Characterization of Microfragmented Adipose Tissue Architecture, Mesenchymal Stromal Cell Content and Release of Paracrine Mediators
Abstract
:1. Introduction
2. Materials and Methods
2.1. Adipose Tissue Harvesting and Processing
2.2. Histology and Immunohistochemistry
2.3. Cell Count and Viability
2.4. Flow Cytometry
2.5. ASCs Isolation and Differentiation
2.6. Secretome Collection
2.7. Proteomic Analysis
2.8. Nanoparticle Tracking Analysis (NTA)
2.9. EV-Associated MicroRNAs (miRNAs) High Throughput Analysis
2.10. Transmission Electron Microscopy (TEM)
2.11. Principal Component Analysis (PCA) and Hierarchical Clustering
2.12. Statistical Analysis
3. Results
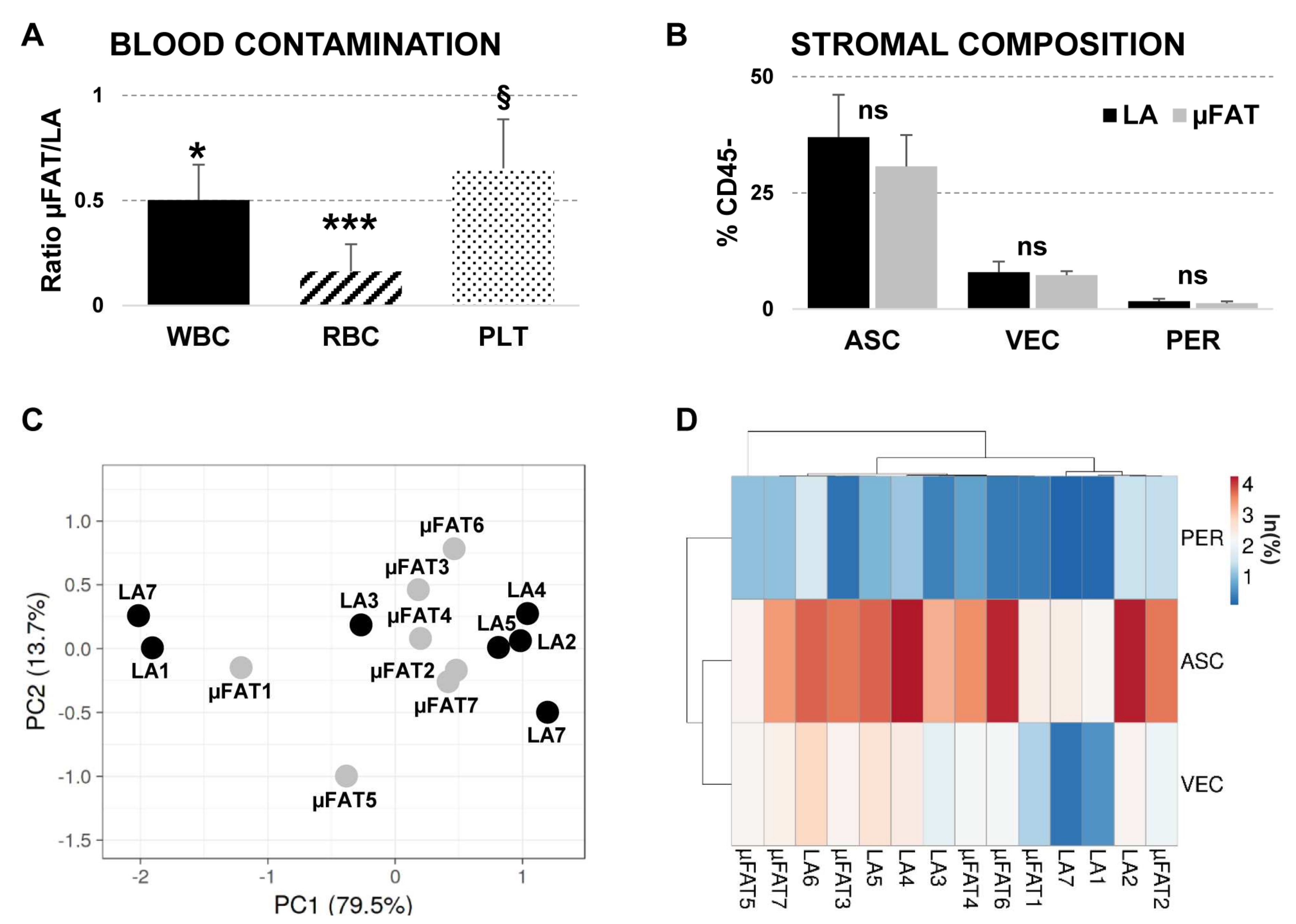
3.1. Tissue Integrity Is Maintained in μFAT
3.2. Microfragmentation Allows for Removal of Blood Contamination
3.3. Population Heterogeneity Is Maintained in μFAT
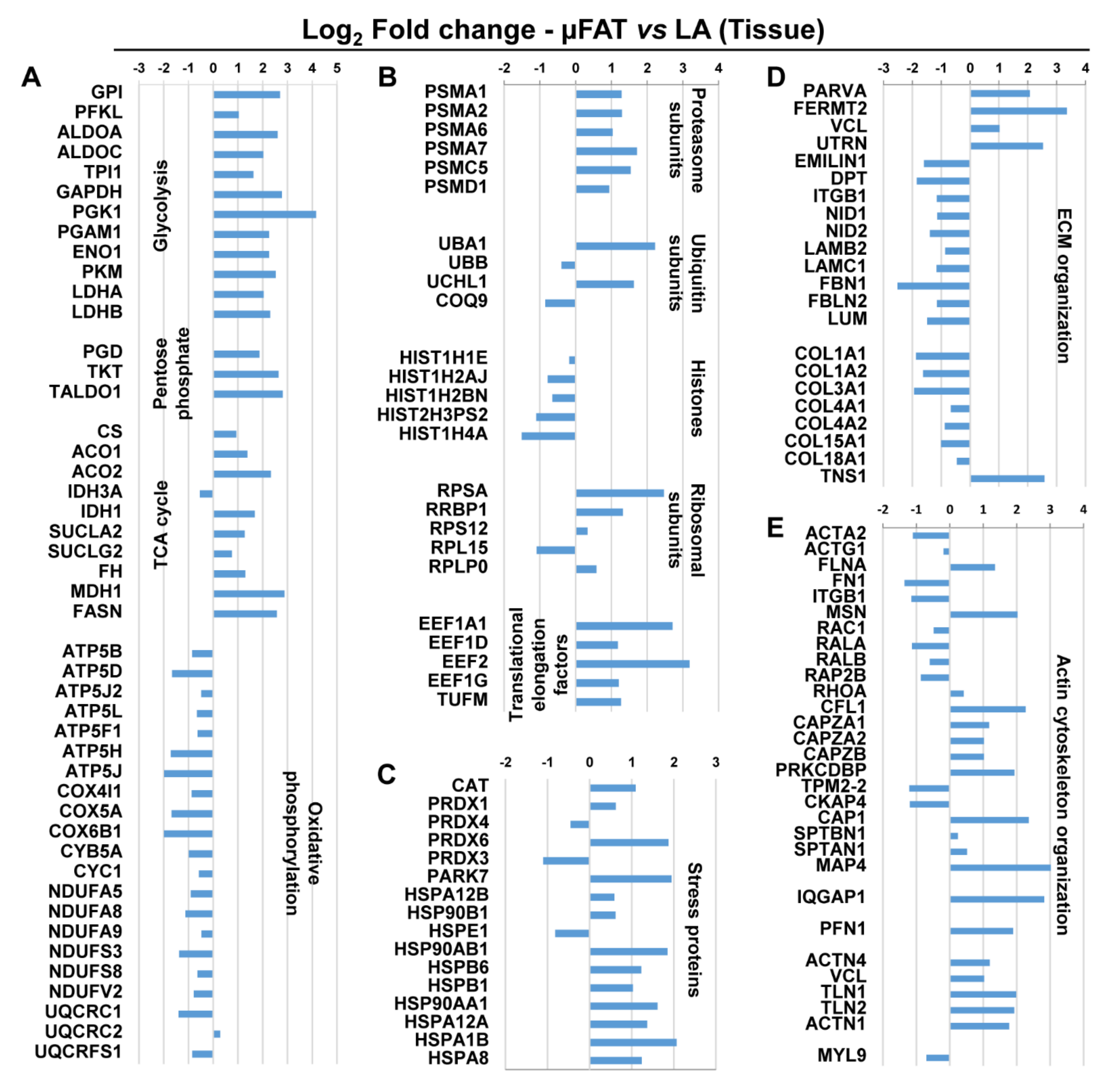
