Dynamic Immune Response to Vibriosis in Pacific Oyster Crassostrea gigas Larvae during the Infection Process as Supported by Accurate Positioning of GFP-Tagged Vibrio Strains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. DNA Transfer and Confirmation of GFP-Tagged Strains
2.3. The Effect of GFP Incorporation on Cell Physiology of Bacteria
2.3.1. Bacterial Growth Curve
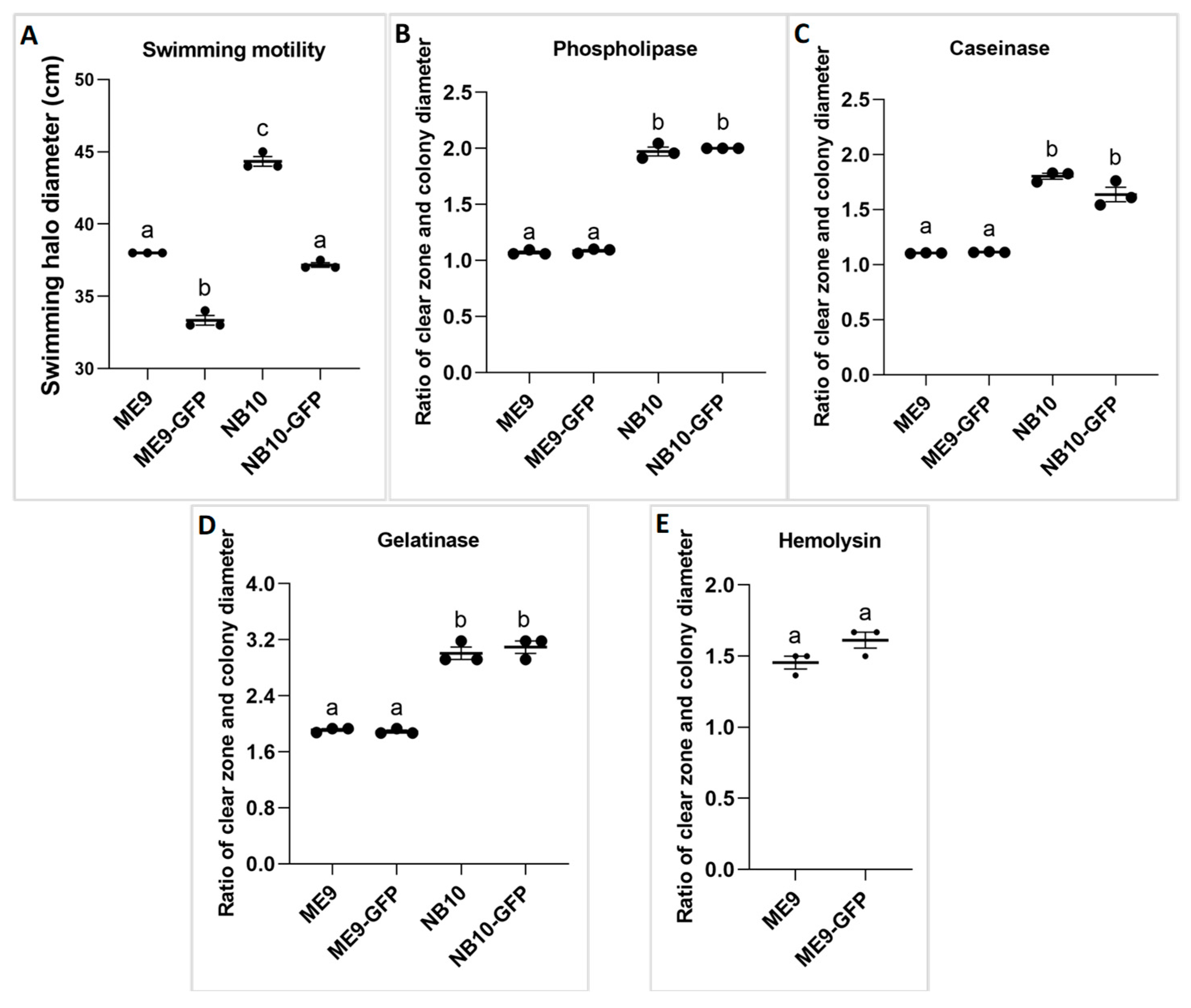
2.3.2. Virulence Factors of the Vibrio Strains
2.4. Gnotobiotic C. gigas Larvae Challenge Test
2.4.1. Rearing of C. gigas D-Veliger Larvae
2.4.2. General Design of the Challenge Tests
2.4.3. Experiments 1, 2, and 3
2.4.4. Experiment 4
2.5. Analytical Work
2.5.1. Observation of Infection Process
2.5.2. H&E Stained Histological Analysis
2.5.3. Modulation of Immune Genes in Bacterial Challenged Larvae
RNA Extraction and cDNA Synthesis
Immune-Related Genes and Primers Used in the Study
Quantitative Real-Time PCR (RT-qPCR) Analysis
2.6. Statistical Analysis
3. Results
3.1. The Effect of GFP Incorporation on Bacterial Growth and Virulence Factors
3.2. Survival Rate of the Oyster Larvae Infected with Vibrio Strains
3.3. Infection Process and Synchronous Histological Changes in Oyster Larvae Challenged by GFP-Tagged Vibrio spp.
3.4. The Effect of Vibrio Infection on Expression of Oyster Immune-Associated Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO: The State of World Fisheries and Aquaculture 2020: Sustainability in Action. 2020. Available online: http://www.fao.org/documents/card/en/c/ca9229en (accessed on 1 October 2020).
- Jeffries, V.E. Three Vibrio strains pathogenic to larvae of Crassostrea gigas and Ostrea edulis. Aquaculture 1982, 29, 201–226. [Google Scholar] [CrossRef]
- Estes, R.M.; Friedman, C.S.; Elston, R.A.; Herwig, R.P. Pathogenicity testing of shellfish hatchery bacterial isolates on Pacific oyster Crassostrea gigas larvae. Dis. Aquat. Organ. 2004, 58, 223–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasegawa, H.; Ha, C.C. TetR-type transcriptional regulator VtpR functions as a global regulator in Vibrio tubiashii. Appl. Environ. Microbiol. 2009, 75, 7602–7609. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, H.; Gharaibeh, D.N.; Lind, E.J.; Häse, C.C. Virulence of metalloproteases produced by Vibrio species on Pacific oyster Crassostrea gigas larvae. Dis. Aquat. Organ. 2009, 85, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Saulnier, D.; Decker, S.D.; Haffner, P.; Cobret, L.; Robert, M.; Garcia, C. A large-scale epidemiological study to identify bacteria pathogenic to Pacific oyster Crassostrea gigas and correlation between virulence and metalloprotease-like activity. Microb. Ecol. 2010, 59, 787–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kesarcodi-watson, A.; Miner, P.; Nicolas, J.; Robert, R. Protective effect of four potential probiotics against pathogen-challenge of the larvae of three bivalves: Pacific oyster (Crassostrea gigas), flat oyster (Ostrea edulis) and scallop (Pecten maximus). Aquaculture 2012, 344–349, 29–34. [Google Scholar] [CrossRef]
