veillonella, Streptococcus, and Prevotella | T. forsythia and T. denticola | ↑ | Oral microbial dysbiosis and periodontitis may exacerbate SLE via immune activation; potential contribution to autoimmune responses. | [214,215]. |
| Cancer | S. oralis, S. mitis, S. sanguinis, Lactobacillus fermentum, Lactobacillus acidophilus, and Bifidobacterium adolescentis | P. gingivalis and F. nucleatum | ↑ | Specific bacteria infiltrate cells, initiate tumor development, and produce cancer-promoting substances like lipopolysaccharides. | [221] |
| Component | Role/Mechanism | Formulation | References |
|---|---|---|---|
| Probiotics | Support balance of oral microbiota, produce specialized metabolites for maintaining microbiota equilibrium, and promote healthy immunity | Probiotic supplements containing strains such as Streptococcus salivarius M18, Streptococcus salivarius K12, Lactobacillus plantarum, Bifidobacterium lactis, Lactobacillus reuteri, and Lactobacillus salivarius help maintain oral microbiota balance, combat bad breath and gum inflammation, and enhance immune responses | [227] |
| Xylitol | Rebalances mouth acidity, reduces S. mutans counts, and disrupts S. mutans energy production | Xylitol mints and gums contain xylitol, natural peppermint flavor, magnesium stearate, and natural menthol | [243] |
| Chlorine dioxide mouth rinse | Neutralizes volatile sulfur compounds (VSCs), kills odor-producing bacteria, and reduces plaque and F. nucleatum counts | Stabilized chlorine dioxide (ClO2), trisodium phosphate, and citric acid; typically found in commercially available mouth rinses | [245] |
| Thymol mouth rinse | Antifungal properties; disrupts C. albicans hyphae production and adhesion to epithelial cells | Thymol, eucalyptol, menthol, and methyl salicylate dissolved in ethanol (27%); available in various commercial mouthwash formulations | [247] |
| Fluoride toothpaste | Strengthens enamel, prevents cavities, and promotes oral hygiene | Sodium fluoride, hydrated silica, cellulose gum, and glycerin, available in various brands and formulations of toothpaste | [222] |
| Hydroxyapatite toothpaste | Replenishes lost minerals in enamel and restores enamel structure; it is an alternative to fluoride-based products | Hydroxyapatite particles, available in various brands and formulations of toothpaste | [253] |
| Cetylpyridinium chloride (CPC) | Has antimicrobial properties, disrupts bacterial membranes and cellular functions, and is effective against plaque-forming bacteria | Found in various oral care formulations such as mouthwashes and toothpaste | [254] |
| Clove oil | Has antibacterial and antifungal effects, inhibits multi-resistant Staphylococcus species growth, and reduces C. albicans ergosterol levels | Clove oil containing eugenol, eugenyl acetate, and carvacrol, available in various oral care products such as mouth rinses and gels | [255] |
| Oil pulling | Generates antioxidants to damage microbial cell walls, removes bacteria by attracting them to oil, and hinders bacterial coaggregation and plaque formation | Coconut, sesame, or sunflower oil, typically used as a standalone oral hygiene technique | [256,257] |
| Antimicrobial peptides | Antimicrobial peptides (AMPs) such as defensins and cathelicidins disrupt microbial cell membranes through electrostatic interactions, forming pores that lead to ion leakage and cell death, providing natural defense against oral pathogens | Antimicrobial peptides (AMPs) like defensins and cathelicidins are integrated into oral care products to combat oral bacteria, fungi, and viruses | [239,240] |
| Enamel strengthening peptides | Enamel strengthening peptides, exemplified by peptide P11-4, mimic the mineral structure of enamel; they facilitate remineralization by attracting calcium ions, promoting the formation of hydroxyapatite essential for enamel regeneration and strengthening | P11-4: dental gels, pastes, and some toothpastes | [240] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajasekaran, J.J.; Krishnamurthy, H.K.; Bosco, J.; Jayaraman, V.; Krishna, K.; Wang, T.; Bei, K. Oral Microbiome: A Review of Its Impact on Oral and Systemic Health. Microorganisms 2024, 12, 1797. https://doi.org/10.3390/microorganisms12091797
Rajasekaran JJ, Krishnamurthy HK, Bosco J, Jayaraman V, Krishna K, Wang T, Bei K. Oral Microbiome: A Review of Its Impact on Oral and Systemic Health. Microorganisms. 2024; 12(9):1797. https://doi.org/10.3390/microorganisms12091797
Chicago/Turabian StyleRajasekaran, John J., Hari Krishnan Krishnamurthy, Jophi Bosco, Vasanth Jayaraman, Karthik Krishna, Tianhao Wang, and Kang Bei. 2024. "Oral Microbiome: A Review of Its Impact on Oral and Systemic Health" Microorganisms 12, no. 9: 1797. https://doi.org/10.3390/microorganisms12091797





