The Contribution of Dairy Bedding and Silage to the Dissemination of Genes Coding for Antimicrobial Resistance: A Narrative Review
Abstract
:1. Introduction
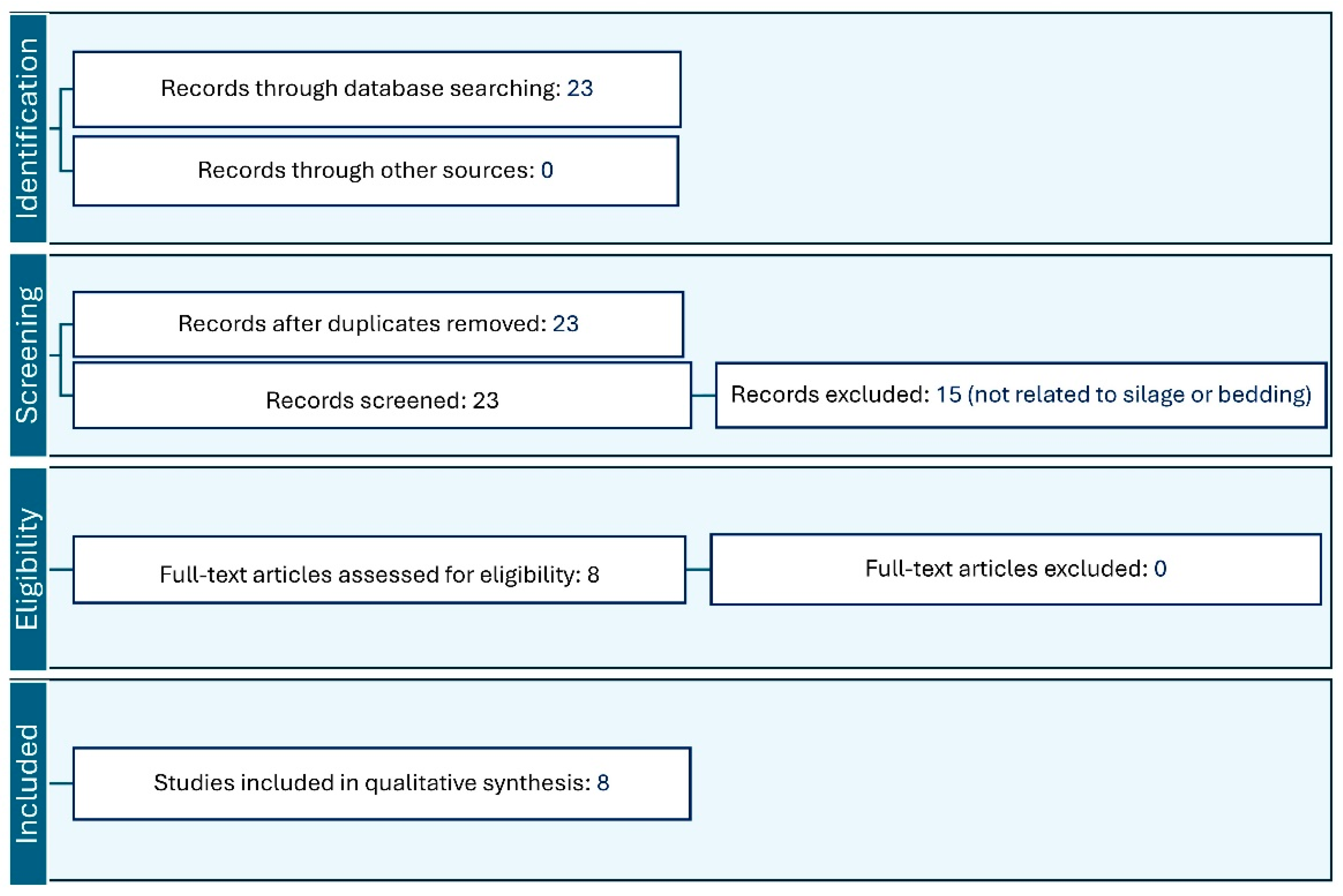
2. Literature Review Methods
3. Bedding for Dairy Cattle and AMR Prevalence
4. AMR Prevalence in Silage
5. Methodological Limitations and Considerations for Silage and Bedding
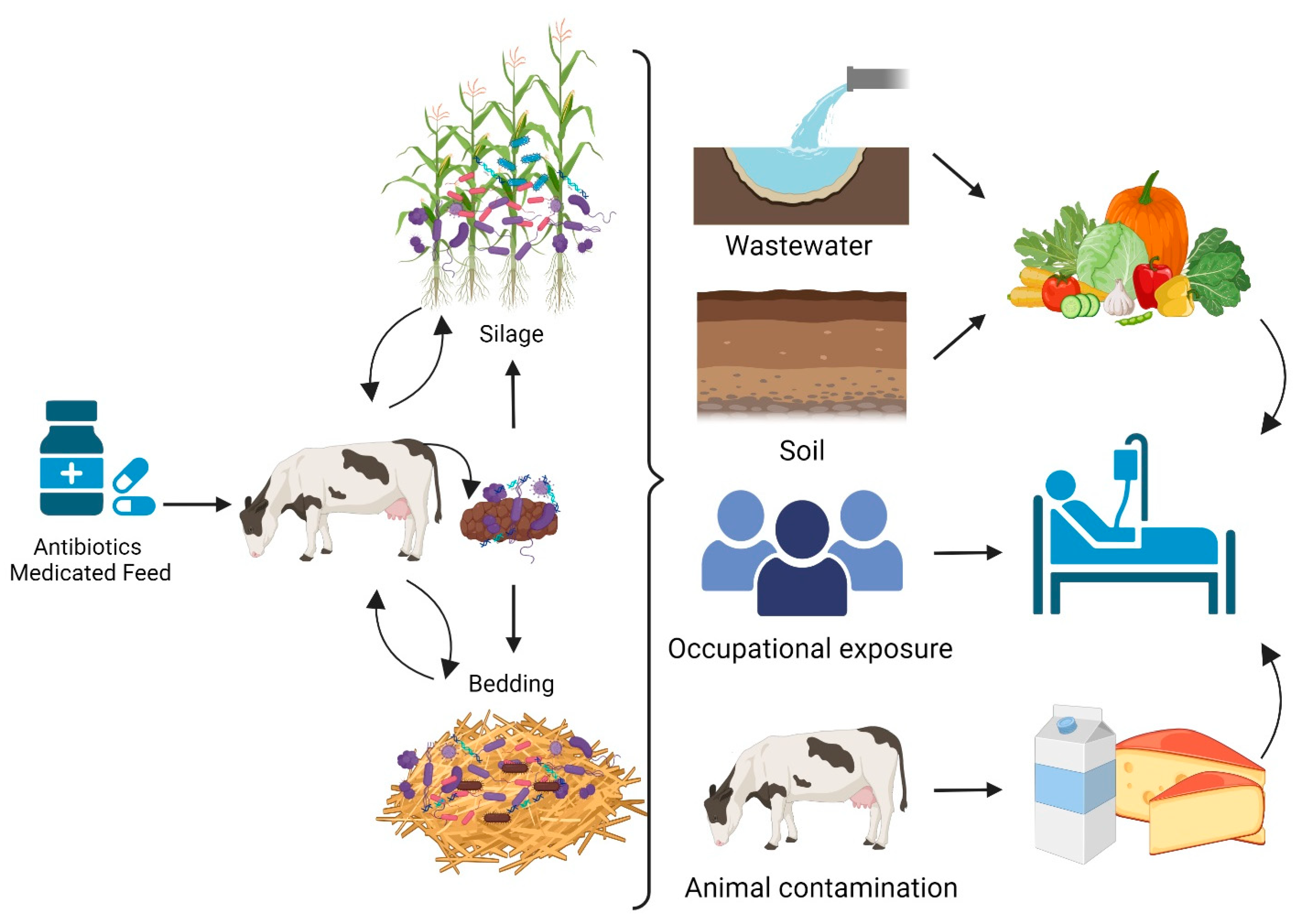
6. AMR Dissemination and Transmission Pathways
7. Critical Surveillance and Practical Management Needs
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic resistance genes from livestock waste: Occurrence, dissemination, and treatment. NPJ Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Tang, K.L.; Caffrey, N.P.; Nóbrega, D.B.; Cork, S.C.; Ronksley, P.E.; Barkema, H.W.; Polachek, A.J.; Ganshorn, H.; Sharma, N.; Kellner, J.D. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: A systematic review and meta-analysis. Lancet Planet. Health 2017, 1, e316–e327. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Sifakis, F.; Harbarth, S.; Schrijver, R.; van Mourik, M.; Voss, A.; Sharland, M.; Rajendran, N.B.; Rodríguez-Baño, J.; Bielicki, J. Surveillance for control of antimicrobial resistance. Lancet Infect. Dis. 2018, 18, e99–e106. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Glennon, E.E.; Chen, D.; Gilbert, M.; Robinson, T.P.; Grenfell, B.T.; Levin, S.A.; Bonhoeffer, S.; Laxminarayan, R. Reducing antimicrobial use in food animals. Science (80-) 2017, 357, 1350–1352. [Google Scholar] [CrossRef] [PubMed]
- Schmerold, I.; van Geijlswijk, I.; Gehring, R. European regulations on the use of antibiotics in veterinary medicine. Eur. J. Pharm. Sci. 2023, 189, 106473. [Google Scholar] [CrossRef]
- Llanos-Soto, S.G.; Vezeau, N.; Wemette, M.; Bulut, E.; Safi, A.G.; Moroni, P.; Shapiro, M.A.; Ivanek, R. Survey of perceptions and attitudes of an international group of veterinarians regarding antibiotic use and resistance on dairy cattle farms. Prev. Vet. Med. 2021, 188, 105253. [Google Scholar] [CrossRef]
- Ferroni, L.; Lovito, C.; Scoccia, E.; Dalmonte, G.; Sargenti, M.; Pezzotti, G.; Maresca, C.; Forte, C.; Magistrali, C.F. Antibiotic consumption on dairy and beef cattle farms of central Italy based on paper registers. Antibiotics 2020, 9, 273. [Google Scholar] [CrossRef]
- Astorga, F.; Navarrete-Talloni, M.J.; Miró, M.P.; Bravo, V.; Toro, M.; Blondel, C.J.; Hervé-Claude, L.P. Antimicrobial resistance in E. coli isolated from dairy calves and bedding material. Heliyon 2019, 5, e02773. [Google Scholar] [CrossRef]
- Subbiah, M.; Shah, D.H.; Besser, T.E.; Ullman, J.L.; Call, D.R. Urine from treated cattle drives selection for cephalosporin resistant Escherichia coli in soil. PLoS ONE 2012, 7, e48919. [Google Scholar] [CrossRef]
- Vinayamohan, P.G.; Pellissery, A.J.; Venkitanarayanan, K. Role of horizontal gene transfer in the dissemination of antimicrobial resistance in food animal production. Curr. Opin. Food Sci. 2022, 47, 100882. [Google Scholar] [CrossRef]
- Heuer, H.; Schmitt, H.; Smalla, K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011, 14, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Tóth, A.G.; Csabai, I.; Krikó, E.; Tőzsér, D.; Maróti, G.; Patai, Á.V.; Makrai, L.; Szita, G.; Solymosi, N. Antimicrobial resistance genes in raw milk for human consumption. Sci. Rep. 2020, 10, 7464. [Google Scholar] [CrossRef] [PubMed]
- Manaia, C.M. Assessing the risk of antibiotic resistance transmission from the environment to humans: Non-direct proportionality between abundance and risk. Trends Microbiol. 2017, 25, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, B.; Greko, C. Antibiotic resistance—Consequences for animal health, welfare, and food production. Upsala J. Med. Sci. 2014, 119, 96–102. [Google Scholar] [CrossRef]
- Zhang, Q.; Zou, X.; Wu, S.; Wu, N.; Chen, X.; Zhou, W. Effects of Pyroligneous Acid on Diversity and Dynamics of Antibiotic Resistance Genes in Alfalfa Silage. Microbiol. Spectr. 2022, 10, e01554-22. [Google Scholar] [CrossRef]
- Tiedje, J.M.; Fang, W.; Manaia, C.M.; Virta, M.; Sheng, H.; Liping, M.A.; Zhang, T.; Edward, T. Antibiotic resistance genes in the human-impacted environment: A One Health perspective. Pedosphere 2019, 29, 273–282. [Google Scholar] [CrossRef]
- Kang, J.; Liu, Y.; Chen, X.; Xu, F.; Wang, H.; Xiong, W.; Li, X. Metagenomic insights into the antibiotic resistomes of typical Chinese dairy farm environments. Front. Microbiol. 2022, 13, 990272. [Google Scholar] [CrossRef]
- Badawy, B.; Gwida, M.; Sadat, A.; El-Toukhy, M.; Sayed-Ahmed, M.; Alam, N.; Ahmad, S.; Ali, M.D.S.; Elafify, M. Prevalence and antimicrobial resistance of virulent Listeria monocytogenes and Cronobacter sakazakii in dairy cattle, the environment, and dried milk with the in vitro application of natural alternative control. Antibiotics 2022, 11, 1087. [Google Scholar] [CrossRef]