- Genard, B.; Miner, P.; Nicolas, J.L.; Moraga, D.; Boudry, P.; Pernet, F.; Tremblay, R. Integrative study of physiological changes associated with bacterial infection in Pacific oyster larvae. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Mersni-Achour, R.; Cheikh, Y.B.; Pichereau, V.; Doghri, I.; Etien, C.; Dégremont, L.; Saulnier, D.; Fruitier-Arnaudin, I.; Travers, M. Factors other than metalloprotease are required for full virulence of french Vibrio tubiashii isolates in oyster larvae. Microbiology 2015, 161, 997–1007. [Google Scholar] [CrossRef]
- Richards, G.P.; Watson, M.A.; Needleman, D.S.; Church, K.M.; Häse, C.C. Mortalities of Eastern and Pacific oyster larvae caused by the pathogens Vibrio coralliilyticus and Vibrio tubiashii. Appl. Environ. Microbiol. 2015, 81, 292–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubert, J.; Barja, J.L.; Romalde, J.L. New insights into pathogenic vibrios affecting bivalves in hatcheries: Present and future prospects. Front. Microbiol. 2017, 8, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Tubiash, H.S.; Chanley, P.E.; Leifson, E. Bacillary necrosis, a disease of larval and juvenile bivalve mollusks. I. Etiology and epizootiology. J. Bacteriol. 1965, 90, 1036–1044. [Google Scholar] [CrossRef] [Green Version]
- Elston, R.; Leibovitz, L. Pathogenesis of experimental vibriosis in larval american oysters, Crassostrea virginica. Can. J. Fish. Aquat. Sci. 1980, 37, 964–978. [Google Scholar] [CrossRef]
- Gómez-León, J.; Villamil, L.; Lemos, M.L.; Novoa, B.; Figueras, A. Isolation of Vibrio alginolyticus and Vibrio splendidus from aquacultured carpet shell clam (Ruditapes decussatus) larvae associated with mass mortalities. Appl. Environ. Microbiol. 2005, 71, 98–104. [Google Scholar] [CrossRef] [Green Version]
- Gómez-León, J.; Villamil, L.; Salger, S.A.; Sallum, R.H.; Remacha-Triviño, A.; Leavitt, D.F.; Gómez-Chiarri, M. Survival of eastern oysters Crassostrea virginica from three lines following experimental challenge with bacterial pathogens. Dis. Aquat. Organ. 2008, 79, 95–105. [Google Scholar] [CrossRef]
- Sandlund, N.; Mortensen, S.; Magnesen, T.; Torkildsen, L.; Bergh, Ø. Immunohistochemistry of great scallop Pecten maximus larvae experimentally challenged with pathogenic bacteria. Dis. Aquat. Organ. 2007, 69, 163–173. [Google Scholar] [CrossRef]