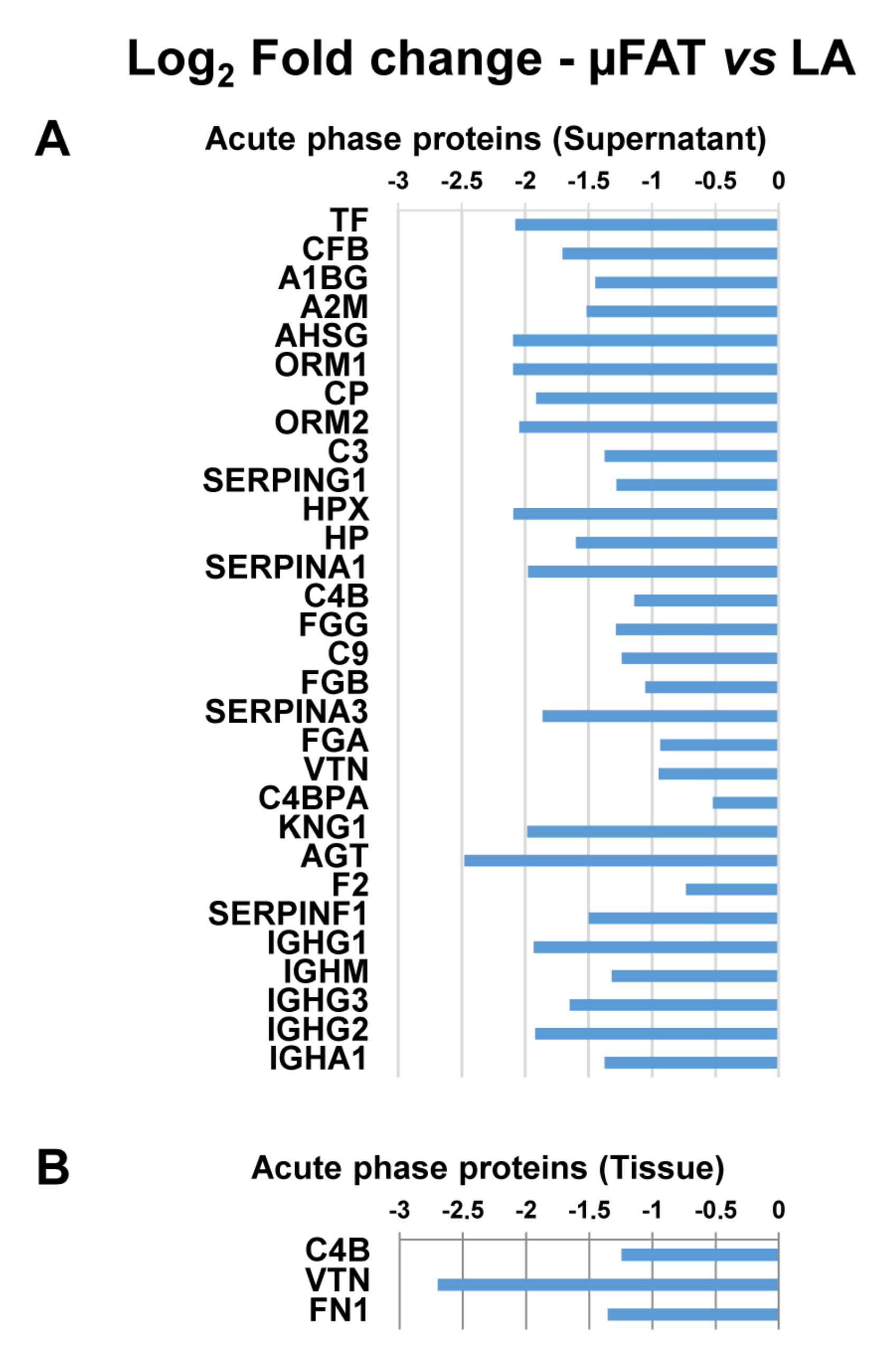
3.4. Microfragmentation Modulates the Protein Fingerprint
3.5. Microfragmentation Allows for Reduction of Peripheral Blood-Derived EV-miRNAs and Increase of Chondro-Protective Ones
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Burden of Disease Collaborative Network. Global Burden of Disease Study 2019 (GBD 2019) Results. Available online: http://ghdx.healthdata.org/gbd-results-tool (accessed on 27 January 2022).
