Ecology of Subglacial Lake Vostok (Antarctica), Based on Metagenomic/Metatranscriptomic Analyses of Accretion Ice
Abstract
:1. Introduction

| Taxon | Unique gene sequences | Unique rRNA gene sequences a | Ecology and physiology b | Species characteristics b | |
|---|---|---|---|---|---|
| ≥200 nt <200 nt | |||||
| BACTERIA | 3495 | 2535 | 460 | ||
| Acidobacteria | 2 | 1 | 0 | acidophilic, soil, adaptable | chemoorganotrophic heterotrophs |
| Actinobacteria | 228 | 151 | 24 | thermophilic, halotolerant, psychrotolerant, alkalaitolerant, psychrophilic, Antarctic, deep sea sediments, lake sediments, some grow on limestone | nitrogen fixation, nitrite oxidation, ammonia oxidation, organic decomposition, heterotrophs |
| Bacterioidetes/Chlorobi | 88 | 61 | 8 | aquatic, sediments, thermophilic, psychrophilic, alkalaiphilic, anaerobic | carbon fixation (use sulfide ions, hydrogen or ferrous ions), reductive TCA cycle |
| Chloroflexi | 1 | 0 | 0 | aerobic, thermophilic | carbon fixation using the 3-hydroxylpropionic bicycle |
| Cyanobacteria | 228 | 144 | 60 | common in Antarctic lakes, at least one is thermophilic (Thermosynecoccus sp.) | carbon fixation using the reductive pentose phosphate cycle, some are from anoxygenic ancestors |
| Deferribacteres | 1 | 1 | 0 | animal intestines, anaerobic | chemoorganotrophic heterotrophic |
| Deinococcus/ Thermus | 5 | 1 | 1 | thermophilic, radiophilic, aerobic some associate with cyanobacteria | chemoorganotrophic heterotrophic |
| Fibrobacteres | 1 | 0 | 1 | anaerobic, inhabit animal intestines | chemoorganotrophic heterotrophic |
| Firmicutes | 602 | 401 | 40 | Spore formers, common in extreme environments, thermophiles, mesophilic, psychrophilic, psychrotolerant, halophilic, hot springs, deep sea thermophilic, anaerobic, aerobic | heterotrophic |
| Fusobacteria | 10 | 8 | 0 | parasitic on animals, anaerobic | chemoorganotrophic heterotrophic |
| Planctomycetes | 6 | 2 | 2 | Fresh, brackish and saline lakes/ponds, anaerobic | chemoautolithotrophic anammox, nitrite reduction using ammonium as electron donor |
| Proteobacteria | 474 | 265 | 46 | ||
| Alphaproteobacteria | 91 | 45 | 7 | Psychrophilic, mesophilic, thermophilic, Antarctic lakes, animal symbionts, aerobic, soil/sediments, aquatic, alkalaitolerant, require calcium, marine, halotolerant | nitrite reduction, nitrifying bacteria, denitrification (nitrate to nitrogen gas), methylotrophic, use inorganic sulfur, oxidize sulfate and thiosulfate, carbon fixation using the reductive pentose phosphate cycle, carbon fixation using the reductive TCA cycle |
| Betaproteobacteria | 105 | 31 | 7 | thermophilic, mesophilic, psychrophilic, aquatic, aerobic, highly adaptable | nitrogen fixation, nitrate reduction, ammonia oxidation, carbon fixation using the reductive pentose phosphate cycle, manganese oxidation, iron oxidation, inorganic sulfur oxidation, arsenic oxidation |
| Deltaproteobacteria | 10 | 5 | 0 | aquatic, soil, mesophilic, anaerobic, aerobic, freshwater debris, predator of Gram-negative bacteria, halotolerant, marine | carbon fixation using the reductive TCA cycle, iron reduction, sulfur reduction, ethanol fermentation |
| Epsilonproteobacteria | 6 | 3 | 2 | Some animal associated, mesophilic, thermophilic, aerobic, anaerobic | carbon fixation using the reductive TCA cycle |