- Eiston, R.A.; Hasegawa, H.; Humphrey, K.L.; Polyak, I.K.; Hase, C.C. Re-emergence of Vibrio tubiashii in bivalve shellfish aquaculture: Severity, environmental drivers, geographic extent and management. Dis. Aquat. Organ. 2008, 82, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Kesarcodi-Watson, A.; Kaspar, H.; Lategan, M.J.; Gibson, L. Two pathogens of greenshellTM mussel larvae, Perna canaliculus: Vibrio splendidus and a V. coralliilyticus/neptunius-like isolate. J. Fish. Dis. 2009, 32, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Travers, M.A.; Barbou, A.; Le Goïc, N.; Huchette, S.; Paillard, C.; Koken, M. Construction of a stable GFP-tagged Vibrio harveyi strain for bacterial dynamics analysis of abalone infection. FEMS Microbiol. Lett. 2008, 289, 34–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rekecki, A.; Gunasekara, R.A.Y.S.A.; Dierckens, K.; Laureau, S.; Boon, N.; Favoreel, H.; Cornelissen, M.; Sorgeloos, P.; Ducatelle, R.; Bossier, P.; et al. Bacterial host interaction of GFP-labelled Vibrio anguillarum HI-610 with gnotobiotic sea bass, Dicentrarchus labrax (L.), larvae. J. Fish. Dis. 2012, 35, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Aboubaker, M.H.; Sabrié, J.; Huet, M.; Koken, M. Establishment of stable GFP-tagged Vibrio aestuarianus strains for the analysis of bacterial infection-dynamics in the Pacific oyster, Crassostrea gigas. Vet. Microbiol. 2013, 164, 392–398. [Google Scholar] [CrossRef]
- Dubert, J.; Nelson, D.R.; Spinard, E.J.; Kessner, L.; Gomez-Chiarri, M.; da Costa, F.; Prado, S.; Barjaa, J.L. Following the infection process of vibriosis in Manila clam (Ruditapes philippinarum) larvae through GFP-tagged pathogenic Vibrio species. J. Invertebr. Pathol. 2016, 133, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Luna-González, A.; Maeda-Martínez, A.N.; Ascencio-Valle, F.; Robles-Mungaray, M. Ontogenetic variations of hydrolytic enzymes in the Pacific oyster Crassostrea gigas. Fish. Shellfish Immunol. 2004, 16, 287–294. [Google Scholar] [CrossRef]
- Tirapé, A.; Bacque, C.; Brizard, R.; Vandenbulcke, F.; Boulo, V. Expression of immune-related genes in the oyster Crassostrea gigas during ontogenesis. Dev. Comp. Immunol. 2007, 31, 859–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genard, B.; Moraga, D.; Pernet, F.; David, É.; Boudry, P.; Tremblay, R. Expression of candidate genes related to metabolism, immunity and cellular stress during massive mortality in the American oyster Crassostrea virginica larvae in relation to biochemical and physiological parameters. Gene 2012, 499, 70–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balseiro, P.; Moreira, R.; Chamorro, R.; Figueras, A.; Novoa, B. Immune responses during the larval stages of Mytilus galloprovincialis: Metamorphosis alters immunocompetence, body shape and behavior. Fish. Shellfish Immunol. 2013, 35, 438–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassim, S.; Genard, B.; Gauthier-Clerc, S.; Moraga, D.; Tremblay, R. Ontogeny of bivalve immunity: Assessing the potential of next-generation sequencing techniques. Rev. Aquac. 2015, 7, 197–217. [Google Scholar] [CrossRef]
