Enterococcus Virulence and Resistant Traits Associated with Its Permanence in the Hospital Environment
Abstract
:1. Introduction
2. Genetic Organization
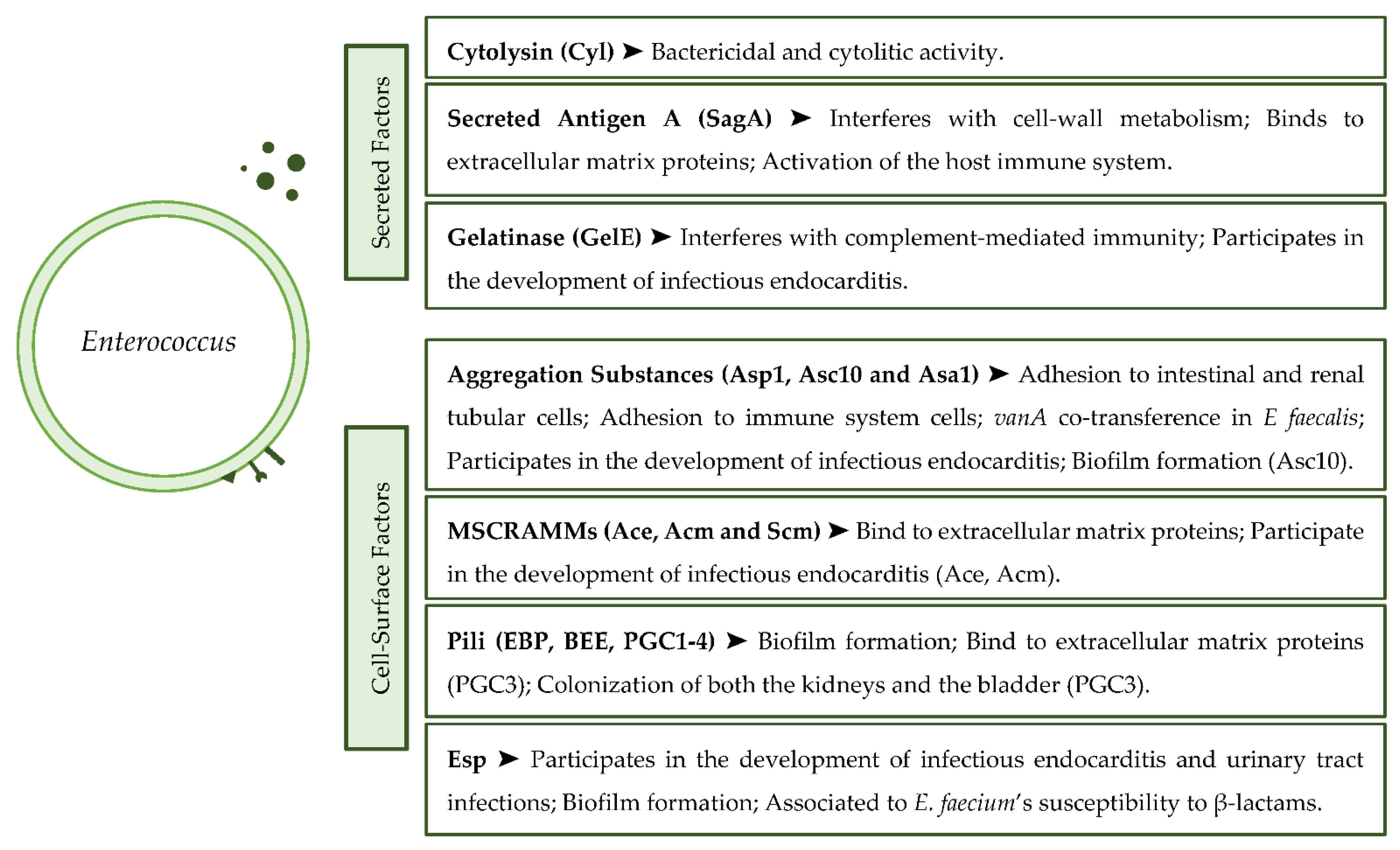
3. Virulence
3.1. Secreted Virulence Factors
3.2. Cell Surface Virulence Factors
4. Antibiotic Resistance
4.1. β-Lactam Resistance
4.2. Aminoglycosides
4.3. Glycopeptides
4.4. Fluoroquinolones
4.5. Tetracyclines
4.6. Oxazolidinones
5. Biocide Tolerance
6. COVID-19 and Enterococcus
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and acquired resistance mechanisms in Enterococcus. Virulence 2012, 3, 421–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebreton, F.; Willems, R.J.L.; Gilmore, M.S. Enterococcus diversity, origins in nature, and gut colonization. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Yasuyoshi, I., Shankar, N., Eds.; Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 5–64. [Google Scholar]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; del Campo, R.; Coque, T.M. Antimicrobial resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018, 6, 185–227. [Google Scholar] [CrossRef] [PubMed]
- Brinkwirth, S.; Ayobami, O.; Eckmanns, T.; Markwart, R. Hospital-acquired infections caused by enterococci: A systematic review and meta-analysis, WHO European Region, 1 January 2010 to 4 February 2020. Euro Surveill. 2021, 26, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Britt, N.S.; Potter, E.M. Clinical epidemiology of vancomycin-resistant Enterococcus gallinarum and Enterococcus casseliflavus bloodstream infections. J. Glob. Antimicrob. Resist. 2017, 5, 57–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toc, D.A.; Pandrea, S.L.; Botan, A.; Mihaila, R.M.; Costache, C.A.; Colosi, I.A.; Junie, L.M. Enterococcus raffinosus, Enterococcus durans and Enterococcus avium Isolated from a Tertiary Care Hospital in Romania—Retrospective Study and Brief Review. Biology 2022, 11, 598. [Google Scholar] [CrossRef] [PubMed]
- Monticelli, J.; Knwzevich, A.; Luzzati, R.; Di Bella, S. Clinical management of non-faecium non-faecalis vancomycin-resistant enterococci infection. Focus on Enterococcus gallinarium and Enterococcus casseliflavus/flavescen. J. Infect. Chemother. 2018, 24, 237–246. [Google Scholar] [CrossRef] [Green Version]
- García-Solache, M.; Rice, L.B. The Enterococcus: A model of adaptability to its environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef] [Green Version]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococcal infection—Treatment and antibiotic resistance. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Yasuyoshi, I., Shankar, N., Eds.; Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 123–184. [Google Scholar]
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 24 January 2022).
- Heller, L.C.; Edelblute, C.M. Long-term metabolic persistence of gram-positive bacteria on health care-relevant plastic. Am. J. Infect. Control 2018, 46, 50–53. [Google Scholar] [CrossRef]
- Grund, A.; Rautenschlein, S.; Jung, A. Tenacity of Enterococcus cecorum at different environmental conditions. J. Appl. Microbiol. 2020, 130, 1494–1507. [Google Scholar] [CrossRef]
- Prieto, A.M.G.; Wijngaarden, J.; Braat, J.C.; Rogers, M.R.C.; Majoor, E.; Brouwer, E.C.; Zhang, X.; Bayjanov, J.R.; Bonten, M.J.M.; Willems, R.J.L.; et al. The two-component system ChtRS contributes to chlorhexidine tolerance in Enterococcus faecium. Antimicrob. Agents Chemother. 2017, 61, e02122-16. [Google Scholar] [CrossRef] [Green Version]
- Pidot, S.; Gao, W.; Buultjens, A.; Monk, I.; Guerillot, R.; Carter, G.; Lee, J.; Lam, M.; Grayson, M.L.; Ballard, S.; et al. Increasing tolerance of hospital Enterococcus faecium to hand-wash alcohols. Sci. Transl. Med. 2018, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobhanipoor, M.H.; Ahmadrajabi, R.; Nave, H.H.; Saffari, F. Reduced susceptibility to biocides among enterococci from clinical and non-clinical sources. Infect. Chemother. 2021, 53, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Geraldes, C.; Verdial, C.; Cunha, E.; Almeida, V.; Tavares, L.; Oliveira, M.; Gil, S. Evaluation of a biocide used in the biological isolation and containment unit of a veterinary teaching hospital. Antibiotics 2021, 10, 639. [Google Scholar] [CrossRef]
- Kalchayanand, N.; Koohmaraie, M.; Wheeler, T.L. Effect of exposure time and organic matter on efficacy of antimicrobial compounds against shiga toxin–producing Escherichia coli and Salmonella. J. Food Prot. 2016, 79, 561–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigel, L.M.; Clewell, D.B.; Gill, S.R.; Clark, N.C.; Mcdougal, L.K.; Flannagan, S.E.; Kolonay, J.F.; Shetty, J.; Killgore, G.E.; Tenover, F.C. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 2003, 302, 1569–1571. [Google Scholar] [CrossRef] [PubMed]
- de Niederhäusern, S.; Sabia, C.; Messi, P.; Guerrieri, E.; Manicardi, G.; Bondi, M. Glycopeptide-resistance transferability from vancomycin-resistant enterococci of human and animal source to Listeria spp. Lett. Appl. Microbiol. 2004, 39, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Clark, N.C.; McDougal, L.K.; Hageman, J.; McDonald, L.C.; Patel, J.B. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob. Agents Chemother. 2008, 52, 452–457. [Google Scholar] [CrossRef] [Green Version]
- Limbago, B.M.; Kallen, A.J.; Zhu, W.; Eggers, P.; McDougal, L.K.; Albrecht, V.S. Report of the 13th vancomycin-resistant Staphylococcus aureus isolate from the United States. J. Clin. Microbiol. 2014, 52, 998–1002. [Google Scholar] [CrossRef] [Green Version]
- van Schaik, W.; Top, J.; Riley, D.R.; Boekhorst, J.; Vrijenhoek, J.E.P.; Schapendonk, C.M.E.; Hendrickx, A.P.A.; Nijman, I.J.; Bonten, M.J.M.; Tettelin, H.; et al. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genom. 2010, 11, 239. [Google Scholar] [CrossRef] [Green Version]
- Paulsen, I.; Banerjei, L.; Myers, G.; Nelson, K.; Seshadri, R.; Read, T.; Fouts, D.; Eisen, J.; Gill, S.; Heidelberg, J.; et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 2003, 299, 2071–2074. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Garbajosa, P.; Bonten, M.J.M.; Robinson, D.A.; Top, J.; Nallapareddy, S.R.; Torres, C.; Coque, T.M.; Canto, R.; Baquero, F.; Murray, B.E.; et al. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 2006, 44, 2220–2228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leavis, H.L.; Willems, R.J.L.; van Wamel, W.J.B.; Schuren, F.H.; Caspers, M.P.M.; Bonten, M.J.M. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog. 2007, 3, 75–96. [Google Scholar] [CrossRef] [PubMed]
- Galloway-Peña, J.; Roh, J.H.; Latorre, M.; Qin, X.; Murray, B.E. Genomic and SNP analyses demonstrate a distant separation of the hospital and community-associated clades of Enterococcus faecium. PLoS ONE 2012, 7, e30187. [Google Scholar] [CrossRef] [PubMed]
- Palmer, K.L.; Godfrey, P.; Griggs, A.; Kos, V.N.; Zucker, J.; Desjardins, C.; Cerqueira, G.; Gevers, D.; Walker, S.; Wortman, J.; et al. Comparative genomics of enterococci: Variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. MBio 2012, 3, e00318-11. [Google Scholar] [CrossRef] [Green Version]
- Lebreton, F.; van Schaik, W.; McGuire, A.M.; Godfrey, P.; Griggs, A.; Mazumdar, V.; Corander, J.; Cheng, L.; Saif, S.; Young, S.; et al. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio 2013, 4, e00534-13. [Google Scholar] [CrossRef] [Green Version]
- Raven, K.E.; Reuter, S.; Reynolds, R.; Brodrick, H.J.; Russell, J.E.; Török, M.E.; Parkhill, J.; Peacock, S.J. A decade of genomic history for healthcare-associated Enterococcus faecium in the United Kingdom and Ireland. Genome Res. 2016, 26, 1388–1396. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.; Kwok, L.Y.; Hou, Q.; Sun, Y.; Li, W.; Zhang, H.; Sun, Z. Comparative genomic analysis revealed great plasticity and environmental adaptation of the genomes of Enterococcus faecium. BMC Genom. 2019, 20, 602. [Google Scholar] [CrossRef]