- Wu, X.; Liu, J.; Feng, J.; Shabbir, M.A.B.; Feng, Y.; Guo, R.; Zhou, M.; Hou, S.; Wang, G.; Hao, H. epidemiology, environmental risks, virulence, and resistance determinants of Klebsiella pneumoniae from dairy cows in Hubei, China. Front. Microbiol. 2022, 13, 858799. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, Y.; Jay-Russell, M.; Lemay, D.G.; Mills, D.A. Reservoirs of antimicrobial resistance genes in retail raw milk. Microbiome 2020, 8, 99. [Google Scholar] [CrossRef]
- Taher, E.M.; Hemmatzadeh, F.; Aly, S.A.; Elesswy, H.A.; Petrovski, K.R. Molecular characterization of antimicrobial resistance genes on farms and in commercial milk with emphasis on the effect of currently practiced heat treatments on viable but nonculturable formation. J. Dairy Sci. 2020, 103, 9936–9945. [Google Scholar] [CrossRef] [PubMed]
- Topp, E.; Larsson, D.G.J.; Miller, D.N.; Van den Eede, C.; Virta, M.P.J. Antimicrobial resistance and the environment: Assessment of advances, gaps and recommendations for agriculture, aquaculture and pharmaceutical manufacturing. FEMS Microbiol. Ecol. 2018, 94, fix185. [Google Scholar] [CrossRef] [PubMed]
- Jayarao, B.; Almeida, R.; Oliver, S.P. Antimicrobial resistance on dairy farms. Foodborne Pathog. Dis. 2019, 16, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Rubiola, S.; Chiesa, F.; Dalmasso, A.; Di Ciccio, P.; Civera, T. Detection of antimicrobial resistance genes in the milk production environment: Impact of host DNA and sequencing depth. Front. Microbiol. 2020, 11, 1983. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L. Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 2021, 19, e06651. [Google Scholar]
- Rowbotham, R.F.; Ruegg, P.L. Bacterial counts on teat skin and in new sand, recycled sand, and recycled manure solids used as bedding in freestalls. J. Dairy Sci. 2016, 99, 6594–6608. [Google Scholar] [CrossRef]
- Pilch, H.E.; Steinberger, A.J.; Sockett, D.C.; Aulik, N.; Suen, G.; Czuprynski, C.J. Assessing the microbiota of recycled bedding sand on a Wisconsin dairy farm. J. Anim. Sci. Biotechnol. 2021, 12, 114. [Google Scholar] [CrossRef]
- Li, P.; Fu, T.; Cai, A.; Descovich, K.; Lian, H.; Gao, T.; Phillips, C.J.C. Effect of peanut shell and rice husk bedding for dairy cows: An analysis of material properties and colostrum microbiota. Animals 2022, 12, 603. [Google Scholar] [CrossRef]
- Bak, A.S.; Herskin, M.S.; Jensen, M.B. Effect of sand and rubber surface on the lying behavior of lame dairy cows in hospital pens. J. Dairy Sci. 2016, 99, 2875–2883. [Google Scholar] [CrossRef]
- Van Gastelen, S.; Westerlaan, B.; Houwers, D.J.; Van Eerdenburg, F. A study on cow comfort and risk for lameness and mastitis in relation to different types of bedding materials. J. Dairy Sci. 2011, 94, 4878–4888. [Google Scholar] [CrossRef]
- Nagy, S.Á.; Tóth, A.G.; Papp, M.; Kaplan, S.; Solymosi, N. Antimicrobial resistance determinants in silage. Sci. Rep. 2022, 12, 5243. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Ying, Y.; Bartell, P.A.; Harvatine, K.J. The effects of feeding rations that differ in fiber and fermentable starch within a day on milk production and the daily rhythm of feed intake and plasma hormones and metabolites in dairy cows. J. Dairy Sci. 2017, 100, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Windig, J.J.; Calus, M.P.L.; Veerkamp, R.F. Influence of herd environment on health and fertility and their relationship with milk production. J. Dairy Sci. 2005, 88, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Cobbold, R.N.; Rice, D.H.; Davis, M.A.; Besser, T.E.; Hancock, D.D. Long-term persistence of multi–drug-resistant Salmonella enterica serovar Newport in two dairy herds. J. Am. Vet. Med. Assoc. 2006, 228, 585–591. [Google Scholar] [CrossRef]
- Davis, M.A.; Sischo, W.M.; Jones, L.P.; Moore, D.A.; Ahmed, S.; Short, D.M.; Besser, T.E. Recent emergence of E. coli carrying blaCTX-M encoded cephalosporin resistance on Washington State dairy farms. Appl. Environ. Microbiol. 2015. [Google Scholar] [CrossRef]
- Cui, Y.; Liu, X.; Dietrich, R.; Märtlbauer, E.; Cao, J.; Ding, S.; Zhu, K. Characterization of Bacillus cereus isolates from local dairy farms in China. FEMS Microbiol. Lett. 2016, 363, fnw096. [Google Scholar] [CrossRef]
- Wu, H.; Wang, Y.; Du, B.; Li, H.; Dong, L.; Hu, H.; Meng, L.; Zheng, N.; Wang, J. Dairy cows bedding material on the microbial structure and antibiotic resistance genes of milk. Front. Microbiol. 2022. [Google Scholar] [CrossRef]