- Lindell, K.; Fahlgren, A.; Hjerde, E.; Willassen, N.P.; Fällman, M.; Milton, D.L. Lipopolysaccharide O-antigen prevents phagocytosis of vibrio anguillarum by rainbow trout (Oncorhynchus mykiss) skin epithelial cells. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [Green Version]
- Bachère, E.; Rosa, R.D.; Schmitt, P.; Poirier, A.C.; Merou, N.; Charrière, G.M.; Destoumieux-Garzón, D. The new insights into the oyster antimicrobial defense: Cellular, molecular and genetic view. Fish. Shellfish Immunol. 2015, 46, 50–64. [Google Scholar] [CrossRef] [Green Version]
- Vieira, G.C.; da Silva, P.M.; Barracco, M.A.; Hering, A.F.; de Albuquerque, M.C.P.; da Rosa Coelho, J.; Schmidt, E.C.; Bouzon, Z.L.; Rosa, R.D.; Perazzolo, L.M. Morphological and functional characterization of the hemocytes from the pearl oyster Pteria hirundo and their immune responses against Vibrio infections. Fish. Shellfish Immunol. 2017, 70, 750–758. [Google Scholar] [CrossRef]
- Wang, L.; Song, X.; Song, L. The oyster immunity. Dev. Comp. Immunol. 2018, 80, 99–118. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [Green Version]
- Thomas-Guyon, H.; Gagnaire, B.; Bado-Nilles, A.; Bouilly, K.; Lapègue, S.; Renault, T. Detection of phenoloxidase activity in early stages of the Pacific oyster Crassostrea gigas (Thunberg). Dev. Comp. Immunol. 2009, 33, 653–659. [Google Scholar] [CrossRef]
- Song, X.; Wang, H.; Xin, L.; Xu, J.; Jia, Z.; Wang, L.; Song, L. The immunological capacity in the larvae of Pacific oyster Crassostrea gigas. Fish. Shellfish Immunol. 2016, 49, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.; Romestand, B.; Fievet, J.; Huvet, A.; Lebart, M.C.; Gueguen, Y.; Bachère, E. Evidence in oyster of a plasma extracellular superoxide dismutase which binds LPS. Biochem. Biophys. Res. Commun. 2005, 338, 1089–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, L.; Wu, L.; Ni, D.; Chang, Y.; Xu, W.; Xing, K. The cDNA cloning and mRNA expression of heat shock protein 70 gene in the haemocytes of bay scallop (Argopecten irradians, Lamarck 1819) responding to bacteria challenge and naphthalin stress. Fish. Shellfish Immunol. 2006, 21, 335–345. [Google Scholar] [CrossRef]
- Sritunyalucksana, K.; Wongsuebsantati, K.; Johansson, M.W.; So, K. Peroxinectin, a cell adhesive protein associated with the proPO system from the black tiger shrimp, Penaeus monodon. Dev. Comp. Immunol. 2001, 25, 353–363. [Google Scholar] [CrossRef]
- Liu, C.; Cheng, W.; Chen, J. The peroxinectin of white shrimp Litopenaeus vannamei is synthesised in the semi-granular and granular cells, and its transcription is up-regulated with Vibrio alginolyticus infection. Fish. Shellfish Immunol. 2005, 18, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Lang, R.P.; Bayne, C.J.; Camara, M.D.; Cunningham, C.; Jenny, M.J.; Langdon, C.J. Transcriptome profiling of selectively bred pacific oyster Crassostrea gigas families that differ in tolerance of heat shock. Mar. Biotechnol. 2009, 11, 650–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Z.; Zhang, T.; Jiang, S.; Wang, M.; Cheng, Q.; Sun, M.; Wang, L.; Song, L. An integrin from oyster Crassostrea gigas mediates the phagocytosis toward Vibrio splendidus through LPS binding activity. Dev. Comp. Immunol. 2015, 53, 253–264. [Google Scholar] [CrossRef]