- Solheim, M.; Aakra, Å.; Snipen, L.G.; Brede, D.A.; Nes, I.F. Comparative genomics of Enterococcus faecalis from healthy Norwegian infants. BMC Genom. 2009, 10, 194. [Google Scholar] [CrossRef] [Green Version]
- Gao, W.; Howden, B.P.; Stinear, T.P. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr. Opin. Microbiol. 2018, 41, 76–82. [Google Scholar] [CrossRef]
- Kawalec, M.; Pietras, Z.; Daniłowicz, E.; Jakubczak, A.; Gniadkowski, M.; Hryniewicz, W.; Willems, R.J.L. Clonal structure of Enterococcus faecalis isolated from Polish hospitals: Characterization of epidemic clones. J. Clin. Microbiol. 2007, 45, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Neumann, B.; Prior, K.; Bender, J.K.; Harmsen, D.; Klare, I.; Fuchs, S.; Bethe, A.; Zühlke, D.; Göhler, A.; Schwarz, S.; et al. A core genome multilocus sequence typing scheme for Enterococcus faecalis. J. Clin. Microbiol. 2019, 57, e01686-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leavis, H.L.; Bonten, M.J.; Willems, R.J. Identification of high-risk enterococcal clonal complexes: Global dispersion and antibiotic resistance. Curr. Opin. Microbiol. 2006, 9, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Segarra, R.A.; Booth, M.C.; Morales, D.A.; Huycke, M.M.; Gilmore, M.S. Molecular characterization of the Enterococcus faecalis hemolysin/bacteriocin determinant. Infect. Immun. 1991, 59, 1239–1246. [Google Scholar] [CrossRef] [Green Version]
- Brock, T.D.; Davie, J.M. Probable identity of a group D hemolysin with a bacteriocine. J. Bacteriol. 1963, 86, 708–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ike, Y.; Hashimoto, H.; Clewell, D.B. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect. Immun. 1984, 45, 528–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilmore, M.S.; Segarra, R.A.; Booth, M.C. An HlyB-type function is required for expression of the Enterococcus faecalis hemolysin/bacteriocin. Infect. Immun. 1990, 58, 3914–3923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyazaki, S.; Ohno, A.; Kobayashi, I.; Uji, T.; Yamaguchi, K.; Goto, S. Cytotoxic effect of hemolytic culture supernatant from Enterococcus faecalis on mouse polymorphonu-neutrophils and macrophages. Microbiol. Immunol. 1993, 37, 265–270. [Google Scholar] [CrossRef]
- Jett, B.D.; Jensen, H.G.; Nordquist, R.E.; Gilmore, M.S. Contribution of the pAD1-encoded cytolysin to the severity of experimental Enterococcus faecalis endophthalmitis. Infect. Immun. 1992, 60, 2445–2452. [Google Scholar] [CrossRef] [Green Version]
- Chow, J.W.; Thal, L.A.; Perri, M.B.; Vazquez, J.A.; Donabedian, S.M.; Clewell, D.B.; Zervos, M.J. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob. Agents Chemother. 1993, 37, 2474–2477. [Google Scholar] [CrossRef] [Green Version]
- Todokoro, D.; Suzuki, T.; Kobayakawa, S.; Tomita, H.; Ohashi, Y.; Akiyama, H. Postoperative Enterococcus faecalis endophthalmitis: Virulence factors leading to poor visual outcome. Jpn. J. Ophthalmol. 2017, 61, 408–414. [Google Scholar] [CrossRef]
- Chilambi, G.S.; Nordstrom, H.R.; Evans, D.R.; Kowalski, R.P.; Dhaliwal, D.K.; Jhanji, V.; Shanks, R.M.Q.; van Tyne, D. Genomic and phenotypic diversity of Enterococcus faecalis isolated from endophthalmitis. PLoS ONE 2021, 16, e0250084. [Google Scholar] [CrossRef]
- Muller, C.; Breton, Y.; Morin, T.; Benachour, A.; Auffray, Y.; Rincé, A.; Auffray, Y.; Rincé, A. The response regulator CroR modulates expression of the secreted stress induced SalB protein in Enterococcus faecalis. J. Bacteriol. 2006, 188, 2636–2645. [Google Scholar] [CrossRef] [Green Version]
- Teng, F.; Kawalec, M.; Weinstock, G.M.; Hryniewicz, W.; Murray, B.E. An Enterococcus faecium secreted antigen, SagA, exhibits broad-spectrum binding to extracellular matrix proteins and appears essential for E. faecium growth. Infect. Immun. 2003, 71, 5033–5041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedicord, V.A.; Lockhart, A.A.K.; Rangan, K.J.; Craig, J.W.; Loschko, J.; Rogoz, A.; Hang, H.; Mucida, D. Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance. Sci. Immunol. 2016, 1, eaai7732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangan, K.J.; Pedicord, V.A.; Wang, Y.-C.; Kim, B.; Lu, Y.; Shaham, S.; Mucida, D.; Hang, H. A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science 2016, 353, 1434–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.; Wang, Y.C.; Hespen, C.W.; Espinosa, J.; Salje, J.; Rangan, K.J.; Oren, D.A.; Kang, J.Y.; Pedicord, V.A.; Hang, H.C. Enterococcus faecium secreted antigen A generates muropeptides to enhance host immunity and limit bacterial pathogenesis. Elife 2019, 8, e45343. [Google Scholar] [CrossRef]
- Paganelli, F.L.; de Been, M.; Braat, J.C.; Hoogenboezem, T.; Vink, C.; Bayjanov, J.; Rogers, M.R.C.; Huebner, J. Distinct SagA from hospital-associated clade A1 Enterococcus faecium strains contributes to biofilm formation. Appl. Environ. Microbiol. 2015, 81, 6873–6882. [Google Scholar] [CrossRef] [Green Version]
- Park, S.Y.; Kim, K.M.; Lee, J.H.; Seo, S.J.; Lee, I.H. Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum. Infect. Immun. 2007, 75, 1861–1869. [Google Scholar] [CrossRef] [Green Version]
- Thurlow, L.R.; Thomas, V.C.; Narayanan, S.; Olson, S.; Fleming, S.D.; Hancock, L.E. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect. Immun. 2010, 78, 4936–4943. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.; Singh, K.; Weinstock, G.M.; Murray, B.E. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 2000, 68, 2579–2586. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.; Singh, K.; Weinstock, G.M.; Murray, B.E. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 2001, 183, 3372–3382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourgogne, A.; Hilsenbeck, S.G.; Dunny, G.M.; Murray, B.E. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: The Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J. Bacteriol. 2006, 188, 2875–2884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinkston, K.L.; Gao, P.; Diaz-Garcia, D.; Sillanpää, J.; Nallapareddy, S.R.; Murray, B.E.; Harvey, B.R. The Fsr quorum-sensing system of Enterococcus faecalis modulates surface display of the collagen-binding MSCRAMM Ace through regulation of gelE. J. Bacteriol. 2011, 193, 4317–4325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, L.B.; Carias, L.; Rudin, S.; Vael, C.; Goossens, H.; Konstabel, C.; Klare, I.; Nallapareddy, S.R.; Huang, W.; Murray, B.E. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J. Infect. Dis. 2003, 187, 508–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Top, J.; Willems, R.; Bonten, M. Emergence of CC17 Enterococcus faecium: From commensal to hospital-adapted pathogen. FEMS Immunol. Med. Microbiol. 2008, 52, 297–308. [Google Scholar] [CrossRef] [Green Version]
- Arias, C.A.; Panesso, D.; Singh, K.; Rice, L.B.; Murray, B.E. Co-transfer of antibiotic resistance genes and a hylEfm-containing virulence plasmid in Enterococcus faecium. Antimicrob. Agents Chemother. 2009, 53, 4240–4246. [Google Scholar] [CrossRef] [Green Version]
- Fabretti, F.; Theilacker, C.; Baldassarri, L.; Kaczynski, Z.; Kropec, A.; Holst, O.; Huebner, J.; Infettive, M.; Istituto, I. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 2006, 74, 4164–4171. [Google Scholar] [CrossRef] [Green Version]
- Geiss-Liebisch, S.; Rooijakkers, S.H.M.; Beczala, A.; Sanchez-Carballo, P.; Kruszynska, K.; Repp, C.; Sakinc, T.; Vinogradov, E.; Holst, O.; Huebner, J.; et al. Secondary cell wall polymers of Enterococcus faecalis are critical for resistance to complement activation via mannose-binding lectin. J. Biol. Chem. 2012, 287, 37769–37777. [Google Scholar] [CrossRef] [Green Version]