- Zhang, X.; Ding, Z.; Usman, S.; Zhang, J.; Chen, M.; Guo, X. Metagenomics insights into the effects of lactic acid bacteria inoculation on the biological reduction of antibiotic resistance genes in alfalfa silage. J. Hazard. Mater. 2023, 443, 130329. [Google Scholar] [CrossRef]
- Wu, Z.; Luo, Y.; Bao, J.; Luo, Y.; Yu, Z. Additives affect the distribution of metabolic profile, microbial communities and antibiotic resistance genes in high-moisture sweet corn kernel silage. Bioresour. Technol. 2020, 315, 123821. [Google Scholar] [CrossRef]
- Uyama, T.; Kelton, D.F.; Morrison, E.I.; de Jong, E.; McCubbin, K.D.; Barkema, H.W.; Dufour, S.; Sanchez, J.; Heider, L.C.; LeBlanc, S.J. Cross-sectional study of antimicrobial use and treatment decision for preweaning Canadian dairy calves. JDS Commun. 2022, 3, 72–77. [Google Scholar] [CrossRef]
- Lardé, H.; Dufour, S.; Archambault, M.; Massé, J.; Roy, J.-P.; Francoz, D. An observational cohort study on antimicrobial usage on dairy farms in Quebec, Canada. J. Dairy Sci. 2021, 104, 1864–1880. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed]
- James, C.; James, S.J.; Onarinde, B.A.; Dixon, R.A.; Williams, N. A critical review of amr risks arising as a consequence of using biocides and certain metals in food animal production. Antibiotics 2023, 12, 1569. [Google Scholar] [CrossRef] [PubMed]
- Guardado Servellón, F.J. Investigation of the Co-occurrence of Zinc and Copper Resistance and Antimicrobial Resistance in Escherichia coli from Beef Cattle Production Systems. Master’s Thesis, University of Calgary, Calgary, AB, Canada, 2020. [Google Scholar]
- Wales, A.D.; Davies, R.H. Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics 2015, 4, 567–604. [Google Scholar] [CrossRef] [PubMed]
- Leach, K.A.; Archer, S.C.; Breen, J.E.; Green, M.J.; Ohnstad, I.C.; Tuer, S.; Bradley, A.J. Recycling manure as cow bedding: Potential benefits and risks for UK dairy farms. Vet. J. 2015, 206, 123–130. [Google Scholar] [CrossRef]
- Agga, G.E.; Couch, M.; Parekh, R.R.; Mahmoudi, F.; Appala, K.; Kasumba, J.; Loughrin, J.H.; Conte, E.D. Lagoon, anaerobic digestion, and composting of animal manure treatments impact on tetracycline resistance genes. Antibiotics 2022, 11, 391. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumari, T.; Rajput, M.S.; Baishya, A.; Bhatt, N.; Roy, S. A review: Effect of bedding material on production, reproduction and health and behavior of dairy animals. Int. J. Livest. Res. 2020, 10, 11–20. [Google Scholar]
- Kalantarzadeh, M. Development of a Heat Treatment to Enhance the Antimicrobial Properties of Wood Based Mulches and Animal Bedding Materials; University of Surrey: Guildford, UK, 2013; ISBN 1088310036. [Google Scholar]
- Hogan, J.S.; Raubenolt, L.; McCormick, J.L.; Weiss, W.P. Evaluation of propane flaming for reducing bacterial counts in sand bedding. J. Dairy Sci. 2012, 95, 6152–6159. [Google Scholar] [CrossRef]
- Bradley, A.J.; Leach, K.A.; Green, M.J.; Gibbons, J.; Ohnstad, I.C.; Black, D.H.; Payne, B.; Prout, V.E.; Breen, J.E. The impact of dairy cows’ bedding material and its microbial content on the quality and safety of milk—A cross sectional study of UK farms. Int. J. Food Microbiol. 2018, 269, 36–45. [Google Scholar] [CrossRef]
- Valdramidis, V.P.; Curran, T.P. Production of Medicated Bedding Straw: Challenges and Perspectives; American Society of Agricultural and Biological Engineers (ASABE): St. Joseph, MI, USA, 2011. [Google Scholar]
- Van Vliet, P.C.J.; Reijs, J.W.; Bloem, J.; Dijkstra, J.; De Goede, R.G.M. Effects of cow diet on the microbial community and organic matter and nitrogen content of feces. J. Dairy Sci. 2007, 90, 5146–5158. [Google Scholar] [CrossRef]
- Allison, M.J.; Robinson, I.M.; Dougherty, R.W.; Bucklin, J.A. Grain overload in cattle and sheep: Changes in microbial populations in the cecum and rumen. Am. J. Vet. Res. 1975, 36, 181–185. [Google Scholar] [PubMed]
- Zhu, L.; Lian, Y.; Lin, D.; Lin, G.; Wang, M. The profile and persistence of clinically critical antibiotic resistance genes and human pathogenic bacteria in manure-amended farmland soils. Front. Cell. Infect. Microbiol. 2022, 12, 1732. [Google Scholar] [CrossRef] [PubMed]
- Pornsukarom, S.; Thakur, S. Horizontal dissemination of antimicrobial resistance determinants in multiple Salmonella serotypes following isolation from the commercial swine operation environment after manure application. Appl. Environ. Microbiol. 2017, 83, e01503-17. [Google Scholar] [CrossRef] [PubMed]