- Montagnani, C.; Kappler, C.; Reichhart, J.M.; Escoubas, J.M. Cg-Rel, the first Rel/NF-κB homolog characterized in a mollusk, the Pacific oyster Crassostrea gigas. FEBS Lett. 2004, 561, 75–82. [Google Scholar] [CrossRef] [Green Version]
- Xiong, X.; Feng, Q.; Chen, L.; Xie, L.; Zhang, R. Cloning and characterization of an IKK homologue from pearl oyster, Pinctada fucata. Dev. Comp. Immunol. 2008, 32, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Eggermont, M.; Bossier, P.; Pande, G.S.J.; Delahaut, V.; Rayhan, A.M.; Gupta, N.; Islam, S.S.; Yumo, E.; Nevejan, N.; Sorgeloos, P.; et al. Isolation of Vibrionaceae from wild blue mussel (Mytilus edulis) adults and their impact on blue mussel larviculture. FEMS Microbiol. Ecol. 2017, 93, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Mbewe, N.; De Bels, L.; Couck, L.; Van Stappen, G.; Van den Broeck, W.; Nevejan, N. Pathogenesis of experimental vibriosis in blue mussel (Mytilus edulis) larvae based on accurate positioning of GFP-tagged Vibrio strains and histopathological and ultrastructural changes of the host. Aquaculture 2021, 535, 736347. [Google Scholar] [CrossRef]
- Rang, C.; Galen, J.E.; Kaper, J.B.; Chao, L. Fitness cost of the green fluorescent protein in gastrointestinal bacteria. Can. J. Microbiol. 2003, 49, 531–537. [Google Scholar] [CrossRef]
- Allison, D.G.; Sattenstall, M.A. The influence of green fluorescent protein incorporation on bacterial physiology: A note of caution. J. Appl. Microbiol. 2007, 103, 318–324. [Google Scholar] [CrossRef]
- Andersen, J.B.; Sternberg, C.; Poulsen, L.K.; Bjørn, S.P.; Givskov, M.; Molin, S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 1998, 64, 2240–2246. [Google Scholar] [CrossRef] [Green Version]
- Natrah, F.M.I.; Ruwandeepika, H.A.D.; Pawar, S.; Karunasagar, I.; Sorgeloos, P.; Bossier, P.; Defoirdt, T. Regulation of virulence factors by quorum sensing in Vibrio harveyi. Vet. Microbiol. 2011, 154, 124–129. [Google Scholar] [CrossRef]
- Yang, Q.; Defoirdt, T. Quorum sensing positively regulates flagellar motility in pathogenic Vibrio harveyi. Environ. Microbiol. 2015, 17, 960–968. [Google Scholar] [CrossRef]
- Langdon, C.J. Growth studies with bacteria-free oyster (Crassostrea gigas) larvae fed on semi-defined artificial diets. Biol. Bull. 1983, 164, 227–235. [Google Scholar] [CrossRef]
