Cloning, Expression, and Characterization of a Metalloprotease from Thermophilic Bacterium Streptomyces thermovulgaris
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Culture Growth
2.2. Cloning and Expression of nprB Gene
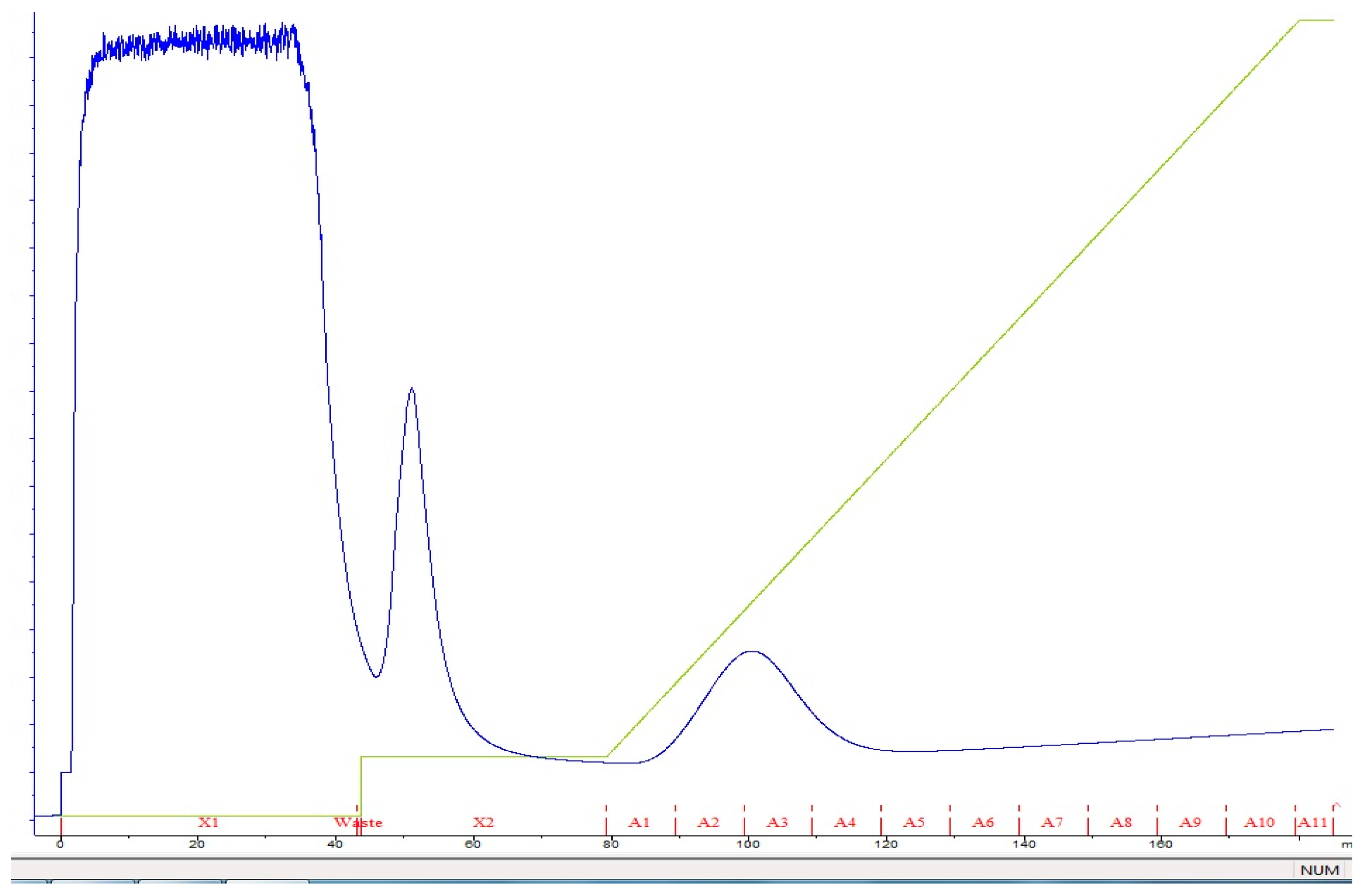
2.3. Purification of Recombinant Thermophilic Neutral Protease
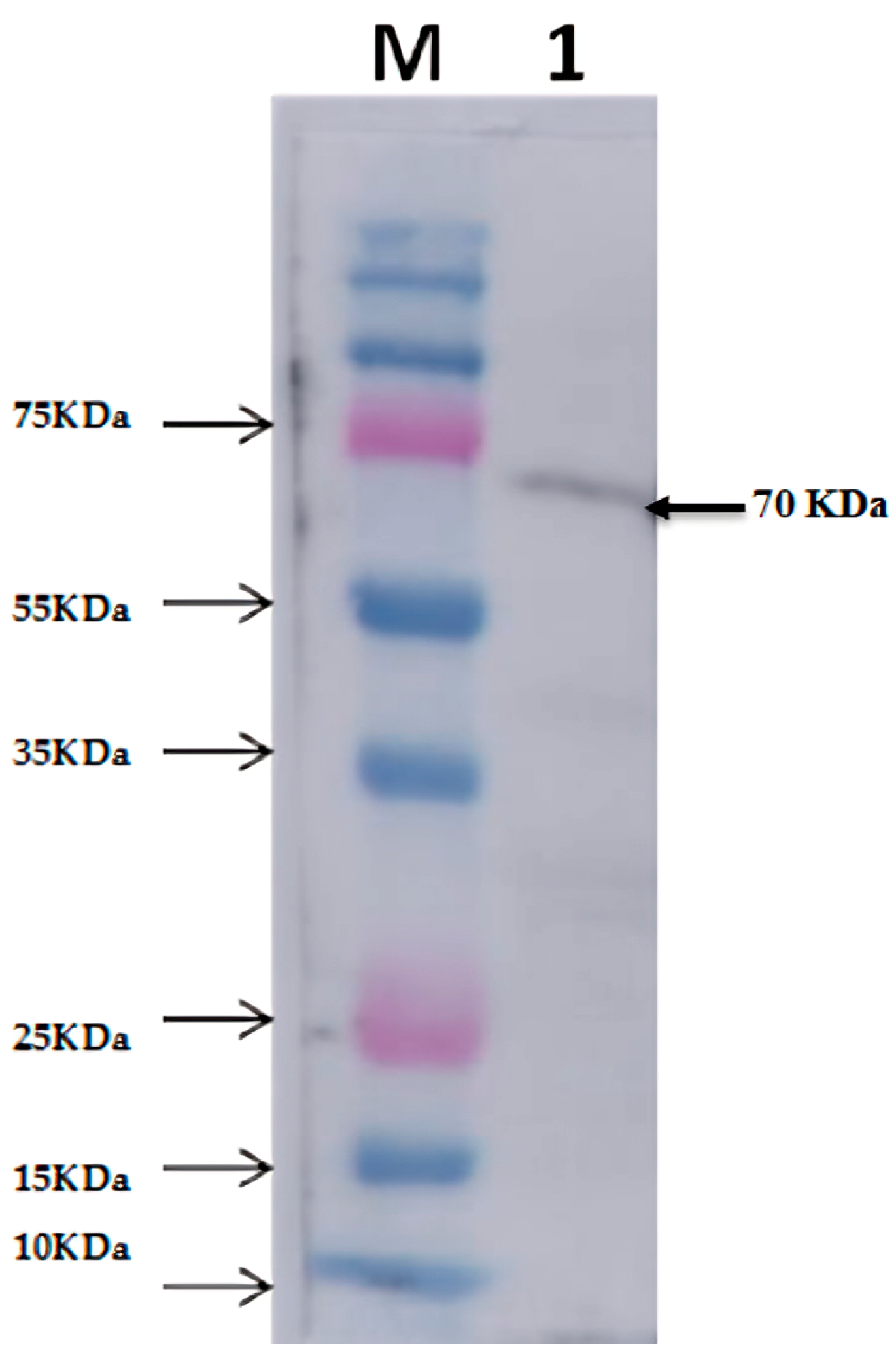
2.4. Western Blot Analysis of Thermophilic Neutral Protease
2.5. Protease Activity Assay
2.6. Characterization of Thermophilic Neutral Protease
2.6.1. Determination of Optimum pH and pH Stability
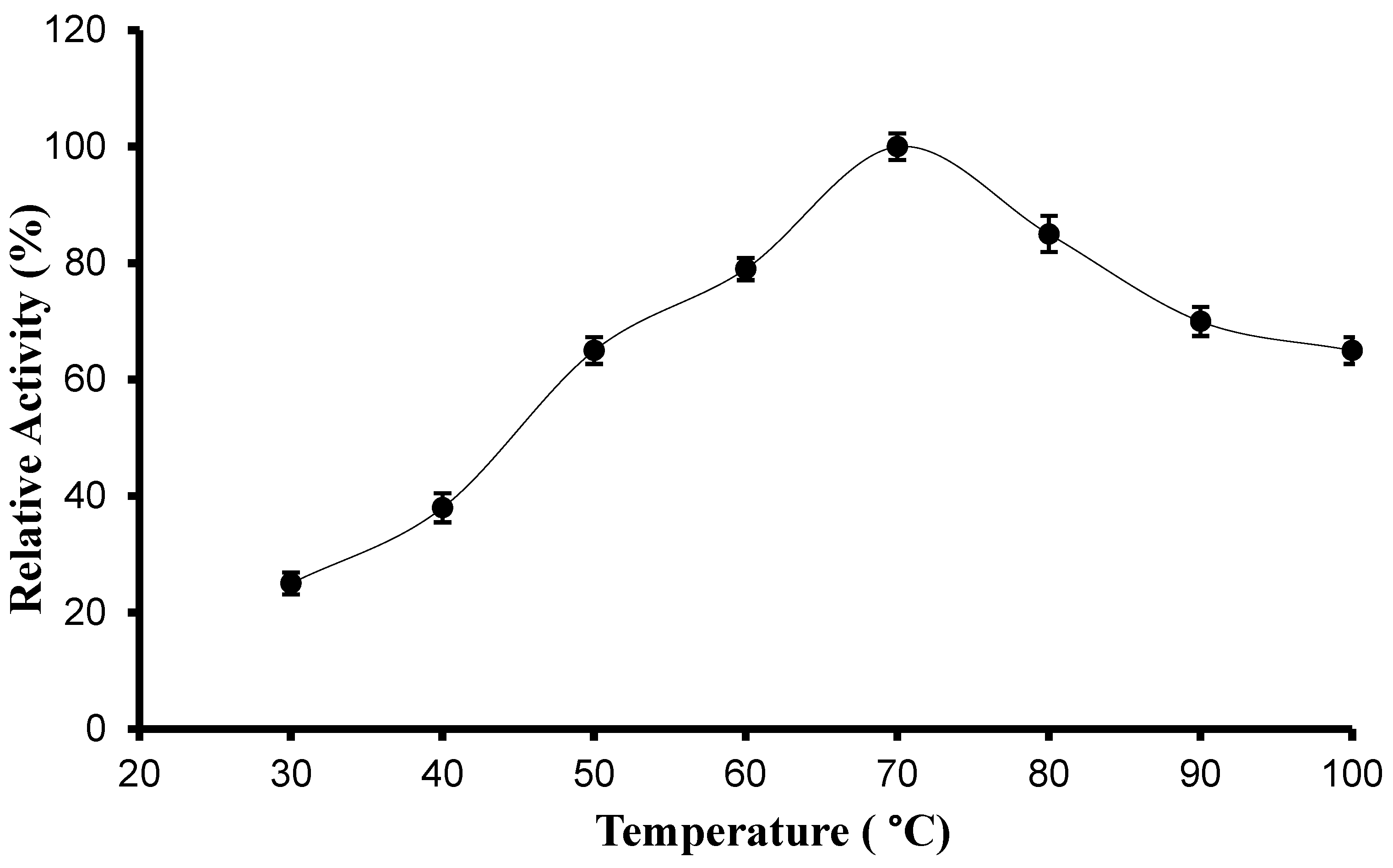
2.6.2. Determination of Optimum Temperature and Thermal Stability
2.6.3. Enzyme Substrate Specificity and Kinetic Parameters (Km and Vmax Determination)
2.6.4. Effect of Various Metals Ions and Organic Solvents
2.6.5. Effect of Detergents and Inhibitors on Protease Activity
3. Results
3.1. Cloning and Sequence Analysis of Neutral Protease B (nprB)
3.2. Expression and Purification of Recombinant Neutral Protease B
3.3. Characterization of Recombinant Neutral Protease B
3.3.1. pH, Temperature Optima, and Stability
3.3.2. Substrate Specificity and Kinetic Properties
3.3.3. Effect of Metal Ions on Recombinant Protease Activity
3.3.4. Effect of Organic Solvents on Recombinant Protease Activity
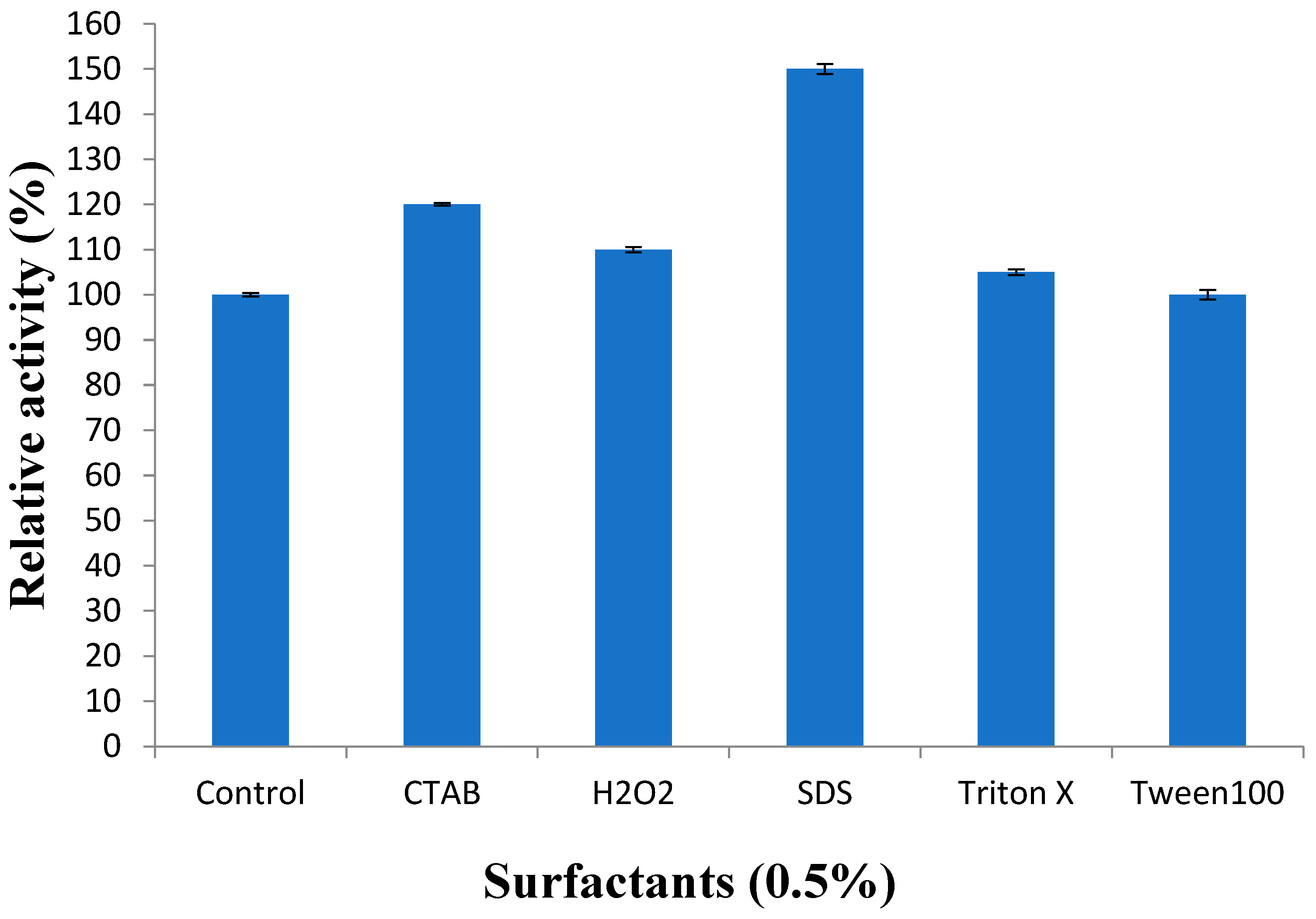
3.3.5. Effect of Surfactants on Recombinant Protease Activity
3.3.6. Effect of Inhibitors on Recombinant Protease Activity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmed, S.; Jabeen, A.; Jamil, A. Xylanase from Trichoderma harzianum: Enzyme characterization and gene isolation. J. Chem. Soc. Pak. 2007, 29, 176–182. [Google Scholar]
- Ahmed, S.; Bashir, A.; Saleem, H.; Saadia, M.; Jamil, A. Production and purification of cellulose-degrading enzymes from a filamentous fungus Trichoderma harzianum. Pak. J. Bot. 2009, 41, 1411–1419. [Google Scholar]