- Chekabab, S.M.; Lawrence, J.R.; Alvarado, A.C.; Predicala, B.Z.; Korber, D.R. Piglet gut and in-barn manure from farms on a raised without antibiotics program display reduced antimicrobial resistance but an increased prevalence of pathogens. Antibiotics 2021, 10, 1152. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, J.; Fréchette, A.; Thériault, W.; Dufour, S.; Fravalo, P.; Thibodeau, A. Comparison of microbiota of recycled manure solids and straw bedding used in dairy farms in eastern Canada. J. Dairy Sci. 2022, 105, 389–408. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, M.; Hamelin, L.; Fréchette, A.; Dufour, S.; Roy, D. Effect of recycled manure solids as bedding on bulk tank milk and implications for cheese microbiological quality. J. Dairy Sci. 2020, 103, 128–140. [Google Scholar] [CrossRef]
- Bewley, J.M.; Robertson, L.M.; Eckelkamp, E.A. A 100-Year Review: Lactating dairy cattle housing management. J. Dairy Sci. 2017, 100, 10418–10431. [Google Scholar] [CrossRef]
- Lange, J.L.; Thorne, P.S.; Kullman, G.J. Determinants of culturable bioaerosol concentrations in dairy barns. Ann. Agric. Environ. Med. 1997, 4, 187–194. [Google Scholar]
- EMA Committee for Medicinal Products for Veterinary Use (CVMP) and EFSA Panel on Biological Hazards (BIOHAZ); Murphy, D.; Ricci, A.; Auce, Z.; Beechinor, J.G.; Bergendahl, H.; Breathnach, R.; Bureš, J.; Duarte Da Silva, J.P.; Hederová, J. EMA and EFSA Joint Scientific Opinion on measures to reduce the need to use antimicrobial agents in animal husbandry in the European Union, and the resulting impacts on food safety (RONAFA). EFSA J. 2017, 15, e04666. [Google Scholar]
- Fregonesi, J.A.; Leaver, J.D. Influence of space allowance and milk yield level on behaviour, performance and health of dairy cows housed in strawyard and cubicle systems. Livest. Prod. Sci. 2002, 78, 245–257. [Google Scholar] [CrossRef]
- Leso, L.; Barbari, M.; Lopes, M.A.; Damasceno, F.A.; Galama, P.; Taraba, J.L.; Kuipers, A. Invited review: Compost-bedded pack barns for dairy cows. J. Dairy Sci. 2020, 103, 1072–1099. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Wang, C.; Zhao, Q.; Wang, Y.; Huo, M.; Wang, J.; Wang, S. Prevalence and dissemination of antibiotic resistance genes and coselection of heavy metals in Chinese dairy farms. J. Hazard. Mater. 2016, 320, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Hurst, J.J.; Oliver, J.P.; Schueler, J.; Gooch, C.; Lansing, S.; Crossette, E.; Wigginton, K.; Raskin, L.; Aga, D.S.; Sassoubre, L.M. Trends in antimicrobial resistance genes in manure blend pits and long-term storage across dairy farms with comparisons to antimicrobial usage and residual concentrations. Environ. Sci. Technol. 2019, 53, 2405–2415. [Google Scholar] [CrossRef]
- Sun, H.-Z.; Peng, K.-L.; Xue, M.-Y.; Liu, J.-X. Metagenomics analysis revealed the distinctive ruminal microbiome and resistive profiles in dairy buffaloes. Anim. Microbiome 2021, 3, 44. [Google Scholar] [CrossRef]
- Sun, M.; Ye, M.; Wu, J.; Feng, Y.; Wan, J.; Tian, D.; Shen, F.; Liu, K.; Hu, F.; Li, H. Positive relationship detected between soil bioaccessible organic pollutants and antibiotic resistance genes at dairy farms in Nanjing, Eastern China. Environ. Pollut. 2015, 206, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Eikmeyer, F.G.; Köfinger, P.; Poschenel, A.; Jünemann, S.; Zakrzewski, M.; Heinl, S.; Mayrhuber, E.; Grabherr, R.; Pühler, A.; Schwab, H. Metagenome analyses reveal the influence of the inoculant Lactobacillus buchneri CD034 on the microbial community involved in grass ensiling. J. Biotechnol. 2013, 167, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Botta, C.; Tsoureki, D.; Gautam, A.; Patz, S.; Pissaridi, K.; Ferrocino, I.; Franciosa, I.; Cocolin, L.; Huson, D.H.; Rantsiou, K. The pros and cons of tracking the microbial contamination in infant food processing chain through amplicon sequencing and metagenomics. In Proceedings of the Food System Microbiomes, Turin, Italy, 17–17 May 2024; p. 50. [Google Scholar]
- Lorenzo, J.M.; Munekata, P.E.; Dominguez, R.; Pateiro, M.; Saraiva, J.A.; Franco, D. Main groups of microorganisms of relevance for food safety and stability: General aspects and overall description. In Innovative Technologies for Food Preservation; Elsevier: Amsterdam, The Netherlands, 2018; pp. 53–107. [Google Scholar]
- Dos Santos, R.A.N.; Abdel-Nour, J.; McAuley, C.; Moore, S.C.; Fegan, N.; Fox, E.M. Clostridium perfringens associated with dairy farm systems show diverse genotypes. Int. J. Food Microbiol. 2022, 382, 109933. [Google Scholar] [CrossRef] [PubMed]
- Kung Jr, L.; Shaver, R.D.; Grant, R.J.; Schmidt, R.J. Silage review: Interpretation of chemical, microbial, and organoleptic components of silages. J. Dairy Sci. 2018, 101, 4020–4033. [Google Scholar] [CrossRef] [PubMed]
- Yue, W.-F.; Du, M.; Zhu, M.-J. High temperature in combination with UV irradiation enhances horizontal transfer of stx 2 gene from E. coli O157: H7 to non-pathogenic E. coli. PLoS ONE 2012, 7, e31308. [Google Scholar] [CrossRef]