- Van Hung, N.; De Schryver, P.; Tam, T.T.; Garcia-Gonzalez, L.; Bossier, P.; Nevejan, N. Application of poly-β-hydroxybutyrate (PHB) in mussel larviculture. Aquaculture 2015, 446, 318–324. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, L.; Xu, F.; Huang, B.; Zhang, G.; Li, L. Validation of housekeeping genes as internal controls for studying gene expression during Pacific oyster (Crassostrea gigas) development by quantitative real-time PCR. Fish. Shellfish Immunol. 2013, 34, 939–945. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vinoj, G.; Vaseeharan, B.; Brennan, G. Green fluorescent protein visualization of Vibrio parahaemolyticus infections in Indian white shrimp Fenneropenaeus indicus (H Milne Edwards). Aquac. Res. 2014, 45, 1989–1999. [Google Scholar] [CrossRef]
- Gobi, N.; Malaikozhundan, B.; Sekar, V.; Shanthi, S.; Vaseeharan, B.; Jayakumar, R.; Khudus Nazar, A. GFP tagged Vibrio parahaemolyticus Dahv2 infection and the protective effects of the probiotic Bacillus licheniformis Dahb1 on the growth, immune and antioxidant responses in Pangasius hypophthalmus. Fish. Shellfish Immunol. 2016, 52, 230–238. [Google Scholar] [CrossRef]
- Girija, V.; Malaikozhundan, B.; Vaseeharan, B.; Vijayakumar, S.; Gobi, N.; Del Valle Herrera, M.; Chen, J.; Santhanam, P. In vitro antagonistic activity and the protective effect of probiotic Bacillus licheniformis Dahb1 in zebrafish challenged with GFP tagged Vibrio parahaemolyticus Dahv2. Microb. Pathog. 2018, 114, 274–280. [Google Scholar] [CrossRef]
- Busschaert, P.; Frans, I.; Crauwels, S.; Zhu, B.; Willems, K.; Bossier, P.; Michiels, C.; Verstrepen, K.; Lievens, B.; Rediers, H. Comparative genome sequencing to assess the genetic diversity and virulence attributes of 15 Vibrio anguillarum isolates. J. Fish. Dis. 2015, 38, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Leung, K.Y. Biochemical characterization of different types of adherence of Vibrio species to fish epithelial cells. Microbiology 2000, 146, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Croxatto, A.; Lauritz, J.; Chen, C.; Milton, D.L. Vibrio anguillarum colonization of rainbow trout integument requires a DNA locus involved in exopolysaccharide transport and biosynthesis. Environ. Microbiol. 2007, 9, 370–382. [Google Scholar] [CrossRef]
- Duperthuy, M.; Schmitt, P.; Garzón, E.; Caro, A.; Rosa, R.D.; Le Roux, F.; Lautrédou-Audouy, N.; Got, P.; Romestand, B.; de Lorgeril, J.; et al. Use of OmpU porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc. Natl. Acad. Sci. USA 2011, 108, 2993–2998. [Google Scholar] [CrossRef] [Green Version]
- Duperthuy, M.; Binesse, J.; Le Roux, F.; Romestand, B.; Caro, A.; Got, P.; Givaudan, A.; Mazel, D.; Bachère, E.; Destoumieux-Garzón, D. The major outer membrane protein OmpU of Vibrio splendidus contributes to host antimicrobial peptide resistance and is required for virulence in the oyster Crassostrea gigas. Environ. Microbiol. 2010, 12, 951–963. [Google Scholar] [CrossRef] [Green Version]