| Gammaproteobacteria | 254 | 176 | 28 | thermophilic, mesophilic, psychrophilic, psychrotolerant, aerobic, anaerobic, peizophilic, deep sea, halophilic, polar ice, soil, sediments, permafrost, 33 distinct sequences from species of Psychrobacter, 10 distinct sequences from species of Halomonas (halophilic), some produce intracellular gas vesicles, some are animal associated | nitrogen fixation, nitrate reduction, nitrite respiration, denitrification, sulfur oxidation, chemolithoautotrophs, iron oxidation, mineralization of aromatics, carbon fixation using the reductive pentose phosphate cycle |
| Uncultured Proteobacteria | 8 | 5 | 2 | unknown | unknown |
| Spirochaetes | 3 | 3 | 0 | animal pathogens | heterotrophic |
| Tenericutes | 4 | 4 | 0 | saprobes and arthropod pathogens/symbionts, anaerobic | heterotrophic |
| Verrucomicrobia | 3 | 1 | 0 | freshwater, soil, symbionts of protists and nematodes, aerobic | heterotrophic |
| Uncultured Bacteria | 1839 | 1492 | 278 | Sequences similar to those from uncultured and unidentified species, many from other environmental metagenomic studies | Unknown |
| ARCHAEA | 2 | 0 | 0 | deep hydrate-bearing sediment, peizotolerant, psychrotolerant | Methanotrophic, carbon fixation using the reductive acetyl-CoA pathway |
| EUKARYA | 221 | 124 | 27 | ||
| Amoebozoa | 1 | 1 | 0 | Nolandella sp.; aquatic; feed on bacteria, diatoms, nematodes, fungi, protozoans and organic matter | Heterotrophic |
| Archaeplastida | 74 | 28 | 9 | ||
| Chlorophyta | 10 | 5 | 4 | Antarctic and polar green algal species | carbon fixation using the reductive pentose phosphate cycle |
| Rhodophyta | 1 | 0 | 0 | Antarctic red alga | carbon fixation using the reductive pentose phosphate cycle |
| Streptophyta | 63 | 23 | 5 | Pollen from lake sediments or from glacial deposition? | (carbon fixation using the reductive pentose phosphate cycle)—non-viable? |
| Chromalveolata | 12 | 6 | 2 | diatoms, heterokonts, predatory protists, dinoflagellates, ciliates, Antarctic, aquatic | carbon fixation using the reductive pentose phosphate cycle, heterotrophic |
| Excavata | 2 | 0 | 0 | freshwater species | heterotrophic |
| Opisthokonta | 115 | 79 | 10 | ||
| Animalia | 24 | 10 | 3 | ||
| Arthropoda | 16 | 8 | 0 | Arctic, Antarctic, aquatic. (e.g., Daphnia sp., Ellipura, Branchiopoda, Entomobryiadae). | heterotrophic |
| Bilateria | 1 | 0 | 1 | Deep sediment environmental sample | unknown |
| Chordata | 3 | 1 | 0 | Aves, from meteoric ice or contaminant? | heterotrophic |
| Cnideria | 1 | 0 | 0 | Small sea anemone, lives in soft sediment with water salinities of 9 to 52 ppt at temperatures from −1 to 28 °C. | heterotrophic |
| Mollusca | 1 | 0 | 1 | Nutricola sp., cold water marine bivalve that burrows into sediments. | heterotrophic |
| Rotifera | 1 | 1 | 0 | Survives under extreme conditions; feed on detritus, bacteria, algae and protists. | heterotrophic |
| Tardigrada | 1 | 0 | 1 | Hardy animal, eats rotifers and algae, can survive from approximately −270 to 150 °C | heterotrophic |
| Fungi | 91 | 69 | 7 | ||
| Ascomycota | 48 | 34 | 4 | Antarctic, polar, aquatic, soil | heterotrophic |
| Basidiomycota | 29 | 24 | 0 | Antarctic, polar, psychrophilic, psychrotolerant | heterotrophic |