- Wang, M.; Wang, L.; Yu, Z. Fermentation dynamics and bacterial diversity of mixed lucerne and sweet corn stalk silage ensiled at six ratios. Grass Forage Sci. 2019, 74, 264–273. [Google Scholar] [CrossRef]
- Gurmessa, B.; Milanovic, V.; Pedretti, E.F.; Corti, G.; Ashworth, A.J.; Aquilanti, L.; Ferrocino, I.; Corvaglia, M.R.; Cocco, S. Post-digestate composting shifts microbial composition and degrades antimicrobial resistance genes. Bioresour. Technol. 2021, 340, 125662. [Google Scholar] [CrossRef] [PubMed]
- Lord, J.; Gikonyo, A.; Miwa, A. Antimicrobial resistance among Enterobacteriaceae, Staphylococcus aureus, and Pseudomonas spp. isolates from clinical specimens from a hospital in Nairobi, Kenya. PeerJ 2021, 9, e11958. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tao, X.; Liu, Q.; Zhang, Y.J.; Xu, J.; Zhang, W.; Wang, J.; Zhang, D.; Li, B.; Wang, L. Succession changes of fermentation parameters, nutrient components and bacterial community of sorghum stalk silage. Front. Microbiol. 2022, 13, 982489. [Google Scholar] [CrossRef] [PubMed]
- Kurle, J.E.; Sheaffer, C.C.; Crookston, R.K.; Peterson, R.H.; Chester-Jones, H.; Lueschen, W.E. Popcorn, sweet corn, and sorghum as alternative silage crops. J. Prod. Agric. 1991, 4, 432–436. [Google Scholar] [CrossRef]
- Miller, S.A.; Ferreira, J.P.; LeJeune, J.T. Antimicrobial use and resistance in plant agriculture: A one health perspective. Agriculture 2022, 12, 289. [Google Scholar] [CrossRef]
- Lombardi, A.; Dal Maistro, L.; De Dea, P.; Gatti, M.; Giraffa, G.; Neviani, E. A polyphasic approach to highlight genotypic and phenotypic diversities of Lactobacillus helveticus strains isolated from dairy starter cultures and cheeses. J. Dairy Res. 2002, 69, 139–149. [Google Scholar] [CrossRef]
- Li, R.; Jiang, D.; Zheng, M.; Tian, P.; Zheng, M.; Xu, C. Microbial community dynamics during alfalfa silage with or without clostridial fermentation. Sci. Rep. 2020, 10, 17782. [Google Scholar] [CrossRef]
- Mo, L.; Yu, J.; Jin, H.; Hou, Q.; Yao, C.; Ren, D.; An, X.; Tsogtgerel, T.; Zhang, H. Investigating the bacterial microbiota of traditional fermented dairy products using propidium monoazide with single-molecule real-time sequencing. J. Dairy Sci. 2019, 102, 3912–3923. [Google Scholar] [CrossRef]
- Tsimenidis, S.; Vrochidou, E.; Papakostas, G.A. Omics data and data representations for deep learning-based predictive modeling. Int. J. Mol. Sci. 2022, 23, 12272. [Google Scholar] [CrossRef]
- Medeiros-Silva, J.; Jekhmane, S.; Paioni, A.L.; Gawarecka, K.; Baldus, M.; Swiezewska, E.; Breukink, E.; Weingarth, M. High-resolution NMR studies of antibiotics in cellular membranes. Nat. Commun. 2018, 9, 3963. [Google Scholar] [CrossRef]
- Kok, M.; Maton, L.; van der Peet, M.; Hankemeier, T.; van Hasselt, J.G.C. Unraveling antimicrobial resistance using metabolomics. Drug Discov. Today 2022, 27, 1774–1783. [Google Scholar] [CrossRef] [PubMed]
- Laskaris, P.; Tolba, S.; Calvo-Bado, L.; Wellington, L. Coevolution of antibiotic production and counter-resistance in soil bacteria. Environ. Microbiol. 2010, 12, 783–796. [Google Scholar] [CrossRef] [PubMed]
- Almansour, A.M.; Alhadlaq, M.A.; Alzahrani, K.O.; Mukhtar, L.E.; Alharbi, A.L.; Alajel, S.M. The silent threat: Antimicrobial-resistant pathogens in food-producing animals and their impact on public health. Microorganisms 2023, 11, 2127. [Google Scholar] [CrossRef] [PubMed]
- Dungan, R.S.; McKinney, C.W.; Leytem, A.B. Tracking antibiotic resistance genes in soil irrigated with dairy wastewater. Sci. Total Environ. 2018, 635, 1477–1483. [Google Scholar] [CrossRef] [PubMed]
- Jain, V.K.; Singh, M.; Joshi, V.G.; Chhabra, R.; Singh, K.; Rana, Y.S. Virulence and antimicrobial resistance gene profiles of Staphylococcus aureus associated with clinical mastitis in cattle. PLoS ONE 2022, 17, e0264762. [Google Scholar]
- Han, G.; Zhang, B.; Luo, Z.; Lu, B.; Luo, Z.; Zhang, J.; Wang, Y.; Luo, Y.; Yang, Z.; Shen, L. Molecular typing and prevalence of antibiotic resistance and virulence genes in Streptococcus agalactiae isolated from Chinese dairy cows with clinical mastitis. PLoS ONE 2022, 17, e0268262. [Google Scholar] [CrossRef]
- Sebastianski, M.; Bridger, N.A.; Featherstone, R.M.; Robinson, J.L. Disease outbreaks linked to pasteurized and unpasteurized dairy products in Canada and the United States: A systematic review. Can. J. Public Health 2022, 113, 569–578. [Google Scholar] [CrossRef]
- Lindgren, S.; Pettersson, K.; Kaspersson, A.; Jonsson, A.; Lingvall, P. Microbial dynamics during aerobic deterioration of silages. J. Sci. Food Agric. 1985, 36, 765–774. [Google Scholar] [CrossRef]