- Ahmed, S.; Imdad, S.S.; Jamil, A. Comparative study for the kinetics of extracellular xylanases from Trichoderma harzianum and Chaetomium thermophilum. Electron. J. Biotechnol. 2012, 15. [Google Scholar] [CrossRef]
- Wu, X.; Ahmed, S.; Cui, X.; Hang, J.; Wang, S.; Liu, S.; Fang, Y. Expression and characterization of a novel organic solvent tolerant protease from Bacillus sphaericus DS11. Prep. Biochem. Biotechnol. 2021, 51, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Li, Z.; Liu, J.; Sun, Y.; Jia, X.; Xuan, Y.H.; Zhang, J.; Jeon, C.O. A Zinc-Dependent Protease AMZ-tk from a Thermophilic Archaeon is a New Member of the Archaemetzincin Protein Family. Front. Microbiol. 2015, 6, 1380. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Patil, R.; Jadhav, B. Isolation and characterization of protease-producing Bacillus species from soil of dairy industry. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 853–860. [Google Scholar] [CrossRef]
- Srivastava, N.; Shiburaj, S.; Khare, S.K. Improved production of alkaline and solvent-stable proteases from a halotolerant Exiguobacterium isolate through heterologous expression. Int. J. Biol. Macromol. 2024, 260 Pt 1, 129507. [Google Scholar] [CrossRef] [PubMed]
- Hou, E.; Xia, T.; Zhang, Z.; Mao, X. Purification and characterization of an alkaline protease from Micrococcus sp. isolated from the South China Sea. J. Ocean. Univ. China 2017, 16, 319–325. [Google Scholar] [CrossRef]
- Kasana, R.C.; Salwan, R.; Yadav, S.K. Microbial proteases: Detection, production, and genetic improvement. Crit. Rev. Microbiol. 2011, 37, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Sharma, N.K.; Thakur, N.; Savitri Bhalla, T.C. Organic solvent tolerant metallo protease of novel isolate Serratia marcescens PPB-26: Production and characterization. 3 Biotech 2016, 6, 180. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yuzuki, M.; Matsushima, K.; Koyama, Y. Expression of key hydrolases for soy sauce fermentation in Zygosaccharomyces rouxii. J. Biosci. Bioeng. 2015, 119, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, B.; Zheng, Y.; Shan, A.; Cheng, B. Neutral protease expression and optimized conditions for the degradation of blood cells using recombinant Pichia pastoris. Int. Biodeterior. Biodegrad. 2014, 93, 235–240. [Google Scholar] [CrossRef]