| Mucorales | 1 | 0 | 1 | Aquatic, parasitic on arthropods | heterotrophic |
| Uncultured fungi | 13 | 11 | 2 | unknown | unknown |
| Rhizaria | 1 | 0 | 0 | Freshwater, Paulinella sp. | heterotrophic |
| Uncultured eukaryotes | 16 | 10 | 6 | unknown | unknown |
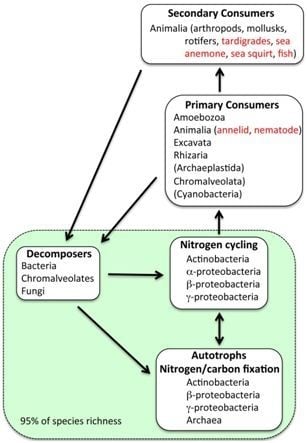
2. Results and Discussion
2.1. Summary of Results
| Taxon | Unique gene sequences | Unique rRNA gene sequences a | Ecology and physiology b | Species characteristics b | |
|---|---|---|---|---|---|
| ≥200 nt <200 nt | |||||
| BACTERIA | 155 | 69 | 21 | ||
| Actinobacteria | 14 | 1 | 4 | fish pathogen, psychrophilic, ocean/lake sediments | chemoorganotrophic heterotrophic |
| Bacterioidetes/Chlorobi | 1 | 0 | 0 | psychrophilic, alkalaiphilic, aerobic | heterotrophic |
| Chloroflexi | 1 | 0 | 1 | ||
| Deinococcus/Thermus | 1 | 0 | 0 | thermophilic, radiophilic, some associate with cyanobacteria | chemoorganotrophic heterotrophic |
| Firmicutes | 16 | 5 | 0 | alkalaiphilic, thermophilic, mesophilic, psychrophilic, soil/sediments, anaerobic, some parasitic/symbiotic on animals | heterotrophic, nitrate reduction |
| Fusobacteria | 1 | 0 | 0 | mesophilic, parasitic on animals, anaerobic | heterotrophic |
| Proteobacteria | 71 | 27 | 6 | ||
| Alphaproteobacteria | 8 | 5 | 0 | mesophilic, psychrophilic, aerobic, acid tolerance, aquatic, sediments, animal symbionts | nitrogen fixation, heterotrophic, carbon fixation using the reductive pentose phosphate cycle |
| Betaproteobacteria | 22 | 6 | 3 | annelid symbiont, annelid associated, Arctic soils, aquatic, Antarctic marine, intracellular gas vacuoles, high amounts of 16:1 ω7c fatty acids, psychrophilic, mesophilic, thermophilic, aerobic, highly adaptable, hot springs, (e.g., Thiobacillus sp., related to Hydrogenophilus thermoluteus, previously reported by Bulat et al. 2004 [20,21] Lake Vostok accretion ice at 3,607 m depth) | nitrogen fixation, chemoorganotrophic heterotrophic, aromatic hydrocarbon degradation, nitrous oxide reduction, arsenic oxidation, arsenic reduction, inorganic sulfur oxidation, chemolithoautotroph,, hydrogen oxidation, carbon fixation using the reductive pentose phosphate cycle |
| Gammaproteobacteria | 39 | 14 | 3 | fish intestinal symbionts (2 species), nematode associated, animal associated, plant associated, aquatic, soil/sediment, thermophilic, mesophilic, psychrophilic, anaerobic, aerobic, halotolerant | nitrogen fixation, nitrate reduction, nitrite respiration, heterotrophic, carbon fixation using the reductive pentose phosphate cycle |
| Uncultured Proteobacteria | 2 | 2 | 0 | ||
| Uncultured Bacteria | 50 | 36 | 10 | sequences similar to those from uncultured and unidentified species, many from other environmental metagenomic studies | unknown |
| EUKARYA | 29 | 12 | 9 | ||
| Archaeplastida | 2 | 1 | 1 | ||
| Streptophyta | 2 | 1 | 1 | pollen from lake sediments or from glacial deposition? | (carbon fixation using the reductive pentose phosphate cycle)—non-viable? |
| Opisthokonta | 26 | 10 | 8 | ||
| Animalia | 5 | 0 | 0 | ||
| Arthropoda | 5 | 0 | 0 | aquatic, Acari, parasitic | heterotrophic |
| Fungi | 22 | 10 | 8 | ||
| Ascomycota | 13 | 7 | 3 | aquatic, one grows on marble and limestone, one isolated from mid-ocean hydrothermal vents, some from sediments, one can use methanol as a carbon source, Antarctic species | heterotrophic |