- Larsen, H.D.; Jørgensen, K. Growth of Bacillus cereus in pasteurized milk products. Int. J. Food Microbiol. 1999, 46, 173–176. [Google Scholar] [CrossRef]
- Dumalisile, P.; Witthuhn, R.C.; Britz, T.J. Impact of different pasteurization temperatures on the survival of microbial contaminants isolated from pasteurized milk. Int. J. Dairy Technol. 2005, 58, 74–82. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L. Maximum levels of cross-contamination for 24 antimicrobial active substances in non-target feed. Part 12: Tetracyclines: Tetracycline, chlortetracycline, oxytetracycline, and doxycycline. EFSA J. 2021, 19, e06864. [Google Scholar] [PubMed]
- Tang, X.; Lou, C.; Wang, S.; Lu, Y.; Liu, M.; Hashmi, M.Z.; Liang, X.; Li, Z.; Liao, Y.; Qin, W. Effects of long-term manure applications on the occurrence of antibiotics and antibiotic resistance genes (ARGs) in paddy soils: Evidence from four field experiments in south of China. Soil Biol. Biochem. 2015, 90, 179–187. [Google Scholar] [CrossRef]
- Hu, H.-W.; Han, X.-M.; Shi, X.-Z.; Wang, J.-T.; Han, L.-L.; Chen, D.; He, J.-Z. Temporal changes of antibiotic-resistance genes and bacterial communities in two contrasting soils treated with cattle manure. FEMS Microbiol. Ecol. 2016, 92, fiv169. [Google Scholar] [CrossRef] [PubMed]
- Gou, M.; Hu, H.-W.; Zhang, Y.-J.; Wang, J.-T.; Hayden, H.; Tang, Y.-Q.; He, J.-Z. Aerobic composting reduces antibiotic resistance genes in cattle manure and the resistome dissemination in agricultural soils. Sci. Total Environ. 2018, 612, 1300–1310. [Google Scholar] [CrossRef] [PubMed]
- Han, X.-M.; Hu, H.-W.; Chen, Q.-L.; Yang, L.-Y.; Li, H.-L.; Zhu, Y.-G.; Li, X.-Z.; Ma, Y.-B. Antibiotic resistance genes and associated bacterial communities in agricultural soils amended with different sources of animal manures. Soil Biol. Biochem. 2018, 126, 91–102. [Google Scholar] [CrossRef]
- Leclercq, S.O.; Wang, C.; Sui, Z.; Wu, H.; Zhu, B.; Deng, Y.; Feng, J. A multiplayer game: Species of Clostridium, Acinetobacter, and Pseudomonas are responsible for the persistence of antibiotic resistance genes in manure-treated soils. Environ. Microbiol. 2016, 18, 3494–3508. [Google Scholar] [CrossRef]
- Sukhum, K.V.; Vargas, R.C.; Boolchandani, M.; D’souza, A.W.; Patel, S.; Kesaraju, A.; Walljasper, G.; Hegde, H.; Ye, Z.; Valenzuela, R.K. Manure microbial communities and resistance profiles reconfigure after transition to manure pits and differ from those in fertilized field soil. MBio 2021, 12, e00798-21. [Google Scholar] [CrossRef]
- Dixon, M.; Flint, S.; Palmer, J.; Love, R.; Biggs, P.; Beuger, A. Analysis of culturable and non-culturable bacteria and their potential to form biofilms in a primary treated dairy wastewater system. Environ. Technol. 2018, 39, 2185–2192. [Google Scholar] [CrossRef]
- Stefańska, I.; Kwiecień, E.; Jóźwiak-Piasecka, K.; Garbowska, M.; Binek, M.; Rzewuska, M. Antimicrobial susceptibility of lactic acid bacteria strains of potential use as feed additives-the basic safety and usefulness criterion. Front. Vet. Sci. 2021, 8, 687071. [Google Scholar] [CrossRef]
- Andersson, D.I.; Balaban, N.Q.; Baquero, F.; Courvalin, P.; Glaser, P.; Gophna, U.; Kishony, R.; Molin, S.; Tønjum, T. Antibiotic resistance: Turning evolutionary principles into clinical reality. FEMS Microbiol. Rev. 2020, 44, 171–188. [Google Scholar] [CrossRef]
- Davies, J. Inactivation of antibiotics and the dissemination of resistance genes. Science (80-) 1994, 264, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Devirgiliis, C.; Barile, S.; Perozzi, G. Antibiotic resistance determinants in the interplay between food and gut microbiota. Genes Nutr. 2011, 6, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Cui, C.; Li, X.; Yan, J.; Sun, E.; Wang, C.; Guo, H.; Hao, Y. Prevalence, antimicrobial susceptibility, and antibiotic resistance gene transfer of Bacillus strains isolated from pasteurized milk. J. Dairy Sci. 2023, 106, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.; Godden, S.M.; Royster, E.; Crooker, B.A.; Timmerman, J.; Fox, L. Relationships among bedding materials, bedding bacteria counts, udder hygiene, milk quality, and udder health in US dairy herds. J. Dairy Sci. 2019, 102, 10213–10234. [Google Scholar] [CrossRef] [PubMed]
- Lima, T.; Domingues, S.; Da Silva, G.J. Manure as a potential hotspot for antibiotic resistance dissemination by horizontal gene transfer events. Vet. Sci. 2020, 7, 110. [Google Scholar] [CrossRef]
- Boerlin, P.; Reid-Smith, R.J. Antimicrobial resistance: Its emergence and transmission. Anim. Health Res. Rev. 2008, 9, 115–126. [Google Scholar] [CrossRef]
- Omisakin, F.; MacRae, M.; Ogden, I.D.; Strachan, N.J.C. Concentration and prevalence of Escherichia coli O157 in cattle feces at slaughter. Appl. Environ. Microbiol. 2003, 69, 2444–2447. [Google Scholar] [CrossRef]