- Hendrickx, A.P.A.; Willems, R.J.L.; Bonten, M.J.M.; van Schaik, W. LPxTG surface proteins of enterococci. Trends Microbiol. 2009, 17, 423–430. [Google Scholar] [CrossRef]
- Isenmann, R.; Schwarz, M.; Rozdzinski, E.; Marre, R.; Beger, H.G. Aggregation substance promotes colonic mucosal invasion of Enterococcus faecalis in an ex vivo model. J. Surg. Res. 2000, 89, 132–138. [Google Scholar] [CrossRef]
- Sartingen, S.; Rozdzinski, E.; Muscholl-Silberhorn, A.; Marre, R. Aggregation substance increases adherence and internalization, but not translocation, of Enterococcus faecalis through different intestinal epithelial cells in vitro. Infect. Immun. 2000, 68, 6044–6047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, C.L.; Moore, E.A.; Hoag, J.A.; Hirt, H.; Dunny, G.M.; Erlandsen, S.L. Inducible expression of Enterococcus faecalis aggregation substance surface protein facilitates bacterial internalization by cultured enterocytes. Infect. Immun. 2000, 68, 7190–7194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreft, B.; Marre, R.; Schramm, U.; Wirth, R. Aggregation substance of Enterococcus faecalis mediates adhesion to cultured renal tubular cells. Infect. Immun. 1992, 60, 25–30. [Google Scholar] [CrossRef] [Green Version]
- Rozdzinski, E.; Marre, R.; Susa, M.; Wirth, R.; Muscholl-Silberhorn, A. Aggregation substance-mediated adherence of Enterococcus faecalis to immobilized extracellular matrix proteins. Microb. Pathog. 2001, 30, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Vanek, N.N.; Simon, S.I.; Jacques-Palaz, K.; Mariscalco, M.M.; Dunny, G.M.; Rakita, R.M. Enterococcus faecalis aggregation substance promotes opsonin-independent binding to human neutrophils via a complement receptor type 3-mediated mechanism. FEMS Immunol. Med. Microbiol. 1999, 26, 49–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Süßmuth, S.D.; Muscholl-Silberhorn, A.; Wirth, R.; Susa, M.; Marre, R.; Rozdzinski, E. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect. Immun. 2000, 68, 4900–4906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakita, R.M.; Vanek, N.N.; Jacques-Palaz, K.; Mee, M.; Mariscalco, M.M.; Dunny, G.M.; Snuggs, M.; van Winkle, W.B.; Simon, S.I. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect. Immun. 1999, 67, 6067–6075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paoletti, C.; Foglia, G.; Princivalli, M.S.; Magi, G.; Guaglianone, E.; Donelli, G.; Pruzzo, C.; Biavasco, F.; Facinelli, B. Co-transfer of vanA and aggregation substance genes from Enterococcus faecalis isolates in intra- and interspecies matings. J. Antimicrob. Chemother. 2007, 59, 1005–1009. [Google Scholar] [CrossRef]
- Schlievert, P.M.; Gahr, P.J.; Assimacopoulos, A.P.; Dinges, M.M.; Stoehr, J.A.; Harmala, J.W.; Hirt, H.; Dunny, G.M. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect. Immun. 1998, 66, 218–223. [Google Scholar] [CrossRef] [Green Version]
- Bhatty, M.; Cruz, M.R.; Frank, K.L.; Laverde-Gomez, J.A.; Andrade, F.; Garsin, D.A.; Dunny, G.M.; Kaplan, H.B.; Christie, P.J. Enterococcus faecalis pCF10-encoded surface proteins PrgA, PrgB (Aggregation Substance), and PrgC contribute to plasmid transfer, biofilm formation, and virulence. Mol. Microbiol. 2015, 95, 660–677. [Google Scholar] [CrossRef] [Green Version]
- Rich, R.L.; Kreikemeyer, B.; Owens, R.T.; LaBrenz, S.; Narayana, S.V.L.; Weinstock, G.M.; Murray, B.E.; Höök, M. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J. Biol. Chem. 1999, 274, 26939–26945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nallapareddy, S.R.; Weinstock, G.M.; Murray, B.E. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol. Microbiol. 2003, 47, 1733–1747. [Google Scholar] [CrossRef] [PubMed]
- Sillanpää, J.; Nallapareddy, S.R.; Prakash, V.P.; Qin, X.; Hook, M.; Weinstock, G.M.; Murray, B.E. Identification and phenotypic characterization of a second collagen adhesin, Scm, and genome-based identification and analysis of 13 other predicted MSCRAMMs, including four distinct pilus loci, in Enterococcus faecium. Microbiology 2008, 154, 3199–3211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nallapareddy, S.R.; Qin, X.; Weinstock, G.M.; Höök, M.; Murray, B.E. Enterococcus faecalis adhesin, Ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect. Immun. 2000, 68, 5218–5224. [Google Scholar] [CrossRef] [Green Version]
- Kowalski, W.J.; Kasper, E.L.; Hatton, J.F.; Murray, B.E.; Nallapareddy, S.R.; Gillespie, M.J. Enterococcus faecalis adhesin, Ace, mediates attachment to particulate dentin. J. Endod. 2006, 32, 634–637. [Google Scholar] [CrossRef]
- Nallapareddy, S.R.; Singh, K.; Murray, B.E. Contribution of the collagen adhesin Acm to pathogenesis of Enterococcus faecium in experimental endocarditis. Infect. Immun. 2008, 76, 4120–4128. [Google Scholar] [CrossRef] [Green Version]
- Singh, K.; Nallapareddy, S.R.; Sillanpää, J.; Murray, B.E. Importance of the collagen adhesin Ace in pathogenesis and protection against Enterococcus faecalis experimental endocarditis. PLoS Pathog. 2010, 6, e1000716. [Google Scholar] [CrossRef]
- Singh, K.; Pinkston, K.L.; Gao, P.; Harvey, B.R.; Murray, B.E. Anti-Ace monoclonal antibody reduces Enterococcus faecalis aortic valve infection in a rat infective endocarditis model. Pathog. Dis. 2018, 76, fty084. [Google Scholar] [CrossRef]
- Nallapareddy, S.R.; Singh, K.; Okhuysen, P.C.; Murray, B.E. A functional collagen adhesin gene, acm, in clinical isolates of Enterococcus faecium correlates with the recent success of this emerging nosocomial pathogen. Infect. Immun. 2008, 76, 4110–4119. [Google Scholar] [CrossRef] [Green Version]
- Tendolkar, P.M.; Baghdayan, A.S.; Shankar, N. Putative surface proteins encoded within a novel transferable locus confer a high-biofilm phenotype to Enterococcus faecalis. J. Bacteriol. 2006, 188, 2063–2072. [Google Scholar] [CrossRef] [Green Version]
- Molinos, A.C.; Abriouel, H.; Omar, N.; López, R.L.; Galvez, A. Detection of ebp (endocarditis- and biofilm-associated pilus) genes in enterococcal isolates from clinical and non-clinical origin. Int. J. Food Microbiol. 2008, 126, 123–126. [Google Scholar]
- Nallapareddy, S.R.; Singh, K.; Sillanpää, J.; Garsin, D.A.; Höök, M.; Erlandsen, S.L.; Murray, B.E. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Investig. 2006, 116, 2799–2807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, X.; Galloway-Peña, J.R.; Sillanpää, J.; Roh, J.H.; Nallapareddy, S.R.; Chowdhury, S.; Bourgogne, A.; Choudhury, T.; Muzny, D.M.; Buhay, C.J.; et al. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 2012, 12, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sillanpää, J.; Nallapareddy, S.R.; Singh, K.; Parakash, V.P.; Fothergill, T.; Ton-That, H.; Murray, B.E. Characterization of the ebpfm pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence 2010, 1, 236–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montealegre, M.C.; Singh, K.; Somarajan, S.R.; Yadav, P.; Chang, C.; Spencer, R.; Sillanpää, J.; Ton-That, H.; Murray, B.E. Role of the Emp pilus subunits of Enterococcus faecium in biofilm formation, adherence to host extracellular matrix components, and experimental infection. Infect. Immun. 2016, 84, 1491–1500. [Google Scholar] [CrossRef] [Green Version]
- Shankar, V.; Baghdayan, A.S.; Huycke, M.M.; Lindahl, G.; Gilmore, M.S. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 1999, 67, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Eaton, T.J.; Gasson, M.J. A variant enterococcal surface protein Espfm in Enterococcus faecium; distribution among food, commensal, medical, and environmental isolates. FEMS Microbiol. Lett. 2002, 216, 269–275. [Google Scholar] [CrossRef] [Green Version]
- Toledo-Arana, A.; Valle, J.; Solano, C.; Arrizubieta, M.J.; Cucarella, C.; Lamata, M.; Amorena, B.; Leiva, J.; Penadés, J.R.; Lasa, I. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 2001, 67, 4538–4545. [Google Scholar] [CrossRef] [Green Version]