- Denkin, S.M.; Nelson, D.R. Regulation of Vibrio anguillarum empA metalloprotease expression and its role in virulence. Appl. Environ. Microbiol. 2004, 70, 4193–4204. [Google Scholar] [CrossRef] [Green Version]
- Binesse, J.; Saulnier, D.; Mazel, D. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl. Environ. Microbiol. 2007, 73, 777–784. [Google Scholar] [CrossRef] [Green Version]
- Mateo, D.R.; Siah, A.; Araya, M.T.; Berthe, F.C.J.; Johnson, G.R.; Greenwood, S.J. Differential in vivo response of soft-shell clam hemocytes against two strains of Vibrio splendidus: Changes in cell structure, numbers and adherence. J. Invertebr. Pathol. 2009, 102, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Qiu, L.; Cheng, Q.; Zhang, H.; Wang, L.; Song, L. Evidence for cleavage of the metalloprotease Vsm from Vibrio splendidus strain JZ6 by an M20 peptidase (PepT-like protein) at low temperature. Front. Microbiol. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojas, R.; Miranda, C.D.; Santander, J.; Romero, J. First report of Vibrio tubiashii associated with a massive larval mortality event in a commercial hatchery of scallop Argopecten purpuratus in Chile. Front. Microbiol. 2016, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Yue, F.; Shi, X.; Zhou, Z.; Wang, L.; Wang, M.; Yang, J.; Qiu, L.; Song, L. The expression of immune-related genes during the ontogenesis of scallop Chlamys farreri and their response to bacterial challenge. Fish. Shellfish Immunol. 2013, 34, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Pateras, I.; Giaginis, C.; Tsigris, C.; Patsouris, E.; Theocharis, S. NF-κB signaling at the crossroads of inflammation and atherogenesis: Searching for new therapeutic links. Expert Opin. Ther. Targets 2014, 18, 1089–1101. [Google Scholar] [CrossRef]
- De Lorgeril, J.; Zenagui, R.; Rosa, R.D.; Piquemal, D.; Bachère, E. Whole transcriptome profiling of successful immune response to Vibrio infections in the oyster Crassostrea gigas by digital gene expression analysis. PLoS ONE 2011, 6, e23142. [Google Scholar] [CrossRef] [Green Version]
- Fleury, E.; Huvet, A. Microarray analysis highlights immune response of Pacific oysters as a determinant of resistance to summer mortality. Mar. Biotechnol. 2012, 14, 203–217. [Google Scholar] [CrossRef] [Green Version]
- Johansson, M.W.; Lind, M.I.; Holmblad, T.; Thornqvist, P.O.; Soderhall, K. Peroxinectin, a novel cell adhesion protein from crayfish blood. Biochem. Biophys. Res. Commun. 1995, 216, 1079–1087. [Google Scholar] [CrossRef]
- Huang, Y.; Zhao, L.L.; Feng, J.L.; Zhu, H.X.; Huang, X.; Ren, Q.; Wang, W. A novel integrin function in innate immunity from Chinese mitten crab (Eriocheir sinensis). Dev. Comp. Immunol. 2015, 52, 155–165. [Google Scholar] [CrossRef]
- Johansson, M.W.; Söderhäll, K. A cell adhesion factor from crayfish haemocytes has degranulating activity towards crayfish granular cells. Insect Biochem. 1989, 19, 183–190. [Google Scholar] [CrossRef]