- Stecher, B.; Denzler, R.; Maier, L.; Bernet, F.; Sanders, M.J.; Pickard, D.J.; Barthel, M.; Westendorf, A.M.; Krogfelt, K.A.; Walker, A.W. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 2012, 109, 1269–1274. [Google Scholar] [CrossRef]
- Oliver, J.P.; Gooch, C.A.; Lansing, S.; Schueler, J.; Hurst, J.J.; Sassoubre, L.; Crossette, E.M.; Aga, D.S. Invited review: Fate of antibiotic residues, antibiotic-resistant bacteria, and antibiotic resistance genes in US dairy manure management systems. J. Dairy Sci. 2020, 103, 1051–1071. [Google Scholar] [CrossRef]
- Nabadda, S.; Kakooza, F.; Kiggundu, R.; Walwema, R.; Bazira, J.; Mayito, J.; Mugerwa, I.; Sekamatte, M.; Kambugu, A.; Lamorde, M. Implementation of the World Health Organization global antimicrobial resistance surveillance system in Uganda, 2015–2020: Mixed-methods study using national surveillance data. JMIR Public Health Surveill. 2021, 7, e29954. [Google Scholar] [CrossRef]
- Fonseca, M.; Heider, L.C.; Léger, D.; Mcclure, J.; Rizzo, D.; Dufour, S.; Kelton, D.F.; Renaud, D.; Barkema, H.W.; Sanchez, J. Canadian Dairy Network for Antimicrobial Stewardship and Resistance (CaDNetASR): An On-Farm Surveillance System. Front. Vet. Sci. 2022, 8, 1653. [Google Scholar] [CrossRef] [PubMed]
- Waglechner, N.; Wright, G.D. Antibiotic resistance: It’s bad, but why isn’t it worse? BMC Biol. 2017, 15, 84. [Google Scholar] [CrossRef] [PubMed]
- Kadri, S.S. Key takeaways from the US CDC’s 2019 antibiotic resistance threats report for frontline providers. Crit. Care Med. 2020, 48, 939–945. [Google Scholar] [CrossRef]
- Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar]
- World Health Organization. 2019 Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Saini, V.; McClure, J.T.; Léger, D.; Dufour, S.; Sheldon, A.G.; Scholl, D.T.; Barkema, H.W. Antimicrobial use on Canadian dairy farms. J. Dairy Sci. 2012, 95, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Scott, H.M.; Acuff, G.; Bergeron, G.; Bourassa, M.W.; Gill, J.; Graham, D.W.; Kahn, L.H.; Morley, P.S.; Salois, M.J.; Simjee, S. Critically important antibiotics: Criteria and approaches for measuring and reducing their use in food animal agriculture. Ann. N. Y. Acad. Sci. 2019, 1441, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Prescott, J.F. Veterinary antimicrobial stewardship in North America. Aust. Vet. J. 2019, 97, 243–248. [Google Scholar] [CrossRef]
- Tsegaye, L.; Huston, P.; Milliken, R.; Hanniman, K.; Nesbeth, C.; Noad, L. Antimicrobial resistance (amr): How is an international public health threat advanced in canada? the case of antimicrobial resistance. Can. Commun. Dis. Rep. 2016, 42, 223. [Google Scholar] [CrossRef]
- Anadón, A. WS14 The EU ban of antibiotics as feed additives (2006): Alternatives and consumer safety. J. Vet. Pharmacol. Ther. 2006, 29, 41–44. [Google Scholar] [CrossRef]
- Roy, J.-P.; Archambault, M.; Desrochers, A.; Dubuc, J.; Dufour, S.; Francoz, D.; Paradis, M.-È.; Rousseau, M. New Quebec regulation on the use of antimicrobials of very high importance in food animals: Implementation and impacts in dairy cattle practice. Can. Vet. J. 2020, 61, 193. [Google Scholar]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Kapley, A. Antibiotic resistance: Global health crisis and metagenomics. Biotechnol. Rep. 2021, 29, e00604. [Google Scholar] [CrossRef] [PubMed]
- Mathew, P.; Sivaraman, S.; Chandy, S. Communication strategies for improving public awareness on appropriate antibiotic use: Bridging a vital gap for action on antibiotic resistance. J. Fam. Med. Prim. Care 2019, 8, 1867–1871. [Google Scholar] [CrossRef] [PubMed]
- Garvey, M. Bacteriophages and food production: Biocontrol and bio-preservation options for food safety. Antibiotics 2022, 11, 1324. [Google Scholar] [CrossRef] [PubMed]
- Masterson, K.; Meade, E.; Garvey, M.; Lynch, M.; Major, I.; Rowan, N.J. Development of a low-temperature extrusion process for production of GRAS bioactive-polymer loaded compounds for targeting antimicrobial-resistant (AMR) bacteria. Sci. Total Environ. 2021, 800, 149545. [Google Scholar] [CrossRef]
- Libante, V.; Nombre, Y.; Coluzzi, C.; Staub, J.; Guédon, G.; Gottschalk, M.; Teatero, S.; Fittipaldi, N.; Leblond-Bourget, N.; Payot, S. Chromosomal conjugative and mobilizable elements in Streptococcus suis: Major actors in the spreading of antimicrobial resistance and bacteriocin synthesis genes. Pathogens 2019, 9, 22. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, Y.; Galgano, S.; Houdijk, J.; Xie, W.; Jin, Y.; Lin, J.; Song, W.; Fu, Y.; Li, X. Recent Progress in Phage Therapy to Modulate Multidrug-Resistant Acinetobacter baumannii, including in Human and Poultry. Antibiotics 2022, 11, 1406. [Google Scholar] [CrossRef]