- Tendolkar, P.M.; Baghdayan, A.S.; Gilmore, M.S.; Shankar, N. Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect. Immun. 2004, 72, 6032–6039. [Google Scholar] [CrossRef] [Green Version]
- Heikens, E.; Bonten, M.J.M.; Willems, R.J.L. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J. Bacteriol. 2007, 189, 8233–8240. [Google Scholar] [CrossRef] [Green Version]
- Taglialegna, A.; Matilla-Cuenca, L.; Dorado-Morales, P.; Navarro, S.; Ventura, S.; Garnett, J.A.; Lasa, I.; Valle, J. The biofilm-associated surface protein Esp of Enterococcus faecalis forms amyloid-like fibers. Nat. Partn. J. 2020, 6, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heikens, E.; Singh, K.; Jacques-Palaz, K.D.; van Luit-Asbroek, M.; Oostdijk, E.A.N.; Bonten, M.J.M.; Murray, B.E.; Willems, R.J.L. Contribution of the enterococcal surface protein Esp to pathogenesis of Enterococcus faecium endocarditis. Microbes Infect. 2011, 13, 1185–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shankar, N.; Lockatell, C.V.; Baghdayan, A.S.; Drachenberg, C.; Gilmore, M.S.; Johnson, D.E. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 2001, 69, 4366–4372. [Google Scholar] [CrossRef] [Green Version]
- Meredith, K.; Bolhuis, A.; O’Neill, M.A.A. Enterococcal surface protein Esp affects antibiotic sensitivity in Enterococcus faecium. Int. J. Antimicrob. Agents 2009, 34, 392–393. [Google Scholar] [CrossRef] [PubMed]
- Weng, P.L.; Ramli, R.; Hamat, R.A. Antibiotic susceptibility patterns, biofilm formation and esp gene among clinical enterococci: Is there any association? Int. J. Environ. Res. Public Health 2019, 16, 3439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prieto, A.M.G.; Urbanus, R.T.; Zhang, X.; Bierschenk, D.; Koekman, C.A.; van Luit-Asbroek, M.; Ouwerkerk, J.P.; Pape, M.; Paganelli, F.L.; Wobser, D.; et al. The N-terminal domain of the thermo-regulated surface protein PrpA of Enterococcus faecium binds to fibrinogen, fibronectin and platelets. Nat. Publ. Group 2015, 5, 18255. [Google Scholar]
- Hendrickx, A.P.A.; van Luit-Asbroek, M.; Schapendonk, C.M.E.; van Wamel, W.J.B.; Braat, J.C.; Wijnands, L.M.; Bonten, M.J.M.; Willems, R.J.L. SgrA, a nidogen-binding LPXTG surface adhesin implicated in biofilm formation, and EcbA, a collagen binding MSCRAMM, are two novel adhesins of hospital-acquired Enterococcus faecium. Infect. Immun. 2009, 77, 5097–5106. [Google Scholar] [CrossRef] [Green Version]
- Weese, J.S.; Blondeau, J.; Boothe, D.; Guardabassi, L.G.; Gumley, N.; Papich, M.; Jessen, L.R.; Lappin, M.; Rankin, S.; Westropp, J.L.; et al. International Society for Companion Animal Infectious Diseases (ISCAID) guidelines for the diagnosis and management of bacterial urinary tract infections in dogs and cats. Vet. J. 2019, 247, 8–25. [Google Scholar] [CrossRef]
- Lima, L.M.; da Silva, B.N.M.; Barbosa, G.; Barreiro, E.J. β-Lactam antibiotics: An overview from a medicinal chemistry perspective. Eur. J. Med. Chem. 2020, 208, 112829. [Google Scholar] [CrossRef]
- Arbeloa, A.; Segal, H.; Hugonnet, J.E.; Josseaume, N.; Dubost, L.; Brouard, J.P.; Gutmann, L.; Mengin-Lecreulx, D.; Arthur, M. Role of class A penicillin-binding proteins in PBP5-mediated β-lactam resistance in Enterococcus faecalis. J. Bacteriol. 2004, 186, 1221–1228. [Google Scholar] [CrossRef] [Green Version]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti-Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- Williamson, R.; Gutmann, L.; Horaud, T.; Delbos, F.; Acar, J.F. Use of penicillin-binding proteins for the identification of enterococci. J. Gen. Microbiol. 1986, 132, 1929–1937. [Google Scholar] [CrossRef] [PubMed]
- Sifaoui, F.; Arthur, M.; Rice, L.; Gutmann, L. Role of penicillin-binding protein 5 in expression of ampicillin resistance and peptidoglycan structure in Enterococcus faecium. Antimicrob. Agents Chemother. 2001, 45, 2594–2597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ossiprandi, M.C.; Bottarelli, E.; Cattabiani, F.; Bianchi, E. Susceptibility to vancomycin and other antibiotics of 165 Enterococcus strains isolated from dogs in Italy. Comp. Immunol. Microbiol. Infect. Dis. 2008, 31, 1–9. [Google Scholar] [CrossRef]
- Ghosh, A.; Dowd, S.E.; Zurek, L. Dogs leaving the ICU carry a very large multi-drug resistant enterococcal population with capacity for biofilm formation and horizontal gene transfer. PLoS ONE 2011, 6, e22451. [Google Scholar] [CrossRef] [Green Version]
- Bang, K.; An, J.U.; Kim, W.; Dong, H.J.; Kim, J.; Cho, S. Antibiotic resistance patterns and genetic relatedness of Enterococcus faecalis and Enterococcus faecium isolated from military working dogs in Korea. J. Vet. Sci. 2017, 18, 229–236. [Google Scholar] [CrossRef]
- Rice, L.B.; Carias, L.L.; Rudin, S.; Laktičová, V.; Wood, A.; Hutton-Thomas, R. Enterococcus faecium low-affinity pbp5 is a transferable determinant. Antimicrob. Agents Chemother. 2005, 49, 5007–5012. [Google Scholar] [CrossRef] [Green Version]
- García-Solache, M.; Lebreton, F.; McLaughlin, R.E.; Whiteaker, J.D.; Gilmore, M.S.; Rice, L.B. Homologous recombination within large chromosomal regions facilitates acquisition of β-lactam and vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 2016, 60, 5777–5786. [Google Scholar] [CrossRef] [Green Version]
- Novais, C.; Tedim, A.P.; Lanza, V.F.; Freitas, A.R.; Silveira, E.; Escada, R.; Roberts, A.P.; Al-Haroni, M.; Baquero, F.; Peixe, L.; et al. Co-diversification of Enterococcus faecium core genomes and PBP5: Evidences of PBP5 horizontal transfer. Front. Microbiol. 2016, 7, 1581. [Google Scholar] [CrossRef]
- Montealegre, M.C.; Roh, J.H.; Rae, M.; Davlieva, M.G.; Singh, K.; Shamoo, Y.; Murray, B.E. Differential penicillin-binding protein 5 (PBP5) levels in the Enterococcus faecium clades with different levels of ampicillin resistance. Antimicrob. Agents Chemother. 2017, 61, e02034-16. [Google Scholar] [CrossRef] [Green Version]
- Darehkordi, H.; Saffari, F.; Mollaei, H.R.; Ahmadrajabi, R. Amino acid substitution mutations and mRNA expression levels of the pbp5 gene in clinical Enterococcus faecium isolates conferring high level ampicillin resistance. J. Pathol. Microbiol. Immunol. 2019, 127, 115–122. [Google Scholar]
- Galloway-Peña, J.R.; Rice, L.B.; Murray, B.E. Analysis of PBP5 of early U.S. isolates of Enterococcus faecium: Sequence variation alone does not explain increasing ampicillin resistance over time. Antimicrob. Agents Chemother. 2011, 55, 3272–3277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietta, E.; Montealegre, M.C.; Roh, J.H.; Cocconcelli, P.S.; Murray, B.E. Enterococcus faecium PBP5-S/R, the missing link between PBP5-S and PBP5-R. Antimicrob. Agents Chemother. 2014, 58, 6978–6981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohn, S.C.; Ulvik, A.; Jureen, R.; Willems, R.J.L.; Top, J.; Leavis, H.; Harthug, S.; Langeland, N. Duplex real-time PCR assay for rapid detection of ampicillin-resistant Enterococcus faecium. Antimicrob. Agents Chemother. 2004, 48, 556–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poeta, P.; Costa, D.; Igrejas, G.; Sáenz, Y.; Zarazaga, M.; Rodrigues, J.; Torres, C. Polymorphisms of the pbp5 gene and correlation with ampicillin resistance in Enterococcus faecium isolates of animal origin. J. Med. Microbiol. 2007, 56, 236–240. [Google Scholar] [CrossRef]
- Belhaj, M.; ben Boubaker, I.B.; Slim, A. Penicillin-binding protein 5 sequence alteration and levels of plp5 mRNA expression in clinical isolates of Enterococcus faecium with different levels of ampicillin resistance. Microb. Drug Resist. 2016, 22, 202–210. [Google Scholar] [CrossRef]
- Murray, B.E.; Mederski-Samaroj, B. A new mechanism for in vitro penicillin resistance in Streptococcus faecalis. J. Clin. Investig. 1983, 72, 1168–1171. [Google Scholar] [CrossRef]