- Umeadi, C.; Kandeel, F.; Al-Abdullah, I.H. Ulinastatin is a novel protease inhibitor and neutral protease activator. Transpl. Proc. 2008, 40, 387–389. [Google Scholar] [CrossRef] [PubMed]
- Neog, P.R.; Yadav, M.; Konwar, B.K. Cloning, expression, and characterization of a surfactant-stable alkaline serine protease (KNBSSP1) from Bacillus safensis PRN1 with remarkable applications in laundry and leather industries. Biocatal. Agric. Biotechnol. 2023, 54, 102935. [Google Scholar] [CrossRef]
- Qureshi, A.S.; Simair, A.A.; Ali, C.H.; Khushk, I.; Khokhar, J.A.; Ahmad, A.; Danish, M.; Lu, C. Production, purification and partial characterization of organo-solvent tolerant protease from newly isolated Bacillus sp. BBXS-2. Ferment. Technol. 2018, 7, 1. [Google Scholar] [CrossRef]
- Wang, J.; Xu, A.; Wan, Y.; Li, Q. Purification and characterization of a new metallo-neutral protease for beer brewing from Bacillus amyloliquefaciens SYB-001. Appl. Biochem. Biotechnol. 2013, 170, 2021–2033. [Google Scholar] [CrossRef] [PubMed]
- Barret, A. Proteolytic enzymes: Aspartic and metallo-peptidases. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1995. [Google Scholar]
- Miyoshi, S.-I.; Shinoda, S. Microbial metalloproteases and pathogenesis. In Microbes and Infection; Elsevier: Amsterdam, The Netherlands, 2000. [Google Scholar]
- Song, P.; Xu, W.; Wang, K.; Zhang, Y.; Wang, F.; Zhou, X.; Shi, H.; Feng, W. Cloning, expression and characterization of metalloproteinase HypZn from Aspergillus niger. PLoS ONE 2021, 16, e0259809. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yeoman, K.H.; Edwards, C. Protease production by Streptomyces thermovulgaris grown on rapemeal-derived media. J. Appl. Bacteriol. 1994, 77, 264–270. [Google Scholar] [CrossRef] [PubMed]