- Grässel, S.; Muschter, D. Recent advances in the treatment of osteoarthritis. F1000Research 2020, 9, 325. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, J.; Fitzpatrick, J.; Perera, N.K.P.; Orchard, J. Osteoarthritis-a systematic review of long-term safety implications for osteoarthritis of the knee. BMC Musculoskelet. Disord. 2019, 20, 151. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.-L.; Zhou, A.-G.; Zhang, H.; Zhang, J. Efficacy of platelet-rich plasma in the treatment of knee osteoarthritis: A meta-analysis of randomized controlled trials. Arthroscopy 2017, 33, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Buendía-López, D.; Medina-Quirós, M.; Fernández-Villacañas Marín, M.Á. Clinical and radiographic comparison of a single LP-PRP injection, a single hyaluronic acid injection and daily NSAID administration with a 52-week follow-up: A randomized controlled trial. J. Orthop. Traumatol. 2018, 19, 3. [Google Scholar] [CrossRef]
- Guillibert, C.; Charpin, C.; Raffray, M.; Benmenni, A.; Dehaut, F.-X.; El Ghobeira, G.; Giorgi, R.; Magalon, J.; Arniaud, D. Single injection of high volume of autologous pure PRP provides a significant improvement in knee osteoarthritis: A prospective routine care study. Int. J. Mol. Sci. 2019, 20, 1327. [Google Scholar] [CrossRef] [Green Version]
- Borić, I.; Hudetz, D.; Rod, E.; Jeleč, Ž.; Vrdoljak, T.; Skelin, A.; Polašek, O.; Plečko, M.; Trbojević-Akmačić, I.; Lauc, G.; et al. A 24-month follow-up study of the effect of intra-articular injection of autologous microfragmented fat tissue on proteoglycan synthesis in patients with knee osteoarthritis. Genes 2019, 10, 1051. [Google Scholar] [CrossRef] [Green Version]
- Ragni, E.; Colombini, A.; Viganò, M.; Libonati, F.; Perucca Orfei, C.; Zagra, L.; De Girolamo, L. Cartilage protective and immunomodulatory features of osteoarthritis synovial fluid-treated adipose-derived mesenchymal stem cells secreted factors and extracellular vesicles-embedded MiRNAs. Cells 2021, 10, 1072. [Google Scholar] [CrossRef]
- Caplan, A.I. Mesenchymal stem cells: Time to change the name! Stem Cells Transl. Med. 2017, 6, 1445–1451. [Google Scholar] [CrossRef] [Green Version]
- Caplan, A.I.; Dennis, J.E. Mesenchymal stem cells as trophic mediators. J. Cell. Biochem. 2006, 98, 1076–1084. [Google Scholar] [CrossRef]
- Maumus, M.; Manferdini, C.; Toupet, K.; Peyrafitte, J.-A.; Ferreira, R.; Facchini, A.; Gabusi, E.; Bourin, P.; Jorgensen, C.; Lisignoli, G.; et al. Adipose mesenchymal stem cells protect chondrocytes from degeneration associated with osteoarthritis. Stem Cell Res. 2013, 11, 834–844. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, N.; Mak, C.; Bojanic, C.; To, K.; Khan, W. Meta-analysis of adipose tissue derived cell-based therapy for the treatment of knee osteoarthritis. Cells 2021, 10, 1365. [Google Scholar] [CrossRef]
- Tremolada, C.; Colombo, V.; Ventura, C. Adipose tissue and mesenchymal stem cells: State of the Art and Lipogems® Technology Development. Curr. Stem Cell Rep. 2016, 2, 304–312. [Google Scholar] [CrossRef] [Green Version]
- Alharbi, Z.; Opländer, C.; Almakadi, S.; Fritz, A.; Vogt, M.; Pallua, N. Conventional vs. micro-fat harvesting: How fat harvesting technique affects tissue-engineering approaches using adipose tissue-derived stem/stromal cells. J. Plast. Reconstr. Aesthet. Surg. 2013, 66, 1271–1278. [Google Scholar] [CrossRef]