- Coudron, P.E.; Markowitz, S.M.; Wong, E.S. Isolation of a beta-lactamase-producing, aminoglycoside-resistant strain of Enterococcus faecium. Antimicrob. Agents Chemother. 1992, 36, 1125–1126. [Google Scholar] [CrossRef] [Green Version]
- Sarti, M.; Campanile, F.; Sabia, C.; Santagati, M.; Gargiulo, R.; Stefani, A. Polyclonal diffusion of beta-lactamase-producing Enterococcus faecium. J. Clin. Microbiol. 2012, 50, 169–172. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.A.; Toth, M.; Stewart, N.K.; Maltz, L.; Vakulenko, S.B. Structural basis for the diversity of the mechanism of nucleotide hydrolysis by the aminoglycoside-2′′-phosphotransferases. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 1129–1137. [Google Scholar] [CrossRef]
- Shaw, K.J.; Rather, P.N.; Hare, R.S.; Miller, G.H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 1993, 57, 138–163. [Google Scholar] [CrossRef] [PubMed]
- Costa, Y.; Galimand, M.; Leclercq, R.; Duval, J.; Courvalin, P. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob. Agents Chemother. 1993, 37, 1896–1903. [Google Scholar] [CrossRef] [Green Version]
- Wright, G.D.; Ladak, P. Overexpression and characterization of the chromosomal aminoglycoside 6′-N-acetyltransferase from Enterococcus faecium. Antimicrob. Agents Chemother. 1997, 41, 956–960. [Google Scholar] [CrossRef] [Green Version]
- del Campo, R.; Galán, J.C.; Tenorio, C.; Ruiz-Garbajosa, P.; Zarazaga, M.; Torres, C.; Baquero, F. New aac(6′)-I genes in Enterococcus hirae and Enterococcus durans: Effect on β-lactam/aminoglycoside synergy. J. Antimicrob. Chemother. 2005, 55, 1053–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trieu-Cuot, P.; Courvalin, P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5″-aminoglycoside phosphotransferase type III. Gene 1983, 23, 331–341. [Google Scholar] [CrossRef]
- Kao, S.J.; You, I.; Clewell, D.B.; Donabedian, S.M.; Zervos, M.J.; Petrin, J.; Shaw, K.J.; Chow, J.W. Detection of the high-level aminoglycoside resistance gene aph(2″)-Ib in Enterococcus faecium. Antimicrob. Agents Chemother. 2000, 44, 2876–2879. [Google Scholar] [CrossRef] [Green Version]
- Chow, J.W.; Zervos, M.J.; Lerner, S.A.; Thal, L.A.; Donabedian, S.M.; Jaworski, D.D.; Tsai, S.; Shaw, K.J.; Clewell, D.B. A novel gentamicin resistance gene in Enterococcus. Antimicrob. Agents Chemother. 1997, 41, 511–514. [Google Scholar] [CrossRef] [Green Version]
- Tsai, S.F.; Zervos, M.J.; Clewell, D.B.; Donabedian, S.M.; Sahm, D.F.; Chow, J.W. A new high-level gentamicin resistance gene, aph(2″)-Id, in Enterococcus spp. Antimicrob. Agents Chemother. 1998, 42, 1229–1232. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.M.; Kobayashi, N.; Ishino, M.; Sumi, A.; Kobayashi, K.-I.; Uehara, N.; Watanabe, N. Detection of a novel aph(2″) allele (aph [2″]-Ie) conferring high-level gentamicin resistance and a spectinomycin resistance gene ant(9)-Ia (aad9) in clinical isolates of enterococci. Microb. Drug Resist. 2005, 11, 239–247. [Google Scholar] [CrossRef]
- Ounissi, H.; Derlot, E.; Carlier, C.; Courvalin, P. Gene homogeneity for aminoglycoside-modifying enzymes in gram-positive cocci. Antimicrob. Agents Chemother. 1990, 34, 2164–2168. [Google Scholar] [CrossRef] [Green Version]
- Carlier, C.; Courvalin, P. Emergence of 4′,4″-aminoglycoside nucleotidyltransferase in enterococci. Antimicrob. Agents Chemother. 1990, 34, 1565–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Courvalin, P.; Carlier, C.; Collatz, E. Plasmid-mediated resistance to aminocyclitol antibiotics in group D streptococci. J. Bacteriol. 1980, 143, 541–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mederski-Samoraj, B.D.; Murray, B.E. High-level resistance to gentamicin in clinical isolates of enterococci. J. Infect. Dis. 1983, 147, 751–757. [Google Scholar] [CrossRef]
- Ferretti, J.J.; Gilmore, K.S.; Courvalin, P. Nucleotide sequence analysis of the gene specifying the phosphotransferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J. Bacteriol. 1986, 167, 631–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López, M.; Rezusta, A.; Seral, C.; Aspiroz, C.; Marne, C.; Aldea, M.J.; Ferrer, I.; Revillo, M.J.; Castillo, F.J.; Torres, C. Detection and characterization of a ST6 clone of vanB2-Enterococcus faecalis from three different hospitals in Spain. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 257–260. [Google Scholar] [CrossRef] [PubMed]
- ben Said, L.; Klibi, N.; Lozano, C.; Dziri, R.; ben Slama, K.; Boudabous, A.; Torres, C. Diversity of enterococcal species and characterization of high-level aminoglycoside resistant enterococci of samples of wastewater and surface water in Tunisia. Sci. Total Environ. 2015, 530–531, 11–17. [Google Scholar] [CrossRef]
- Niu, H.; Yu, H.; Hu, T.; Tian, G.; Zhang, L.; Guo, X.; Hu, H.; Wang, Z. The prevalence of aminoglycoside-modifying enzyme and virulence genes among enterococci with high-level aminoglycoside resistance in Inner Mongolia, China. Braz. J. Microbiol. 2016, 47, 691–696. [Google Scholar] [CrossRef] [Green Version]
- Ngbede, E.O.; Raji, M.A.; Kwanashie, C.N.; Kwaga, J.K.P.; Adikwu, A.A.; Maurice, N.A.; Adamu, A.M. Characterization of high-level ampicillin-and aminoglycoside resistant enterococci isolated from non-hospital sources. J. Med. Microbiol. 2017, 66, 1027–1032. [Google Scholar] [CrossRef]
- bin Kim, Y.; Seo, K.W.; Son, S.H.; Noh, E.B.; Lee, Y.J. Genetic characterization of high-level aminoglycoside-resistant Enterococcus faecalis and Enterococcus faecium isolated from retail chicken meat. Poult. Sci. 2019, 98, 5981–5988. [Google Scholar]
- Harada, S.; Shibue, Y.; Aoki, K.; Ishii, Y.; Tateda, K. Prevalence of high-level aminoglycoside resistance and genes encoding aminoglycoside-modifying enzymes in Enterococcus faecalis and Enterococcus faecium isolated in a University Hospital in Tokyo. Jpn. J. Infect. Dis. 2020, 73, 476–480. [Google Scholar] [CrossRef]
- Peyvasti, V.S.; Mobarez, A.M.; Shahcheraghi, F.; Khoramabadi, N.; Rahmati, N.R.; Doust, R.H. High-level aminoglycoside resistance and distribution of aminoglycoside resistance genes among Enterococcus spp. clinical isolates in Tehran, Iran. J. Glob. Antimicrob. Resist. 2020, 20, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Adamecz, Z.; Nielsen, K.L.; Kirkby, N.S.; Frimodt-Møller, N. Aminoglycoside resistance genes in Enterococcus faecium: Mismatch with phenotype. J. Antimicrob. Chemother. 2021, 76, 2215–2217. [Google Scholar] [CrossRef] [PubMed]
- McMurtry, T.A.; Barekat, A.; Rodriguez, F.; Purewal, P.; Bulman, Z.P.; Lenhard, J.R. Capability of Enterococcus faecalis to shield Gram-negative pathogens from aminoglycoside exposure. J. Antimicrob. Chemother. 2021, 76, 2610–2614. [Google Scholar] [CrossRef] [PubMed]
- Galimand, M.; Schmitt, E.; Panvert, M.; Desmolaize, B.; Douthwaite, S.; Mechulam, Y.; Courvalin, P. Intrinsic resistance to aminoglycosides in Enterococcus faecium is conferred by the 16S rRNA m5C1404-specific methyltransferase EfmM. RNA 2011, 17, 251–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eliopoulos, G.M.; Farber, B.F.; Murray, B.E.; Wennersten, C.; Moellering, R.C. Ribosomal resistance of clinical enterococcal to streptomycin isolates. Antimicrob. Agents Chemother. 1984, 25, 398–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerma, L.L.; Benomar, N.; Valenzuela, A.S.; del Muñoz, M.C.C.; Gálvez, A.; Abriouel, H. Role of EfrAB efflux pump in biocide tolerance and antibiotic resistance of Enterococcus faecalis and Enterococcus faecium isolated from traditional fermented foods and the effect of EDTA as EfrAB inhibitor. Food Microbiol. 2014, 44, 249–257. [Google Scholar] [CrossRef]