| Basidiomycota | 4 | 0 | 4 | Antarctic, marine, aquatic | heterotrophic |
| Uncultured fungi | 4 | 3 | 1 | unknown | unknown |
| Uncultured eukaryote | 1 | 1 | 0 | unknown | unknown |
2.2. Extremophiles

2.3. Metabolic Classification

2.4. Eukaryotes
2.5. Possible Marine Environment in Lake Vostok

3. Experimental Section
3.1. Acquisition and Processing of Ice Core Sections
3.2. DNA and RNA Extraction
3.3. cDNA Synthesis and Amplification of cDNA and DNA
3.4. Addition of 454 A and B Sequences by PCR Amplification
3.5. Sequence Analysis
3.6. Metabolic Analysis
4. Conclusions
Acknowledgments
References
- Kapista, A.; Ridley, J.F.; Robin, G.Q.; Siegert, M.J.; Zotikov, I. Large deep freshwater lake beneath the ice of central Antarctica. Nature 1996, 381, 684–686. [Google Scholar] [CrossRef]
- MacGregor, J.A.; Matsuoka, K.; Studinger, M. Radar detection of accreted ice over Lake Vostok, Antarctica. Earth Planet Sci. Lett. 2009, 282, 222–233. [Google Scholar] [CrossRef]
- Masalov, V.N.; Lukin, V.V.; Shermetiev, A.N.; Popov, S.V. Geophysical investigation of the subglacial Lake Vostok in Eastern Antarctica. Dokl. Earth Sci. 2001, 379A, 734–738. [Google Scholar]
- Studinger, M.; Karner, G.D.; Bell, R.E.; Levin, V.; Raymond, C.A.; Tikku, A. Geophysical models for the tectonic framework of the Lake Vostok region East Antarctica. Earth Planet Sci. Lett. 2003, 216, 663–677. [Google Scholar] [CrossRef]
- Wright, A.; Siegert, M.J. The identification and physiographical setting of Antarctic subglacial lakes: An update based on recent discoveries. Geophys. Monogr. Ser. 2011, 192, 9–26. [Google Scholar] [CrossRef]
- Jouzel, J.; Petit, J.R.; Souchez, R.; Barkov, N.I.; Lipenkov, V.Y.; Raynaud, D.; Stievenard, M.; Vassiliev, N.; Verbeke, V.; Vimeux, F. More than 200 meters of lake ice above subglacial Lake Vostok, Antarctica. Science 1999, 286, 2138–2141. [Google Scholar] [CrossRef]
- Bell, R.; Studinger, M.; Tikku, A.; Castello, J.D. Comparative biological analyses of accretion ice from subglacial Lake Vostok. In Life in Ancient Ice; Castello, J.D., Rogers, S.O., Eds.; Princeton University Press: Princeton, NJ, USA, 2005; pp. 251–267. [Google Scholar]
- Gramling, C. A tiny window opens into Lake Vostok, while a vast continent awaits. Science 2012, 335, 788–789. [Google Scholar] [CrossRef]
- Siegert, M.J.; Ellis-Evans, J.C.; Tranter, M.; Mayer, C.; Petit, J.; Salamatin, A.; Priscu, J.C. Physical, chemical and biological processes in Lake Vostok and other Antarctic subglacial lakes. Nature 2001, 414, 603–609. [Google Scholar]
- Siegert, M.J.; Tranter, M.; Ellis-Evans, J.C.; Priscu, J.C.; Lyons, W.B. The hydrochemistry of Lake Vostok and the potential for life in Antarctic subglacial lakes. Hydrol. Process. 2003, 17, 795–814. [Google Scholar] [CrossRef]
- Bulat, S.A.; Alekhina, I.A.; Lipenkov, V.Y.; Lukin, V.V.; Marie, D.; Petit, J.R. Cell concentrations of microorganisms in glacial and lake ice of the Vostok ice core, East Antarctica. Microbiology 2009, 78, 808–810. [Google Scholar] [CrossRef]
- Christner, B.C.; Royston-Bishop, G.; Foreman, C.M.; Arnold, B.R.; Tranter, M.; Welch, K.A.; Lyons, W.B.; Tsapin, A.I.; Studinger, M.; Priscu, J.C. Limnological conditions in subglacial Lake Vostok, Antarctica. Limnol. Oceanogr. 2006, 51, 2485–2501. [Google Scholar] [CrossRef]