- James, P.D.; Edwards, C. The effects of temperature on growth and production of the antibiotic granaticin by a thermotolerant streptomycete. J. Gen. Microbiol. 1989, 135, 1997–2003. [Google Scholar] [CrossRef] [PubMed]
- Bar-Shimon, M.; Yehuda, H.; Cohen, L.; Weiss, B.; Kobeshnikov, A.; Daus, A.; Goldway, M.; Wisniewski, M.; Droby, S. Characterization of extracellular lytic enzymes produced by the yeast biocontrol agent Candida oleophila. Curr. Genet. 2004, 45, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Arulmani, M.; Aparanjini, K.; Vasanthi, K.; Arumugam, P.; Arivuchelvi, M.P. Thangavelu Kalaichelvanet TP. Purification and partial characterization of serine protease from thermostable alkalophilic Bacillus laterosporus-AK1. World J. Microbiol. Biotechnol. 2007, 23, 475–481. [Google Scholar] [CrossRef]
- Bollag, D.M.; Edelstein, S. Protein Methods; A John Wiley & Sons. Inc. Publication: Hoboken, NJ, USA, 1991. [Google Scholar]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef]
- Fricke, B.; Drössler, K.; Willhardt, I.; Schierhorn, A.; Menge, S.; Rücknagel, P. The cell envelope-bound metalloprotease (camelysin) from Bacillus cereus is a possible pathogenic factor. Biochim. Biophys. Acta 2001, 28, 132–146. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, F.; Tayyab, M.; Mehmood, T.; Naseer, R.; Iqbal, S. Optimization of Fermentative Parameters for Hyperproduction of Protease from Aspergillus viridi using Lignocellulosic Byproducts as Sole Substrate. Waste Biomass Valorization 2024, 15, 3761–3771. [Google Scholar] [CrossRef]
- Thebti, W.; Riahi, Y.; Belhadj, O. Purification and characterization of a new thermostable, haloalkaline, solvent stable, and detergent compatible serine protease from Geobacillus toebii strain LBT 77. BioMed Res. Int. 2016, 2016, 9178962. [Google Scholar] [CrossRef]
- Lineweaver, H.; Burk, D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]