- Caplan, A.I. All MSCs are pericytes? Cell Stem Cell 2008, 3, 229–230. [Google Scholar] [CrossRef] [Green Version]
- Bianchi, F.; Maioli, M.; Leonardi, E.; Olivi, E.; Pasquinelli, G.; Valente, S.; Mendez, A.J.; Ricordi, C.; Raffaini, M.; Tremolada, C.; et al. A new nonenzymatic method and device to obtain a fat tissue derivative highly enriched in pericyte-like elements by mild mechanical forces from human lipoaspirates. Cell Transpl. 2013, 22, 2063–2077. [Google Scholar] [CrossRef]
- Eyroh. The Effect of Micro Fragmented Adipose Tissue (MFAT) on Knee Osteoarthritis; ClinicalTrials.gov: 2020. Available online: https://www.clinicaltrials.gov/ct2/show/NCT03467919?term=mfat&draw=2&rank=3 (accessed on 27 January 2022).
- Coleman, S.R. Structural fat grafting: More than a permanent filler. Plast. Reconstr. Surg. 2006, 118, 108S–120S. [Google Scholar] [CrossRef] [Green Version]
- Stanco, D.; Viganò, M.; Perucca Orfei, C.; Di Giancamillo, A.; Peretti, G.M.; Lanfranchi, L.; de Girolamo, L. Multidifferentiation potential of human mesenchymal stem cells from adipose tissue and hamstring tendons for musculoskeletal cell-based therapy. Regen. Med. 2015, 10, 729–743. [Google Scholar] [CrossRef] [Green Version]
- Shah, D.; Naciri, M.; Clee, P.; Al-Rubeai, M. NucleoCounter—An efficient technique for the determination of cell number and viability in animal cell culture processes. Cytotechnology 2006, 51, 39–44. [Google Scholar] [CrossRef] [Green Version]
- Van Dongen, J.A.; Tuin, A.J.; Spiekman, M.; Jansma, J.; Van Der Lei, B.; Harmsen, M.C. Comparison of intraoperative procedures for isolation of clinical grade stromal vascular fraction for regenerative purposes: A systematic review. J. Tissue Eng. Regen. Med. 2018, 12, e261–e274. [Google Scholar] [CrossRef] [Green Version]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Carlson, A.; Sinitcyn, P.; Mann, M.; Cox, J. Visualization of LC-MS/MS proteomics data in MaxQuant. Proteomics 2015, 15, 1453–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ragni, E.; Perucca Orfei, C.; De Luca, P.; Mondadori, C.; Viganò, M.; Colombini, A.; De Girolamo, L. Inflammatory priming enhances mesenchymal stromal cell secretome potential as a clinical product for regenerative medicine approaches through secreted factors and EV-MiRNAs: The example of joint disease. Stem Cell Res. Ther. 2020, 11, 165. [Google Scholar] [CrossRef]
- D’haene, B.; Mestdagh, P.; Hellemans, J.; Vandesompele, J. MiRNA expression profiling: From reference genes to global mean normalization. Methods Mol. Biol. 2012, 822, 261–272. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Bourin, P.; Bunnell, B.A.; Casteilla, L.; Dominici, M.; Katz, A.J.; March, K.L.; Redl, H.; Rubin, J.P.; Yoshimura, K.; Gimble, J.M. Stromal cells from the adipose tissue-derived stromal vascular fraction and culture expanded adipose tissue-derived stromal/stem cells: A joint statement of the international federation for adipose therapeutics and science (IFATS) and the international society for cellular therapy (ISCT). Cytotherapy 2013, 15, 641–648. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Zhu, J.; Fan, L.; Lin, Q.; Fu, D.; Wei, B.; Wei, S. MicroRNA profiling of exosomes derived from red blood cell units: Implications in transfusion-related immunomodulation. Biomed. Res. Int. 2019, 2019, 2045915. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [Green Version]