- Abyzov, S.S.; Poglazova, M.N.; Mitskevich, J.N.; Ivanov, M.V. Common features of microorganisms in ancient layers of the Antarctic ice sheet. In Life in Ancient Ice; Castello, J.D., Rogers, S.O., Eds.; Princeton University Press: Princeton, NJ, USA, 2005; pp. 240–250. [Google Scholar]
- Christner, B.C.; Mosley-Thompson, E.; Thompson, L.G.; Reeve, J.N. Isolation of bacteria and 16S rDNAs from Lake Vostok accretion ice. Environ. Micrbiol. 2001, 3, 570–577. [Google Scholar] [CrossRef]
- Christner, B.C.; Mosley-Thompson, E.; Thompson, L.G.; Reeve, J.N. Classification of bacteria in polar and nonpolar global ice. In Life in Ancient Ice; Castello, J.D., Rogers, S.O., Eds.; Princeton University Press: Princeton, NJ, USA, 2005; pp. 227–239. [Google Scholar]
- D’Elia, T.; Veerapaneni, R.; Rogers, S.O. Isolation of microbes from Lake Vostok accretion ice. Appl. Environ. Microbiol. 2008, 74, 4962–4965. [Google Scholar] [CrossRef]
- D’Elia, T.; Veerapaneni, R.; Theraisnathan, V.; Rogers, S.O. Isolation of fungi from Lake Vostok accretion ice. Mycologia 2009, 101, 751–763. [Google Scholar] [CrossRef]
- Karl, D.M.; Bird, D.F.; Björkman, K.; Houlihan, T.; Shakelford, R.; Tupas, L. Microorganisms in the accreted ice of Lake Vostok, Antarctica. Science 1999, 286, 2144–2147. [Google Scholar] [CrossRef]
- Priscu, J.C.; Adams, E.E.; Lyons, W.B.; Voytek, M.A.; Mogk, D.W.; Brown, R.L.; McKay, C.P.; Takacs, C.D.; Welch, K.A.; Wolf, C.F. Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science 1999, 286, 2141–2144. [Google Scholar] [CrossRef]
- Bulat, S.A.; Alekhina, I.A.; Blot, M.; Petit, J.R.; Waggenbach, D.; Lipenkov, V.Y.; Vasilyeva, L.P.; Wloch, D.M.; Raynaud, D. DNA signature of thermophilic bacteria from the aged accretion ice of Lake Vostok, Antarctica: Implications for searching for life in extreme icy environments. Int. J. Astobiol. 2004, 1, 1–12. [Google Scholar]
- Lavire, C.; Normand, P.; Alekhina, I.; Bulat, S.; Prieur, D.; Birrien, J.L.; Fournier, P.; Hänni, C.; Petit, J.R. Presence of Hydrogenophilus thermoluteolus DNA in accretion ice in the subglacial Lake Vostok, Antarctica, assessed using rrs, cbb and hox. Environ. Microbiol. 2006, 8, 2106–2114. [Google Scholar] [CrossRef]
- Shtarkman, Y.M.; Koçer, Z.A.; Edgar, R.; Veerapaneni, R.; D’Elia, T.; Morris, P.F.; Rogers, S.O. Subglacial Lake Vostok (Antarctica) accretion ice contains a diverse set of sequences from aquatic, marine and sediment-inhabiting Bacteria and Eukarya. PLoS One 2012. in revision. [Google Scholar]
- Ferracciolli, F.; Finn, C.A.; Jordan, T.A.; Bell, R.E.; Anderson, L.M.; Damaske, D. East Antarctic rifting triggers uplift of the Gamburtsev Mountains. Nature 2011, 479, 388–392. [Google Scholar] [CrossRef]
- Young, D.A.; Wright, A.P.; Roberts, J.L.; Warner, R.C.; Young, N.W.; Greenbaum, J.S.; Schroeder, D.M.; Holt, J.W.; Sugden, D.E.; Blankenship, D.D. A dynamic early East Antarctic Ice Sheet suggested by ice-covered fjord landscapes. Nature 2011, 474, 72–75. [Google Scholar]