- Kobayashi, M.; Johansson, M.W.; Söderhäll, K. The 76 kD cell-adhesion factor from crayfish haemocytes promotes encapsulation in vitro. Cell Tissue Res. 1990, 260, 13–18. [Google Scholar] [CrossRef]
- Thörnqvist, P.O.; Johansson, M.W.; Söderhäll, K. Opsonic activity of cell adhesion proteins and β-1,3-glucan binding proteins from two crustaceans. Dev. Comp. Immunol. 1994, 18, 3–12. [Google Scholar] [CrossRef]
- Johansson, M.W.; Söderhäll, K. Isolation and purification of a cell adhesion factor from crayfish blood cells. Cell 1988, 106, 1795–1803. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Genes a | Gene Description b | Primer Sequences (5′-3′) | Reference |
|---|---|---|---|
| RS18 | Ribosomal protein S18 | F: GCCATCAAGGGTATCGGTAGAC R: CTGCCTGTTAAGGAACCAGTCAG | [53] |
| RL7 | Ribosomal protein L7 | F: TCCCAAGCCAAGGAAGGTTATGC R: CAAAGCGTCCAAGGTGTTTCTCAA | |
| SOD | The antioxidant enzymes | F: TGAAGGCCGTCTGTGTATTG R: TCCATGCTGTCCTGGTGTTA | AJ496219 c |
| HSP70 | Acute phase proteins | F: CCAGTTGAGGATACTCTTGAGTGC R: ATGTCGATAACGGTCCCTTTCT | [35] |
| IKK | Immunity signaling pathways | F: TCTCACACCCACACACCTATGC R: AGTAGTTTTCCACCAGGGGATAAG | |
| Rel/NF-κB | F: GAAGGCAAAGGGAGGTGATGAG R: GGTGTGCGGAAGACAATGGC | ||
| Integrin β-1 | Cell adhesion molecule | F: TCATCTGTGGAGGTCTGAGTCG R: TGTACATGCAGGGGCTTTTGTC | |
| Peroxinectin | F: GCCAAACCTCGCCTACCTTC R: GTGGAGTTGACGCGTGACATA | [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Loor, A.; Bels, L.D.; Stappen, G.V.; Broeck, W.V.d.; Nevejan, N. Dynamic Immune Response to Vibriosis in Pacific Oyster Crassostrea gigas Larvae during the Infection Process as Supported by Accurate Positioning of GFP-Tagged Vibrio Strains. Microorganisms 2021, 9, 1523. https://doi.org/10.3390/microorganisms9071523
Wang D, Loor A, Bels LD, Stappen GV, Broeck WVd, Nevejan N. Dynamic Immune Response to Vibriosis in Pacific Oyster Crassostrea gigas Larvae during the Infection Process as Supported by Accurate Positioning of GFP-Tagged Vibrio Strains. Microorganisms. 2021; 9(7):1523. https://doi.org/10.3390/microorganisms9071523
Chicago/Turabian StyleWang, Dongdong, Alfredo Loor, Lobke De Bels, Gilbert Van Stappen, Wim Van den Broeck, and Nancy Nevejan. 2021. "Dynamic Immune Response to Vibriosis in Pacific Oyster Crassostrea gigas Larvae during the Infection Process as Supported by Accurate Positioning of GFP-Tagged Vibrio Strains" Microorganisms 9, no. 7: 1523. https://doi.org/10.3390/microorganisms9071523
APA StyleWang, D., Loor, A., Bels, L. D., Stappen, G. V., Broeck, W. V. d., & Nevejan, N. (2021). Dynamic Immune Response to Vibriosis in Pacific Oyster Crassostrea gigas Larvae during the Infection Process as Supported by Accurate Positioning of GFP-Tagged Vibrio Strains. Microorganisms, 9(7), 1523. https://doi.org/10.3390/microorganisms9071523