- Bhatt, H.B.; Singh, S.P. Cloning, Expression, and Structural Elucidation of a Biotechnologically Potential Alkaline Serine Protease From a Newly Isolated Haloalkaliphilic Bacillus lehensis JO-26. Front. Microbiol. 2020, 11, 941. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mushtaq, A.; Ansari, T.M.; Mustafa, G.; Shad, M.A.; Cruz-Reyes, J.; Jamil, A. Isolation and characterization of nprB, a novel protease from Streptomyces thermovulgaris. Pak. J. Pharm. Sci. 2020, 33, 2361–2369. [Google Scholar] [PubMed]
- Ahmed, S.; Mustafa, G.; Arshad, M.; Rajoka, M.I. Fungal Biomass Protein Production from Trichoderma harzianum Using Rice Polishing. Biomed Res. Int. 2017, 2017, 6232793. [Google Scholar] [CrossRef]
- Ahmed, S.; Riaz, S.; Jamil, A. Molecular cloning of fungal xylanases: An overview. Appl. Microbiol. Biotechnol. 2009, 84, 19–35. [Google Scholar] [CrossRef]
- Laili, N.; Antonius, S. Production and characterization of extracellular protease from Bacillus sp. 140-B isolated from a pineapple plantation in Lampung, Indonesia. KnE Life Sci. 2017, 3, 170–176. [Google Scholar] [CrossRef]
- Mergulhão, F.J.; Summers, D.K.; Monteiro, G.A. Recombinant protein secretion in Escherichia coli. Biotechnol. Adv. 2005, 23, 177–202. [Google Scholar] [CrossRef] [PubMed]
- Nag, N.; Khan, H.; Tripathi, T. Strategies to improve the expression and solubility of recombinant proteins in E. coli. In Advances in Protein Molecular and Structural Biology Methods; Academic Press: Cambridge, MA, USA, 2022; pp. 1–12. [Google Scholar]
- Valasaki, K.; Staikou, A.; Theodorou, L.G.; Charamopoulou, V.; Zacharaki, P.; Papamichael, E.M. Purification and kinetics of two novel thermophilic extracellular proteases from Lactobacillus helveticus, from kefir with possible biotechnological interest. Bioresour. Technol. 2008, 99, 5804–5813. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Hakim, A.; Hossain, M.S.; Rahman, M.R.; Islam, K.; Azim, M.F.; Ahmed, J.; Assaduzzaman, M.; Hoq, M.M.; Azad, A.K. Partial purification and characterization of serine protease produced through fermentation of organic municipal solid wastes by Serratia marcescens A3 and Pseudomonas putida A2. J. Genet. Eng. Biotechnol. 2018, 16, 29–37. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Allison, S.D.; AdeelaYasid, N.; Shariff, F.M.; Abdul Rahman, N. Molecular Cloning, Characterization, and Application of Organic Solvent-Stable and Detergent-Compatible Thermostable Alkaline Protease from Geobacillus thermoglucosidasius SKF4. J. Microbiol. Biotechnol. 2024, 34, 436–456. [Google Scholar] [CrossRef] [PubMed]
- Hutadilok-Towatana, N.; Painupong, A.; Suntinanalert, P. Purification and characterization of an extracellular protease from alkaliphilic and thermophilic Bacillus sp. PS719. J. Biosci. Bioeng. 1999, 87, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.S.; Goodfellow, B.J.; Benesch, J.; Rocha, J.; Mano, J.; Reis, R.L. Morphology and miscibility of chitosan/soy protein blended membranes. Carbohydr. Polym. 2007, 70, 25–31. [Google Scholar] [CrossRef]
- Akel, H.; Al-Quadan, F.; Yousef, T.K. Characterization of a purified thermostable protease from hyperthermophilic Bacillus strain HUTBS71. Eur. J. Sci. Res. 2009, 31, 280–288. [Google Scholar]
- Zhang, S.; Lv, J. Purification and properties of heat-stable extracellular protease from Pseudomonads fluorescens BJ-10. J. Food Sci. Technol. 2014, 51, 1185–1190. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jayakumar, R.; Jayashree, S.; Annapurna, B.; Seshadri, S. Characterization of thermostable serine alkaline protease from an alkaliphilic strain Bacillus pumilus MCAS8 and its applications. Appl. Biochem. Biotechnol. 2012, 168, 1849–1866. [Google Scholar] [CrossRef] [PubMed]
- Anandharaj, M.; Sivasankari, B.; Siddharthan, N.; Rani, R.P.; Sivakumar, S. Production, Purification, and Biochemical Characterization of Thermostable Metallo-Protease from Novel Bacillus alkalitelluris TWI3 Isolated from Tannery Waste. Appl. Biochem. Biotechnol. 2016, 178, 1666–1686. [Google Scholar] [CrossRef] [PubMed]