- Laffont, B.; Corduan, A.; Rousseau, M.; Duchez, A.-C.; Lee, C.H.C.; Boilard, E.; Provost, P. Platelet microparticles reprogram macrophage gene expression and function. Thromb. Haemost. 2016, 115, 311–323. [Google Scholar] [CrossRef]
- Arraud, N.; Gounou, C.; Turpin, D.; Brisson, A.R. Fluorescence triggering: A general strategy for enumerating and phenotyping extracellular vesicles by flow cytometry. Cytom. A 2016, 89, 184–195. [Google Scholar] [CrossRef]
- Ragni, E.; Perucca Orfei, C.; De Luca, P.; Colombini, A.; Viganò, M.; De Girolamo, L. Secreted factors and EV-MiRNAs orchestrate the healing capacity of adipose mesenchymal stem cells for the treatment of knee osteoarthritis. Int. J. Mol. Sci. 2020, 21, 1582. [Google Scholar] [CrossRef] [Green Version]
- Endisha, H.; Rockel, J.; Jurisica, I.; Kapoor, M. The complex landscape of MicroRNAs in articular cartilage: Biology, pathology, and therapeutic targets. JCI Insight 2018, 3, 121630. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.J.; Hu, H.T.; Li, H.L.; Chang, S. The role of MiRNAs in immune cell development, immune cell activation, and tumor immunity: With a focus on macrophages and natural killer cells. Cells 2019, 8, 1140. [Google Scholar] [CrossRef] [Green Version]
- Gruys, E.; Toussaint, M.J.M.; Niewold, T.A.; Koopmans, S.J. Acute phase reaction and acute phase proteins. J. Zhejiang Univ. Sci. B 2005, 6, 1045–1056. [Google Scholar] [CrossRef] [Green Version]
- Ritter, S.Y.; Subbaiah, R.; Bebek, G.; Crish, J.; Scanzello, C.R.; Krastins, B.; Sarracino, D.; Lopez, M.F.; Crow, M.K.; Aigner, T.; et al. Proteomic analysis of synovial fluid from the osteoarthritic knee: Comparison with transcriptome analyses of joint tissues. Arthritis Rheum. 2013, 65, 981–992. [Google Scholar] [CrossRef] [Green Version]
- Sipe, J.D. Acute-phase proteins in osteoarthritis. Semin. Arthritis Rheum. 1995, 25, 75–86. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, K.; Wang, A.; Liu, S.; Wang, F.; Li, J. Correlation between sialic acid levels in the synovial fluid and the radiographic severity of knee osteoarthritis. Exp. Ther. Med. 2014, 8, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Pearle, A.D.; Scanzello, C.R.; George, S.; Mandl, L.A.; DiCarlo, E.F.; Peterson, M.; Sculco, T.P.; Crow, M.K. Elevated high-sensitivity c-reactive protein levels are associated with local inflammatory findings in patients with osteoarthritis. Osteoarthr. Cartil. 2007, 15, 516–523. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.D.; Triantafillou, S.; Parker, A.; Youssef, P.P.; Coleman, M. Synovial membrane inflammation and cytokine production in patients with early osteoarthritis. J. Rheumatol. 1997, 24, 365–371. [Google Scholar]