- Bender, J.K.; Cattoir, V.; Hegstad, K.; Sadowy, E.; Coque, T.M.; Westh, H.; Hammerum, A.M.; Schaffer, K.; Burns, K.; Murchan, S.; et al. Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: Towards a common nomenclature. Drug Resist. Updates 2018, 40, 25–39. [Google Scholar] [CrossRef]
- Courvalin, P. Vancomycin resistance in Gram-positive cocci. Clin. Infect. Dis. 2006, 42, 25–34. [Google Scholar] [CrossRef]
- Xu, X.; Lin, D.; Yan, G.; Ye, X.; Wu, S.; Guo, Y.; Zhu, D.; Hu, F.; Zhang, Y.; Wang, F.; et al. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob. Agents Chemother. 2010, 54, 4643–4647. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, M.O.; Baptiste, K.E. Vancomycin-resistant enterococci: A review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microb. Drug Resist. 2018, 24, 590–606. [Google Scholar] [CrossRef] [Green Version]
- Mühlberg, E.; Umstätter, F.; Kleist, C.; Domhan, C.; Mier, W.; Uhl, P. Renaissance of vancomycin: Approaches for breaking antibiotic resistance in multidrug-resistant bacteria. Can. J. Microbiol. 2020, 66, 11–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyd, D.A.; Willey, B.M.; Fawcett, D.; Gillani, N.; Mulvey, M.R. Molecular characterization of Enterococcus faecalis N06-0364 with low-level vancomycin resistance harboring a novel D-Ala-D-Ser gene cluster, vanL. Antimicrob. Agents Chemother. 2008, 52, 2667–2672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebreton, F.; Depardieu, F.; Bourdon, N.; Fines-Guyon, M.; Berger, P.; Camiade, S.; Leclercq, R.; Courvalin, P.; Cattoir, V. D-Ala-D-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 2011, 55, 4606–4612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sassi, M.; Guérin, F.; Lesec, L.; Isnard, C.; Fines-Guyon, M.; Cattoir, V.; Giard, J.C. Genetic characterization of a VanG-type vancomycin-resistant Enterococcus faecium clinical isolate. J. Antimicrob. Chemother. 2018, 73, 852–855. [Google Scholar] [CrossRef]
- Arthur, M.; Depardieu, F.; Gerbaud, G.; Galimand, M.; Leclercq, R.; Courvalin, P. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J. Bacteriol. 1997, 179, 97–106. [Google Scholar] [CrossRef] [Green Version]
- Arthur, M.; Molinas, C.; Courvalin, P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 1992, 174, 2582–2591. [Google Scholar] [CrossRef] [Green Version]
- Arthur, M.; Molinas, C.; Courvalin, P. Sequence of the vanY gene required for production of a vancomycin-inducible D,D-carboxypeptidase in Enterococcus faecium BM4147. Gene 1992, 120, 111–114. [Google Scholar] [CrossRef]
- Reynolds, P.E.; Depardieu, F.; Dutka-Malen, S.; Arthur, M.; Courvalin, P. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of D-alanyl-D-alanine. Mol. Microbiol. 1994, 13, 1065–1070. [Google Scholar] [CrossRef]
- Arthur, M.; Depardieu, F.; Molinas, C.; Reynolds, P.; Courvalin, P. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene 1995, 154, 87–92. [Google Scholar] [CrossRef]
- Leclercq, R.; Dutka-Malen, S.; Duval, J.; Courvalin, P. Vancomycin resistance gene vanC is specific to Enterococcus gallinarum. Antimicrob. Agents Chemother. 1992, 36, 2005–2008. [Google Scholar] [CrossRef] [Green Version]
- Navarro, F.; Courvalin, P. Analysis of genes encoding D-alanine-D-alanine ligase-related enzymes in Enterococcus casseliflavus and Enterococcus flavescens. Antimicrob. Agents Chemother. 1994, 38, 1788–1793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stogios, P.J.; Savchenko, A. Molecular mechanisms of vancomycin resistance. Protein Sci. 2020, 29, 654–669. [Google Scholar] [CrossRef] [PubMed]
- Hawkey, P.M. Mechanisms of quinolone action and microbial response. J. Antimicrob. Chemother. 2003, 51, 29–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arsène, S.; Leclercq, R. Role of a qnr-like, gene in the intrinsic resistance of Enterococcus faecalis to fluoroquinolones. Antimicrob. Agents Chemother. 2007, 51, 3254–3258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakanishi, N.; Yoshida, S.; Wakebe, H.; Inoue, M.; Mitsuhashi, S. Mechanisms of clinical resistance to fluoroquinolones in Enterococcus faecalis. Antimicrob. Agents Chemother. 1991, 35, 1053–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korten, V.; Huang, M.U.N.; Murray, B.E. Analysis by PCR and direct DNA sequencing of gyrA mutations associated with fluoroquinolone resistance in Enterococcus faecalis. Antimicrob. Agents Chemother. 1994, 38, 2091–2094. [Google Scholar] [CrossRef] [Green Version]
- Tankovic, J.; Mahjoubi, F.; Courvalin, P.; Duval, J.; Leclercq, R. Development of fluoroquinolone resistance in Enterococcus faecalis and role of mutations in the DNA Gyrase gyrA Gene. Antimicrob. Agents Chemother. 1996, 40, 2558–2561. [Google Scholar] [CrossRef] [Green Version]
- Kanematsu, E.; Deguchi, T.; Yasuda, M.; Kawamura, T.; Nishino, Y.; Kawada, Y. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of DNA topoisomerase IV associated with quinolone resistance in Enterococcus faecalis. Antimicrob. Agents Chemother. 1998, 42, 433–435. [Google Scholar] [CrossRef] [Green Version]
- el Amin, N.; Jalal, S.; Wretlind, B. Alterations in gyrA and parC associated with fluoroquinolone resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 1999, 43, 947–949. [Google Scholar] [CrossRef] [Green Version]
- López, M.; Tenorio, C.; del Campo, R.; Zarazaga, M.; Torres, C. Characterization of the mechanisms of fluoroquinolone resistance in vancomycin-resistant enterococci of different origins. J. Chemother. 2011, 23, 87–91. [Google Scholar] [CrossRef]
- Yasufuku, T.; Shigemura, K.; Shirakawa, T.; Matsumoto, M.; Nakano, Y.; Tanaka, K.; Arakawa, S.; Kawabata, M.; Fujisawa, M. Mechanisms of and risk factors for fluoroquinolone resistance in clinical Enterococcus faecalis isolates from patients with urinary tract infections. J. Clin. Microbiol. 2011, 49, 3912–3916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- bin Kim, Y.; Seo, H.J.; Seo, K.W.; Jeon, H.Y.; Kim, D.K.; Kim, S.W.; Lim, S.K.; Lee, Y.J. Characteristics of high-level ciprofloxacin-resistant Enterococcus faecalis and Enterococcus faecium from retail chicken meat in Korea. J. Food Prot. 2018, 81, 1357–1363. [Google Scholar]
- Yamanaka, H.; Kadomatsu, R.; Takagi, T.; Ohsawa, M.; Yamamoto, N.; Kubo, N.; Takemoto, T.; Ohsawa, K. Antimicrobial resistance profiles of vancomycin-resistant Enterococcus species isolated from laboratory mice. J. Vet. Sci. 2019, 20, e13. [Google Scholar] [CrossRef]
- Oyamada, Y.; Ito, H.; Fujimoto, K.; Asada, R.; Niga, T.; Okamoto, R.; Inoue, M.; Yamagishi, J.I. Combination of known and unknown mechanisms confers high-level resistance to fluoroquinolones in Enterococcus faecium. J. Med. Microbiol. 2006, 55, 729–736. [Google Scholar] [CrossRef]
- Hooper, D.C. Fluoroquinolone resistance among Gram-positive cocci. Lancet Infect. Dis. 2002, 2, 530–538. [Google Scholar] [CrossRef]
- Jonas, B.M.; Murray, B.E.; Weinstock, G.M. Characterization of emeA, a norA homolog and multidrug resistance efflux pump, in Enterococcus faecalis. Antimicrob. Agents Chemother. 2001, 45, 3574–3579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, J.H.; Jacoby, G.A. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 2002, 99, 5638–5642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, J.H.; Jacoby, G.A.; Hooper, D.C. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 2005, 49, 118–125. [Google Scholar] [CrossRef] [Green Version]
- Tran, J.H.; Jacoby, G.A.; Hooper, D.C. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli Topoisomerase IV. Antimicrob. Agents Chemother. 2005, 49, 3050–3052. [Google Scholar] [CrossRef] [Green Version]
- Agersø, Y.; Pedersen, A.G.; Aarestrup, F.M. Identification of Tn5397-like and Tn916-like transposons and diversity of the tetracycline resistance gene tet(M) in enterococci from humans, pigs and poultry. J. Antimicrob. Chemother. 2006, 57, 832–839. [Google Scholar] [CrossRef] [Green Version]