- Sookkheo, B.; Sinchaikul, S.; Phutrakul, S.; Chen, S.T. Purification and characterization of the highly thermostable proteases from Bacillus stearothermophilus TLS33. Protein Expr. Purif. 2000, 20, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.; Dodia, M.; Joshi, R.; Singh, S. Production of Extracellular Halo-alkaline Protease from a Newly Isolated Haloalkaliphilic Bacillus sp. Isolated from Seawater in Western India. World J. Microbiol. Biotechnol. 2006, 22, 375–382. [Google Scholar] [CrossRef]
- Esakkiraj, P.; Meleppat, B.; Lakra, A.K.; Ayyanna, R.; Aru, V. Cloning, expression, characterization, and application of protease produced by Bacillus cereus PMW8. RSC Adv. 2016, 6, 8611–38616. [Google Scholar] [CrossRef]
- Chopra, A.K.; Mathur, D.K. Factors Affecting Protease Production by Bacillus stearothermophilus RM-67. J. Food Prot. 1983, 46, 1020–1025. [Google Scholar] [CrossRef] [PubMed]
- Guangrong, H.; Tiejing, Y.; Po, H.; Jiaxing, J. Purification and characterization of a protease from thermophilic Bacillus strain HS08. Afr. J. Biotechnol. 2006, 5, 2433–2438. [Google Scholar] [CrossRef]
- Johnvesly, B.; Naik, G.R. Production of Bleach-Stable and Halo-Tolerant Alkaline Protease by an Alkalophilic Bacillus pumilus JB05 Isolated from Cement Industry Effluents. J. Microbiol. Biotechnol. 2001, 11, 558–563. [Google Scholar]
- Gupta, A.; Roy, I.; Khare, S.K.; Gupta, M.N. Purification and characterization of a solvent stable protease from Pseudomonas aeruginosa PseA. J. Chromatogr. A 2005, 1069, 155–161. [Google Scholar] [CrossRef] [PubMed]
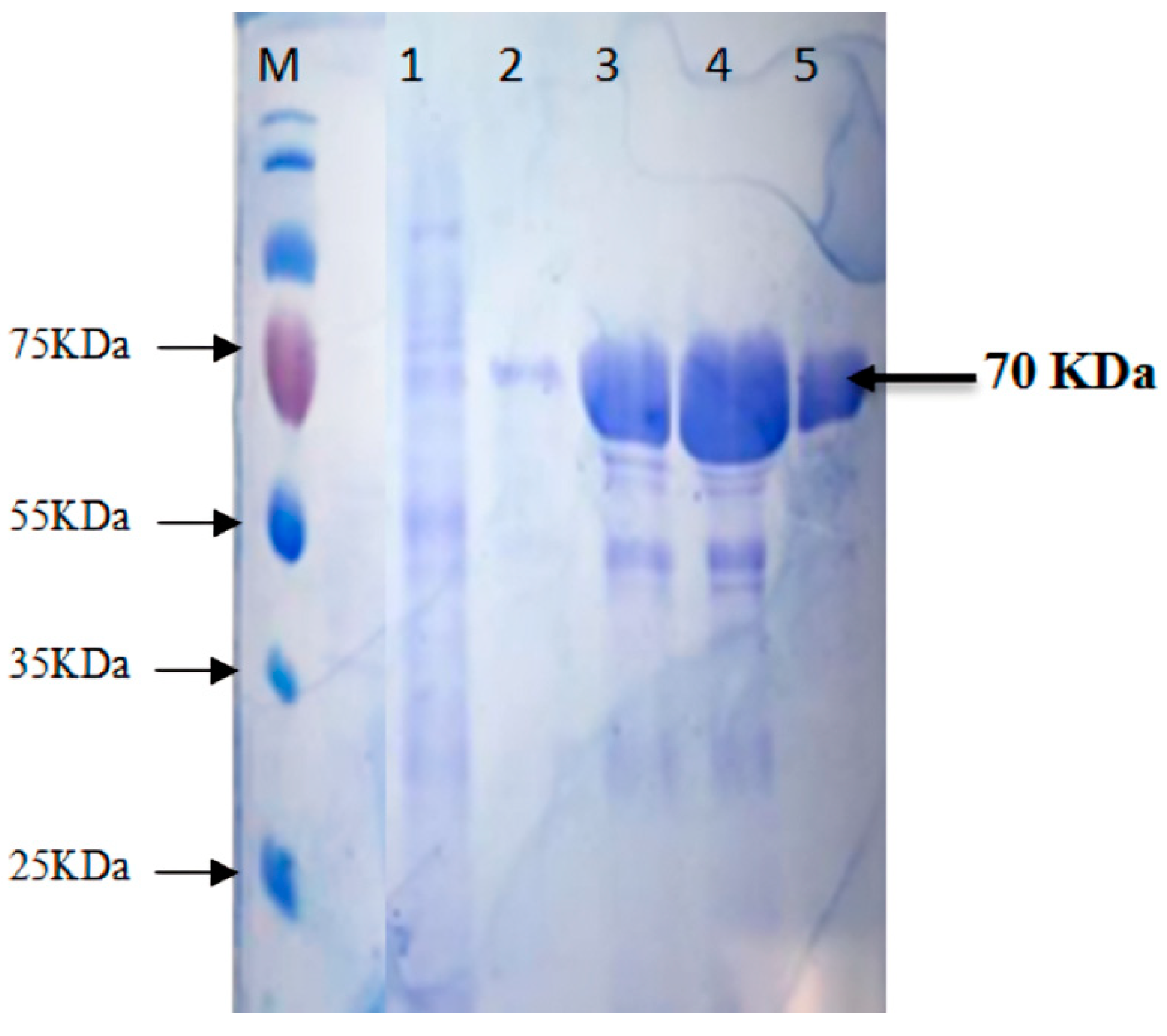

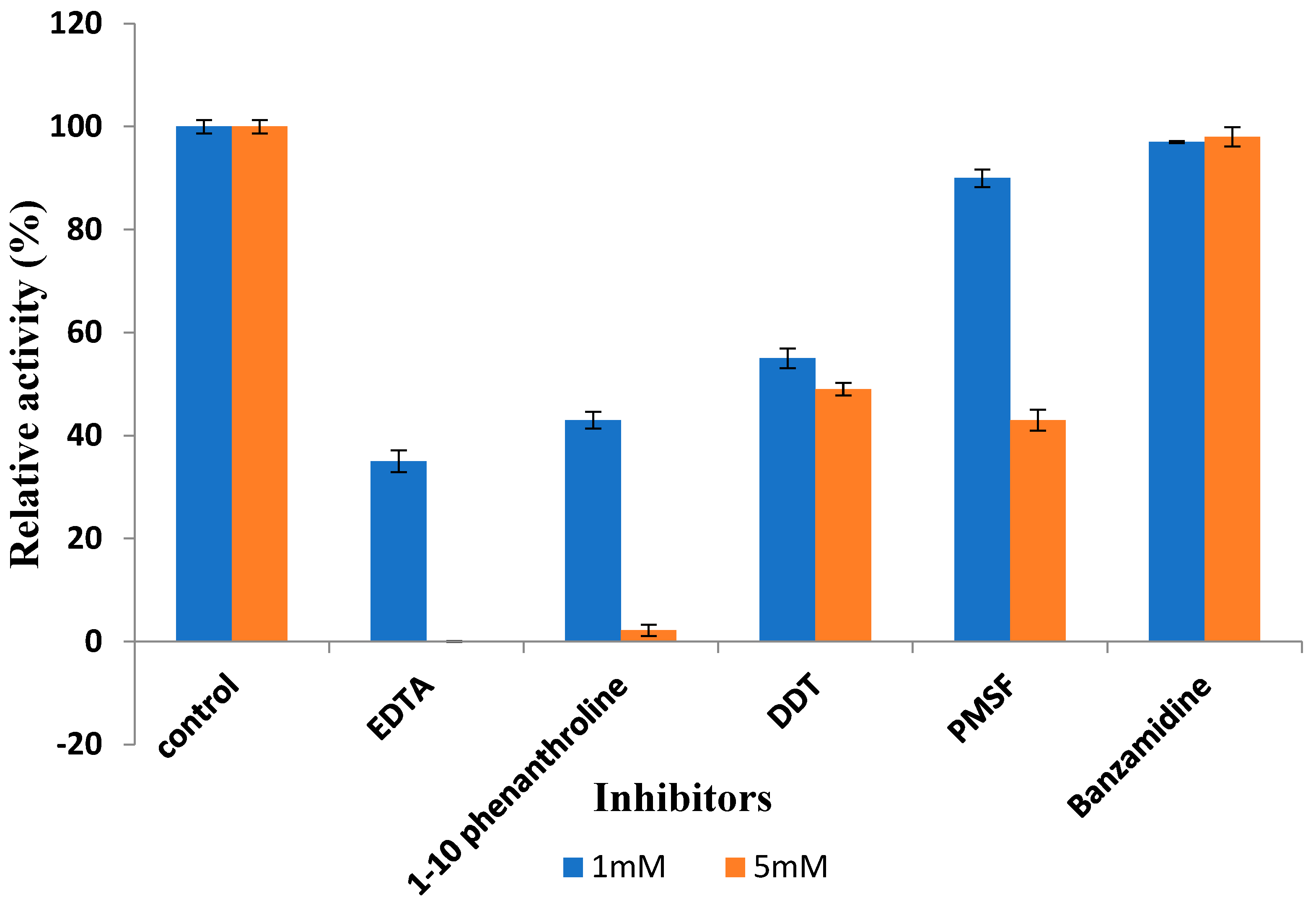
| Purification Steps | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Purification Fold | Recovery Rate (%) |
|---|---|---|---|---|---|
| Crude extract | 82 | 99,867.5 | 2155.6 | - | 100 |
| Ammonium sulfate precipitation (80%) | 65.3 | 90,875.4 | 2570.8 | 2.5 | 80.5 |