- Markowiak, P.; Śliżewska, K. The role of probiotics, prebiotics and synbiotics in animal nutrition. Gut Pathog. 2018, 10, 21. [Google Scholar] [CrossRef]
- Cameron, A.; Zaheer, R.; Adator, E.H.; Barbieri, R.; Reuter, T.; McAllister, T.A. Bacteriocin occurrence and activity in Escherichia coli isolated from bovines and wastewater. Toxins 2019, 11, 475. [Google Scholar] [CrossRef]
- Svircev, A.; Roach, D.; Castle, A. Framing the future with bacteriophages in agriculture. Viruses 2018, 10, 218. [Google Scholar] [CrossRef]
- Dias, R.S.; Eller, M.R.; Duarte, V.S.; Pereira, A.L.; Silva, C.C.; Mantovani, H.C.; Oliveira, L.L.; de, A. M Silva, E.; De Paula, S.O. Use of phages against antibiotic-resistant Staphylococcus aureus isolated from bovine mastitis. J. Anim. Sci. 2013, 91, 3930–3939. [Google Scholar] [CrossRef] [PubMed]
- Varela-Ortiz, D.F.; Barboza-Corona, J.E.; González-Marrero, J.; León-Galván, M.F.; Valencia-Posadas, M.; Lechuga-Arana, A.A.; Sánchez-Felipe, C.G.; Ledezma-García, F.; Gutiérrez-Chávez, A.J. Antibiotic susceptibility of Staphylococcus aureus isolated from subclinical bovine mastitis cases and in vitro efficacy of bacteriophage. Vet. Res. Commun. 2018, 42, 243–250. [Google Scholar] [CrossRef] [PubMed]
- De Lucia, A.; Card, R.M.; Duggett, N.; Smith, R.P.; Davies, R.; Cawthraw, S.A.; Anjum, M.F.; Rambaldi, M.; Ostanello, F.; Martelli, F. Reduction in antimicrobial resistance prevalence in Escherichia coli from a pig farm following withdrawal of group antimicrobial treatment. Vet. Microbiol. 2021, 258, 109125. [Google Scholar] [CrossRef] [PubMed]
- Dorado-García, A.; Mevius, D.J.; Jacobs, J.J.H.; Van Geijlswijk, I.M.; Mouton, J.W.; Wagenaar, J.A.; Heederik, D.J. Quantitative assessment of antimicrobial resistance in livestock during the course of a nationwide antimicrobial use reduction in the Netherlands. J. Antimicrob. Chemother. 2016, 71, 3607–3619. [Google Scholar] [CrossRef]
- Howard, S.J.; Catchpole, M.; Watson, J.; Davies, S.C. Antibiotic resistance: Global response needed. Lancet Infect. Dis. 2013, 13, 1001–1003. [Google Scholar] [CrossRef]
| Type | Antimicrobial Classes | Microorganisms | Genes | Analysis Type | Country | References |
|---|---|---|---|---|---|---|
| Bedding | ||||||
| NI * | Beta-lactams, lincosamide, pleuromutilin | B. cereus | NI | Culture dependent (disc diffusion) and Culture independent (PCR) | China | [36] |
| NI | Beta-lactams, cephalosporin sulfonamide, and aminoglycoside | K. pneumoniae | blaTEM, blaSHV, strA, strB, aadA1, and aac(60)-Ib-cr | Culture dependent (microbroth dilution) and Culture independent (PCR) | China | [19] |
| Straw | Beta-lactams, fluoroquinolone, sulfonamide, aminoglycoside, macrolide, and carbapenem | L. monocytogenes | NI | Culture dependent (disc diffusion) | Egypt | [18] |
| Recycled Sand | Beta-lactams, amphenicol, aminoglycoside, sulfonamides, and tetracycline | S. enterica serovar Newport | CMY-2 | Culture dependent (disc diffusion) and Culture independent (PCR) | USA | [34] |
| Silage | ||||||
| Alfalfa | aminoglycoside and diaminopyrimidine | NA ** | aadA2, ant(6)-Ia, ant(9)-Ia, aph(3’)-IIa, aph(3’)-IIIa, dfrG | Culture independent (shotgun metagenome) | China | [31] |
| Alfalfa | vancomycin, aminoglycoside, tetracycline, and fosmidomycin | NA | arlR, emrB-qacA, cpxR, and penA | Culture independent (shotgun metagenome) | China | [38] |
| Alfalfa | oxazolidinone, tetracycline, polypeptide, and amphenicol | NA | macB, optrA, tetA, bcrA, efrA, patB, tetM and tetW | Culture independent (shotgun metagenome and 16S metabarcoding) | China | [15] |
| Corn | fluoroquinolone, beta-lactam, aminoglycoside, polypeptide, and fosfomycin | NA | tetW.N.W, tetT, tetA46, tlrC, and erm41 | Culture independent (shotgun metagenome) | China | [39] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarrah, A.; Zhang, D.; Darvishzadeh, P.; LaPointe, G. The Contribution of Dairy Bedding and Silage to the Dissemination of Genes Coding for Antimicrobial Resistance: A Narrative Review. Antibiotics 2024, 13, 905. https://doi.org/10.3390/antibiotics13090905
Tarrah A, Zhang D, Darvishzadeh P, LaPointe G. The Contribution of Dairy Bedding and Silage to the Dissemination of Genes Coding for Antimicrobial Resistance: A Narrative Review. Antibiotics. 2024; 13(9):905. https://doi.org/10.3390/antibiotics13090905
Chicago/Turabian StyleTarrah, Armin, Dong Zhang, Pariya Darvishzadeh, and Gisèle LaPointe. 2024. "The Contribution of Dairy Bedding and Silage to the Dissemination of Genes Coding for Antimicrobial Resistance: A Narrative Review" Antibiotics 13, no. 9: 905. https://doi.org/10.3390/antibiotics13090905