- Cauwerts, K.; Decostere, A.; de Graef, E.M.; Haesebrouck, F.; Pasmans, F. High prevalence of tetracycline resistance in Enterococcus isolates from broilers carrying the erm(B) gene. Avian Pathol. 2007, 36, 395–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chotinantakul, K.; Chansiw, N.; Okada, S. Antimicrobial resistance of Enterococcus spp. isolated from Thai fermented pork in Chiang Rai Province, Thailand. J. Glob. Antimicrob. Resist. 2018, 12, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Woźniak-Biel, A.; Bugla-Płoskońska, G.; Burdzy, J.; Korzekwa, K.; Ploch, S.; Wieliczko, A. Antimicrobial resistance and biofilm formation in Enterococcus spp. isolated from humans and turkeys in Poland. Microb. Drug Resist. 2019, 25, 277–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, L.B. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 1998, 42, 1871–1877. [Google Scholar] [CrossRef] [Green Version]
- Sharkey, L.K.R.; Edwards, T.A.; O’Neill, A.J. ABC-F proteins mediate antibiotic resistance through ribosomal protection. MBio 2016, 7, e01975-15. [Google Scholar] [CrossRef] [Green Version]
- Antonelli, A.; D’Andrea, M.M.; Brenciani, A.; Galeotti, C.L.; Morroni, G.; Pollini, S.; Varaldo, P.E.; Rossolini, G.M. Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J. Antimicrob. Chemother. 2018, 73, 1763–1769. [Google Scholar] [CrossRef] [Green Version]
- Brenciani, A.; Fioriti, S.; Morroni, G.; Cucco, L.; Morelli, A.; Pezzotti, G.; Paniccià, M.; Antonelli, A.; Magistrali, C.F.; Rossolini, G.M.; et al. Detection in Italy of a porcine Enterococcus faecium isolate carrying the novel phenicol-oxazolidinone-tetracycline resistance gene poxtA. J. Antimicrob. Chemother. 2019, 74, 817–818. [Google Scholar] [CrossRef]
- Lei, C.W.; Kang, Z.Z.; Wu, S.K.; Chen, Y.P.; Kong, L.H.; Wang, H.N. Detection of the phenicol–oxazolidinone–tetracycline resistance gene poxtA in Enterococcus faecium and Enterococcus faecalis of food-producing animal origin in China. J. Antimicrob. Chemother. 2019, 74, 2459–2461. [Google Scholar] [CrossRef]
- Fioriti, S.; Coccitto, S.N.; Cedraro, N.; Simoni, S.; Morroni, G.; Brenciani, A.; Mangiaterra, G.; Vignaroli, C.; Vezzulli, L.; Biavasco, F.; et al. Linezolid resistance genes in enterococci isolated from sediment and zooplankton in two Italian coastal areas. Appl. Environ. Microbiol. 2021, 87, e02958-20. [Google Scholar] [CrossRef]
- Wang, J.L.; Hsueh, P.R. Therapeutic options for infections due to vancomycin-resistant enterococci. Expert Opin. Pharmacother. 2009, 10, 785–796. [Google Scholar] [CrossRef]
- Lee, B.J.; Vu, B.N.; Seddon, A.N.; Hodgson, H.A.; Wang, S.K. Treatment considerations for CNS infections caused by vancomycin-resistant Enterococcus faecium: A focused review of linezolid and daptomycin. Ann. Pharmacother. 2020, 54, 1243–1251. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, N.; Monticelli, J.; Antonello, R.M.; di Lallo, G.; Frezza, D.; Luzzati, R.; di Bella, S. Therapeutic options for infections due to vanB genotype vancomycin-resistant enterococci. Microb. Drug Resist. 2021, 27, 536–545. [Google Scholar] [CrossRef]
- Wingler, M.J.; Patel, N.R.; King, S.T.; Wagner, J.L.; Barber, K.E.; Stover, K.R. Linezolid for the treatment of urinary tract infections caused by vancomycin-resistant enterococci. Pharmacy 2021, 9, 175. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; He, T.; et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, R.; Qin, T.; Fan, W.; Ma, P.; Gu, B. The emerging problem of linezolid-resistant enterococci. J. Glob. Antimicrob. Resist. 2018, 13, 11–19. [Google Scholar] [CrossRef]
- Càmara, J.; Camoez, M.; Tubau, F.; Pujol, M.; Ayats, J.; Ardanuy, C.; Domínguez, M.Á. Detection of the novel optrA gene among linezolid-resistant enterococci in Barcelona, Spain. Microb. Drug Resist. 2019, 25, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Elghaieb, H.; Freitas, A.R.; Abbassi, M.S.; Novais, C.; Zouari, M.; Hassen, A.; Peixe, L. Dispersal of linezolid-resistant enterococci carrying poxtA or optrA in retail meat and food-producing animals from Tunisia. J. Antimicrob. Chemother. 2019, 74, 2865–2869. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ripa, L.; Feßler, A.T.; Hanke, D.; Eichhorn, I.; Azcona-Gutiérrez, J.M.; Pérez-Moreno, M.O.; Seral, C.; Aspiroz, C.; Alonso, C.A.; Torres, L.; et al. Mechanisms of linezolid resistance among enterococci of clinical origin in Spain—Detection of optrA- and cfr(D)-carrying E. faecalis. Microorganisms 2020, 8, 1155. [Google Scholar] [CrossRef]
- Freitas, A.R.; Finisterra, L.; Tedim, A.P.; Duarte, B.; Novais, C.; Peixe, L. Linezolid- and multidrug-resistant enterococci in raw commercial dog food, Europe, 2019–2020. Emerg. Infect. Dis. 2021, 27, 2221–2224. [Google Scholar] [CrossRef] [PubMed]
- Kloss, P.; Xiong, L.; Shinabarger, D.L.; Mankin, A.S. Resistance mutations in 23S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J. Mol. Biol. 1999, 294, 93–101. [Google Scholar] [CrossRef] [Green Version]
- Mališová, L.; Jakubů, V.; Pomorská, K.; Musílek, M.; Žemličková, H. Spread of linezolid-resistant Enterococcus spp. in human clinical isolates in the Czech Republic. Antibiotics 2021, 10, 219. [Google Scholar] [CrossRef] [PubMed]
- Marshall, S.H.; Donskey, C.J.; Hutton-Thomas, R.; Salata, R.A.; Rice, L.B. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 2002, 46, 3334–3336. [Google Scholar] [CrossRef] [Green Version]
- Kehrenberg, C.; Schwarz, S.; Jacobsen, L.; Hansen, L.H.; Vester, B. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: Methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 2005, 57, 1064–1073. [Google Scholar] [CrossRef]
- Long, K.S.; Poehlsgaard, J.; Kehrenberg, C.; Schwarz, S.; Vester, B. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 2006, 50, 2500–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshpande, L.M.; Ashcraft, D.S.; Kahn, H.P.; Pankey, G.; Jones, R.N.; Farrell, D.J.; Mendes, R.E. Detection of a new cfr-like gene, cfr(B), in Enterococcus faecium isolates recovered from human specimens in the United States as part of the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 2015, 59, 6256–6261. [Google Scholar] [CrossRef] [Green Version]
- Bender, J.K.; Fleige, C.; Klare, I.; Fiedler, S.; Mischnik, A.; Mutters, N.T.; Dingle, K.E.; Werner, G. Detection of a cfr(B) variant in German Enterococcus faecium clinical isolates and the impact on linezolid resistance in Enterococcus spp. PLoS ONE 2016, 11, e0167042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, S.; Boan, P.; Lee, T.; Gangatharan, S.; Tan, S.J.; Daley, D.; Lee, Y.T.; Coombs, G.W. Linezolid-resistant ST872 Enteroccocus faecium harbouring optrA and cfr(D) oxazolidinone resistance genes. Int. J. Antimicrob. Agents 2019, 55, 105831. [Google Scholar] [CrossRef] [PubMed]
- Guerin, F.; Sassi, M.; Dejoies, L.; Zouari, A.; Schutz, S.; Potrel, S.; Auzou, M.; Collet, A.; Lecointe, D.; Auger, G.; et al. Molecular and functional analysis of the novel cfr(D) linezolid resistance gene identified in Enterococcus faecium. J. Antimicrob. Chemother. 2020, 75, 1699–1703. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wang, Y.; Lv, Y.; Wang, S.; Song, Y.; Li, Y.; Liu, J.; Xue, F.; Yang, W.; Zhang, J. Nationwide surveillance of novel oxazolidinone resistance gene optrA in Enterococcus isolates in China from 2004 to 2014. Antimicrob. Agents Chemother. 2016, 60, 7490–7493. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Yang, Y.; Ding, L.; Xu, X.; Lin, D. Molecular investigations of linezolid resistance in enterococci optrA variants from a hospital in Shanghai. Infect. Drug Resist. 2020, 13, 2711–2716. [Google Scholar] [CrossRef]
- Regulation (UE), No. 528/2012 of 22 of May 2012; Official Journal of the European Union—L167/1; European Parliament and Council: Brussels, Belgium, 2012.