| FPLC Ni-coated Hi-trap DEAE Sephadex | 45.9 | 81,641.6 | 5698.5 | 15 | 90.5 |
| Metal Ion | Relative Protease Activity (%) | |
|---|---|---|
| Concentration (mM) | ||
| 2 mM | 10 mM | |
| Cu2+ | 95.0 ± 0.1 | 90.0 ± 1.2 |
| Na+2 | 90.4 ± 0.15 | 91 ± 0.8 |
| Mg2+ | 90.1 ± 0.18 | 89.0 ± 0.09 |
| Ni2+ | 58.5 ± 0.19 | 50.2 ± 1.7 |
| Hg2+ | 78.9 ± 0.2 | 70.0 ± 0.3 |
| Ca2+ | 85.2 ± 1.6 | 100 ± 0.01 |
| Mn2+ | 170.2 ± 1.13 | 184.0 ± 0.2 |
| Fe2+ | 190.6 ± 1.3 | 198.0 ± 1.6 |
| Zn2+ | 75.5 ± 0.02 | 105.0 ± 0.11 |
| Organic Solvents | Concentration | Relative Protease Activity (%) |
|---|---|---|
| Control | 0 | 100 |
| Ethanol | 10% | 90.5 ± 0.9 |
| Methanol | 10% | 95.4 ± 1.1 |
| Hexane | 10% | 91.5 ± 0.8 |
| Benzene | 10% | 105 ± 1.0 |
| DMSO | 10% | 80 ± 0.9 |
| Chloroform | 10% | 105 ± 1.1 |
| Toluene | 10% | 88 ± 0.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mushtaq, A.; Ahmed, S.; Mehmood, T.; Cruz-Reyes, J.; Jamil, A.; Nawaz, S. Cloning, Expression, and Characterization of a Metalloprotease from Thermophilic Bacterium Streptomyces thermovulgaris. Biology 2024, 13, 619. https://doi.org/10.3390/biology13080619
Mushtaq A, Ahmed S, Mehmood T, Cruz-Reyes J, Jamil A, Nawaz S. Cloning, Expression, and Characterization of a Metalloprotease from Thermophilic Bacterium Streptomyces thermovulgaris. Biology. 2024; 13(8):619. https://doi.org/10.3390/biology13080619
Chicago/Turabian StyleMushtaq, Amna, Sibtain Ahmed, Tahir Mehmood, Jorge Cruz-Reyes, Amer Jamil, and Shafaq Nawaz. 2024. "Cloning, Expression, and Characterization of a Metalloprotease from Thermophilic Bacterium Streptomyces thermovulgaris" Biology 13, no. 8: 619. https://doi.org/10.3390/biology13080619
APA StyleMushtaq, A., Ahmed, S., Mehmood, T., Cruz-Reyes, J., Jamil, A., & Nawaz, S. (2024). Cloning, Expression, and Characterization of a Metalloprotease from Thermophilic Bacterium Streptomyces thermovulgaris. Biology, 13(8), 619. https://doi.org/10.3390/biology13080619