- Scanzello, C.R.; McKeon, B.; Swaim, B.H.; DiCarlo, E.; Asomugha, E.U.; Kanda, V.; Nair, A.; Lee, D.M.; Richmond, J.C.; Katz, J.N.; et al. Synovial inflammation in patients undergoing arthroscopic meniscectomy: Molecular characterization and relationship to symptoms. Arthritis Rheum. 2011, 63, 391–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, D.J. Serpins in cartilage and osteoarthritis: What do we know? Biochem. Soc. Trans. 2021, 49, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, B.; Horwitz, M.S.; Jenne, D.E.; Gauthier, F. Neutrophil elastase, proteinase 3, and cathepsin g as therapeutic targets in human diseases. Pharmacol. Rev. 2010, 62, 726–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haraden, C.A.; Huebner, J.L.; Hsueh, M.-F.; Li, Y.-J.; Kraus, V.B. Synovial fluid biomarkers associated with osteoarthritis severity reflect macrophage and neutrophil related inflammation. Arthritis Res. Ther. 2019, 21, 146. [Google Scholar] [CrossRef] [Green Version]
- Gorelik, A.; Sapir, T.; Woodruff, T.M.; Reiner, O. Serping1/C1 inhibitor affects cortical development in a cell autonomous and non-cell autonomous manner. Front. Cell Neurosci. 2017, 11, 169. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Rozelle, A.L.; Lepus, C.M.; Scanzello, C.R.; Song, J.J.; Larsen, D.M.; Crish, J.F.; Bebek, G.; Ritter, S.Y.; Lindstrom, T.M.; et al. Identification of a central role for complement in osteoarthritis. Nat. Med. 2011, 17, 1674–1679. [Google Scholar] [CrossRef] [Green Version]
- So, A.K.; Varisco, P.-A.; Kemkes-Matthes, B.; Herkenne-Morard, C.; Chobaz-Péclat, V.; Gerster, J.-C.; Busso, N. Arthritis is linked to local and systemic activation of coagulation and fibrinolysis pathways. J. Thromb. Haemost. 2003, 1, 2510–2515. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Zhang, Z.; Chang, Z.; Mao, G.; Hu, S.; Zeng, A.; Fu, M. MiR-193b-5p regulates chondrocytes metabolism by directly targeting histone deacetylase 7 in interleukin-1β-induced osteoarthritis. J. Cell Biochem. 2019, 120, 12775–12784. [Google Scholar] [CrossRef]
- Philipot, D.; Guérit, D.; Platano, D.; Chuchana, P.; Olivotto, E.; Espinoza, F.; Dorandeu, A.; Pers, Y.-M.; Piette, J.; Borzi, R.M.; et al. P16INK4a and its regulator MiR-24 link senescence and chondrocyte terminal differentiation-associated matrix remodeling in osteoarthritis. Arthritis Res. Ther. 2014, 16, R58. [Google Scholar] [CrossRef] [Green Version]
- Ramos, Y.F.; Coutinho De Almeida, R.; Mahfouz, A.; Den Hollander, W.; Lakenberg, N.; Houtman, E.; Van Hoolwerff, M.; Suchiman, E.; Rodríguez-Ruiz, A.; Slagboom, P.; et al. Circulating micro RNAs reflecting ongoing osteoarthritis pathophysiology in cartilage as applicable biomarkers. Osteoarthr. Cartil. 2019, 27, S69. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Guo, K.; Su, S.; Li, J.; Li, C. MiR-486-5p is upregulated in osteoarthritis and inhibits chondrocyte proliferation and migration by suppressing SMAD2. Mol. Med. Rep. 2018, 18, 502–508. [Google Scholar] [CrossRef] [Green Version]
- Jingjing, Z.; Nan, Z.; Wei, W.; Qinghe, G.; Weijuan, W.; Peng, W.; Xiangpeng, W. MicroRNA-24 modulates staphylococcus aureus-induced macrophage polarization by suppressing CHI3L1. Inflammation 2017, 40, 995–1005. [Google Scholar] [CrossRef]
- Bi, J.; Zeng, X.; Zhao, L.; Wei, Q.; Yu, L.; Wang, X.; Yu, Z.; Cao, Y.; Shan, F.; Wei, M. MiR-181a induces macrophage polarized to M2 phenotype and promotes M2 macrophage-mediated tumor cell metastasis by targeting KLF6 and C/EBPα. Mol. Ther. Nucleic Acids 2016, 5, e368. [Google Scholar] [CrossRef] [Green Version]