- Maillard, J.-Y. Resistance of bacteria to biocides. Microbiol. Spectr. 2018, 6, 109–126. [Google Scholar] [CrossRef] [PubMed]
- Roedel, A.; Dieckmann, R.; Brendebach, H.; Hammerl, J.A.; Kleta, S.; Noll, M.; al Dahouk, S.; Vinczea, S. Biocide-tolerant Listeria monocytogenes isolates from German food production plants do not show cross-resistance to clinically relevant antibiotics. Appl. Environ. Microbiol. 2019, 85, e01253-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maillard, J.-Y. Factors Affecting the Activities of Microbicides. In Russell, Hugo & Ayliffe’s Principles and Practice of Disinfection, Preservation & Sterilization, 5th ed.; Fraise, A.P., Lambert, P.A., Maillard, J.-Y., Eds.; Blackwell Publishing: London, UK, 2013; pp. 71–86. [Google Scholar]
- Sinel, C.; Augagneur, Y.; Sassi, M.; Bronsard, J.; Cacaci, M.; Guérin, F.; Sanguinetti, M.; Meignen, P.; Cattoir, V.; Felden, B. Small RNAs in vancomycin-resistant Enterococcus faecium involved in daptomycin response and resistance. Sci. Rep. 2017, 7, 11067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaux, C.; Hartke, A.; Martini, C.; Reiss, S.; Albrecht, D.; Budin-Verneuil, A.; Sanguinetti, M.; Engelmann, S.; Hain, T.; Verneuil, N.; et al. Involvement of Enterococcus faecalis small RNAs in stress response. Infect. Immun. 2014, 82, 3599–3611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shioya, K.; Michaux, C.; Kuenne, C.; Hain, T.; Verneuil, N.; Hartsch, T.; Hartke, A.; Giard, J. Genome-wide identification of small RNAs in the opportunistic pathogen Enterococcus faecalis V583. PLoS ONE 2017, 6, e23948. [Google Scholar] [CrossRef] [PubMed]
- Dejoies, L.; le Neindre, K.; Reissier, S.; Felden, B.; Cattoir, V. Distinct expression profiles of regulatory RNAs in the response to biocides in Staphylococcus aureus and Enterococcus faecium. Sci. Rep. 2021, 11, 6892. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Ehrmann, M.A.; Waldhuber, A.; Niemeyer, C.; Miethke, T.; Frick, J.; Xiong, T.; Vogel, R.F. Phosphotransferase systems in Enterococcus faecalis OG1RF enhance anti- stress capacity in vitro and in vivo. Res. Microbiol. 2017, 168, 558–566. [Google Scholar] [CrossRef]
- Rizzotti, L.; Rossi, F.; Torriani, S. Biocide and antibiotic resistance of Enterococcus faecalis and Enterococcus faecium isolated from the swine meat chain. Food Microbiol. 2016, 60, 160–164. [Google Scholar] [CrossRef]
- Suchomel, M.; Lenhardt, A.; Kampf, G.; Grisold, A. Enterococcus hirae, Enterococcus faecium and Enterococcus faecalis show different sensitivities to typical biocidal agents used for disinfection. J. Hosp. Infect. 2019, 103, 435–440. [Google Scholar] [CrossRef]
- Schwaiger, K.; Harms, K.S.; Bischoff, M.; Preikschat, P.; Mölle, G.; Bauer-Unkauf, I.; Lindorfer, S.; Thalhammer, S.; Bauer, J.; Hölzel, C.S. Insusceptibility to disinfectants in bacteria from animals, food and humans—Is there a link to antimicrobial resistance? Front. Microbiol. 2014, 5, 88. [Google Scholar] [CrossRef] [Green Version]
- Wieland, N.; Boss, J.; Lettmann, S.; Fritz, B.; Schwaiger, K.; Bauer, J.; Hölzel, C.S. Susceptibility to disinfectants in antimicrobial-resistant and -susceptible isolates of Escherichia coli, Enterococcus faecalis and Enterococcus faecium from poultry–ESBL/AmpC-phenotype of E. coli is not associated with resistance to a quaternary ammonium. J. Appl. Microbiol. 2017, 122, 1508–1517. [Google Scholar] [CrossRef] [PubMed]
- Esmaeilishirazifard, E.; Usher, L.; Trim, C.; Denise, H.; Sangal, V.; Tyson, G.H.; Barlow, A.; Redway, K.F.; Taylor, J.D.; Kremyda-Vlachou, M.; et al. Bacterial adaptation to venom in snakes and arachnida. Microbiol. Spectr. 2022, e02408-21. [Google Scholar] [CrossRef] [PubMed]
- Bengoechea, J.A.; Bamford, C.G.G. SARS-CoV-2, bacterial co-infections, and AMR: The deadly trio in COVID-19? EMBO Mol. Med. 2020, 12, e12560. [Google Scholar] [CrossRef]
- DeVoe, C.; Segal, M.R.; Wang, L.; Stanley, K.; Madera, S.; Fan, J.; Schouest, J.; Graham-Ojo, R.; Nichols, A.; Prasad, P.A.; et al. Increased rates of secondary bacterial infections, including Enterococcus bacteremia, in patients hospitalized with coronavirus disease 2019 (COVID-19). Infect. Control Epidemiol. 2021, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Toc, D.A.; Mihaila, R.M.; Botan, A.; Bobohalma, C.N.; Risteiu, G.A.; Simut-Cacuci, B.N.; Steorobelea, B.; Troanca, S.; Junie, L.M. Enterococcus and COVID-19: The Emergence of a Perfect Storm? Int. J. Transl. Med. 2022, 2, 220–229. [Google Scholar] [CrossRef]
- Kampmeier, S.; Tönnies, H.; Correa-Martinez, C.L.; Mellmann, A.; Schwierzeck, V. A nosocomial cluster of vancomycin resistant enterococci among COVID-19 patients in an intensive care unit. Antimicrob. Resist. Infect. Control 2020, 9, 154. [Google Scholar] [CrossRef]
- Gaibani, P.; D’Amico, F.; Bartoletti, M.; Lombardo, D.; Rampelli, S.; Fornaro, G.; Coladonato, S.; Siniscalchi, A.; Re, M.C.; Viale, P.; et al. The Gut Microbiota of Critically Ill Patients With COVID-19. Front. Cell. Infect. Microbiol. 2021, 11, 670424. [Google Scholar] [CrossRef]
- Toc, D.A.; Butiuc-Keul, A.L.; Iordache, D.; Botan, A.; Mihaila, R.M.; Costache, C.A.; Colosi, I.A.; Chiorean, C.; Neagoe, D.S.; Gheorghiu, L.; et al. Descriptive Analysis of Circulating Antimicrobial Resistance Genes in Vancomycin-Resistant Enterococcus (VRE) during the COVID-19 Pandemic. Biomedicines 2022, 10, 1122. [Google Scholar] [CrossRef]
- Jeon, K.; Jeong, S.; Lee, N.; Park, M.-J.; Song, W.; Kim, H.-S.; Kim, H.S.; Kim, J.-S. Impact of COVID-19 on Antimicrobial Consumption and Spread of Multidrug-Resistance in Bacterial Infections. Antibiotics 2022, 11, 535. [Google Scholar] [CrossRef]
- Bienzle, D.; Rousseau, J.; Marom, D.; MacNicol, J.; Jacobson, L.; Sparling, S.; Prystajecky, N.; Fraser, E.; Weese, J.S. Risk Factors for SARS-CoV-2 Infection and Illness in Cats and Dogs. Emerg. Infect. Dis. 2022, 28, 1154–1162. [Google Scholar] [CrossRef]

| Enzyme | Type of Resistance Conferred | Reference | ||
|---|---|---|---|---|
| AACs | AAC(6′)-Ii | Intrinsic | Low- to moderate-level resistance to tobramycin and kanamycin | [125,126] |
| AAC(6′)-Id | [127] | |||
| AAC(6′)-Ih | [127] | |||
| APHs | APH(3′)-IIIa | Extrinsic | Low-level resistance to kanamycin and amikacin | [128] |
| APH(2″)-Ib | Extrinsic | High-level resistance to gentamicin | [129] | |
| APH(2″)-Ic | [130] | |||
| APH(2″)-Id | [131] | |||
| APH(2″)-Ie | [132] | |||
| ANTs | ANT(6′)-Ia | Extrinsic | High-level resistance to streptomycin | [3,133] |
| ANT(3″)-Ia or ANT(3″)(9) | [3] | |||
| ANT(4″)-Ia | [134] | |||
| Bifunctional Enzyme (AAC + APH) | AAC-6′-Ie-APH-2 | Extrinsic | High-level resistance to gentamicin | [135,136,137] |
| Antibiotics | |||
|---|---|---|---|
| Group of Antibiotics | Resistance Type | Mechanism of Resistance | Associated Genes |
| β-Lactam | Intrinsic | Low affinity PBPs that do not allow for easy antibiotic binding | pbp5/pbp4 |
| Acquired | Mutations that lead to alteration in PBPs’ molecular structure and cause an even lower affinity | - | |
| Aminoglycosides | Intrinsic | Poor antibiotic uptake trough the cell wall | - |
| Intrinsic | Modification of the antibiotic molecule | aac | |
| Acquired | Modification of the antibiotic molecule | aph, ant, aac-aph | |
| Intrinsic | Target-site modification through rRNA methyltransferase | efmM | |
| Acquired | Target-site modification through point mutations | - | |
| - | Efflux of the antibiotic | efrAB | |
| Glycopeptides | Intrinsic/Acquired | Target-site modification | van operons |
| Fluoroquinolones | Acquired | Target-site modification through gene mutation | gyrA, pacC |
| - | Efflux of the antibiotic | emeA | |
| - | Target-site protection | qnrE.faecalis | |
| Tetracyclines | - | Efflux of the antibiotic | tet(K), tet(L) |
| - | Target-site protection | tet(M), tet(O), tet(S) | |
| Intrinsic/Acquired | Target-site protection | poxtA | |
| Oxazolidinones | Acquired | Target-site modification through point mutations | - |
| Intrinsic/Acquired | Target-site protection | poxtA | |
| Acquired | Target-site protection | optrA | |
| Biocides | |||
| sRNAs | Bacterial survival in stressful environmental conditions such as in the presence of biocides | ||
| PTS Systems | |||
| ChtRS | |||
| Efflux Pumps | QacA/B and EfrAB—efflux of different biocides | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geraldes, C.; Tavares, L.; Gil, S.; Oliveira, M. Enterococcus Virulence and Resistant Traits Associated with Its Permanence in the Hospital Environment. Antibiotics 2022, 11, 857. https://doi.org/10.3390/antibiotics11070857
Geraldes C, Tavares L, Gil S, Oliveira M. Enterococcus Virulence and Resistant Traits Associated with Its Permanence in the Hospital Environment. Antibiotics. 2022; 11(7):857. https://doi.org/10.3390/antibiotics11070857
Chicago/Turabian StyleGeraldes, Catarina, Luís Tavares, Solange Gil, and Manuela Oliveira. 2022. "Enterococcus Virulence and Resistant Traits Associated with Its Permanence in the Hospital Environment" Antibiotics 11, no. 7: 857. https://doi.org/10.3390/antibiotics11070857
APA StyleGeraldes, C., Tavares, L., Gil, S., & Oliveira, M. (2022). Enterococcus Virulence and Resistant Traits Associated with Its Permanence in the Hospital Environment. Antibiotics, 11(7), 857. https://doi.org/10.3390/antibiotics11070857