- Bosetti, M.; Borrone, A.; Follenzi, A.; Messaggio, F.; Tremolada, C.; Cannas, M. Human lipoaspirate as autologous injectable active scaffold for one-step repair of cartilage defects. Cell Transpl. 2016, 25, 1043–1056. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Yu, X.; Yang, Q.; Liu, X.; Fang, J.; Dai, X. Autologous micro-fragmented adipose tissue as stem cell-based natural scaffold for cartilage defect repair. Cell Transpl. 2019, 28, 1709–1720. [Google Scholar] [CrossRef]
- Paolella, F.; Manferdini, C.; Gabusi, E.; Gambari, L.; Filardo, G.; Kon, E.; Mariani, E.; Lisignoli, G. Effect of microfragmented adipose tissue on osteoarthritic synovial macrophage factors. J. Cell Physiol. 2019, 234, 5044–5055. [Google Scholar] [CrossRef]
- Guo, B.; Sawkulycz, X.; Heidari, N.; Rogers, R.; Liu, D.; Slevin, M. Characterisation of novel angiogenic and potent anti-inflammatory effects of micro-fragmented adipose tissue. Int. J. Mol. Sci. 2021, 22, 3271. [Google Scholar] [CrossRef]



| miRNA | µFAT vs. LA | Role |
|---|---|---|
| miR-193b-5p | 1.90 | Inhibit degradation of ECM components and moderate inflammation |
| miR-24-3p | 1.93 | Inhibit senescence, cartilage catabolism and chondrocyte apoptosis |
| miR-320a-3p | 0.43 | Promote chondrocyte viability and chondrogenesis |
| miR-92a-3p | 0.61 | Inhibit cartilage catabolism and promote collagen deposition |
| miR-222-3p | 0.73 | Inhibit cartilage degradation |
| miR-17-5p | 1.03 | Induce autophagy |
| miR-30a-5p | 1.28 | Promote cartilage homeostasis |
| miR-152-3p | 1.03 | Decrease synovial fibroblast proliferation |
| miR-199a-3p | 1.74 | Anti-catabolic |
| miR-130a-3p | 0.89 | Anti-inflammatory |
| miR-210-3p | 0.73 | Inhibit apoptosis and promote chondrocyte proliferation and ECM deposition |
| miR-26a-5p | 3.32 | Promote cartilage homeostasis |
| miR-29a-3p | 3.63 | Inhibit excessive cartilage remodeling |
| miR-27a-3p | 1.59 | Prevent synovial fibroblast migration and invasion |
| miR-148a-3p | 1.56 | Promote hyaline cartilage production |
| miR-26b-5p | 2.55 | Promote cartilage homeostasis |
| miR-27b-3p | 1.72 | Anti-catabolic |
| miRNA | µFAT vs. LA | Role |
|---|---|---|
| miR-24-3p | 1.93 | Anti M1—Pro M2 |
| miR-30d-5p | 1.03 | Anti M1—Pro M2 |
| miR-146a-5p | 0.55 | Anti M1—Pro M2 |
| miR-146b-5p | 2.34 | Anti M1—Pro M2 |
| miR-34a-5p | 0.84 | Pro M2 |
| miR-222-3p | 0.73 | Pro M2 |
| let-7b-5p | 1.59 | Pro M2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragni, E.; Viganò, M.; Torretta, E.; Perucca Orfei, C.; Colombini, A.; Tremolada, C.; Gelfi, C.; de Girolamo, L. Characterization of Microfragmented Adipose Tissue Architecture, Mesenchymal Stromal Cell Content and Release of Paracrine Mediators. J. Clin. Med. 2022, 11, 2231. https://doi.org/10.3390/jcm11082231
Ragni E, Viganò M, Torretta E, Perucca Orfei C, Colombini A, Tremolada C, Gelfi C, de Girolamo L. Characterization of Microfragmented Adipose Tissue Architecture, Mesenchymal Stromal Cell Content and Release of Paracrine Mediators. Journal of Clinical Medicine. 2022; 11(8):2231. https://doi.org/10.3390/jcm11082231
Chicago/Turabian StyleRagni, Enrico, Marco Viganò, Enrica Torretta, Carlotta Perucca Orfei, Alessandra Colombini, Carlo Tremolada, Cecilia Gelfi, and Laura de Girolamo. 2022. "Characterization of Microfragmented Adipose Tissue Architecture, Mesenchymal Stromal Cell Content and Release of Paracrine Mediators" Journal of Clinical Medicine 11, no. 8: 2231. https://doi.org/10.3390/jcm11082231