- Ferrer, M.; Werner, J.; Chernikova, T.N.; Barjiela, R.; Fernández, L.; La Cono, V.; Waldmann, J.; Teeling, H.; Golyshina, O.V.; Glöckner, F.O. Unveiling microbial life in the new deep-sea hypersaline Lake Thetis. Part II: A metagenomic study. Environ. Microbiol. 2011, 14, 268–281. [Google Scholar]
- Toth, D.J.; Lerman, A. Stratified lake and ocean brines: Salt movement and time limits of existence. Limnol. Oceanog. 1975, 20, 715–728. [Google Scholar] [CrossRef]
- Bar-Even, A.; Noor, E.; Milo, R. A survey of carbon fixation pathways through a quantitative lens. J. Exp. Bot. 2012, 63, 2325–2342. [Google Scholar] [CrossRef]
- Burgaud, G.; Le Calvez, T.; Arzur, T.D.; Vandenkoornhuyse, P.; Barbier, G. Diversity of culturable marine filamentous fungi from deep-sea hydrothermal vents. Environ. Microbiol. 2009, 11, 1588–1600. [Google Scholar] [CrossRef]
- Bell, E. Life at Extremes: Environments, Organisms and Strategies for Survival; CABI: Cambridge, MA, USA, 2012; p. 576. [Google Scholar]
- Daniel, M.; Cohen, D.M.; Richard, H.; Rosenblatt, R.H.; Moser, H.G. Biology and description of a bythitid fish from deep-sea thermal vents in the tropical eastern Pacific. Deep Sea Res. 1990, 37, 267–283. [Google Scholar] [CrossRef]
- Gaill, F.; Mann, K.; Wiedemann, H.; Engel, J.; Timpl, R. Structural comparison of cuticle and interstitial collagens from annelids living in shallow sea-water and at deep-sea hydrothermal vents. J. Mol. Biol. 1995, 246, 284–294. [Google Scholar] [CrossRef]
- Shank, T.M.; Black, M.B.; Halanych, K.M.; Lutz, R.A.; Vrijenhoek, R.C. Miocene radiation of deep-sea hydeothermal vent shrimp (Caridea: Bresiliidae): Evidence from mitochondrial cytochrome oxidase subunit I. Mol. Phylogenet. Evol. 1999, 13, 244–254. [Google Scholar] [CrossRef]
- Tunnicliffe, V. The biology of hydrothermal vents: Ecology and evolution. Oceanogr. Mar. Biol. 1991, 29, 319–407. [Google Scholar]
- Tunnicliffe, V.; McArthur, A.G.; McHugh, D. A biogeographical perspective of the deep-sea hydrothermal vent fauna. Adv. Mar. Biol. 1998, 34, 353–442. [Google Scholar] [CrossRef]
- Vishnivetskaya, T.A.; Erokhina, L.G.; Spirina, E.V.; Shatilovich, A.V.; Vorobyova, E.A.; Tsapin, A.; Gilichinsky, D. Viable phototrophs: Cyanobacteria and green algae from the permafrost darkness. In Life in Ancient Ice; Castello, J.D., Rogers, S.O., Eds.; Princeton University Press: Princeton, NJ, USA, 2005; pp. 140–158. [Google Scholar]
- Vrijenhoek, R.C. Gene flow and genetic diversity in naturally fragmented metapopulations of deep-sea thermal vent animals. J. Hered. 1997, 88, 285–293. [Google Scholar] [CrossRef]
- Tarasov, V.G.; Gebruk, A.V.; Mironov, A.N.; Moskalev, L.I. Deep-sea and shallow-water hydrothermal vent communities: Two different phenomena? Chem. Geol. 2005, 224, 5–39. [Google Scholar] [CrossRef]
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, Rhythms, and Aberrations in Global Climate 65 Ma to Present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef]
- Brambilla, E.; Hippe, H.; Hagelstein, A.; Tindall, B.J.; Stackebrandt, E. 16S rDNA diversity of cultured and uncultured prokaryotes of a mat sample from Lake Fryxell, McMurdo Dry Valleys, Antarctica. Extremophiles 2001, 5, 23–33. [Google Scholar] [CrossRef]
- Clocksin, K.M.; Jung, D.O.; Madigan, M.T. Cold-Active Chemoorganotrophic Bacteria from Permanently Ice-Covered Lake Hoare, McMurdo Dry Valleys, Antarctica. Appl. Environ. Microb. 2007, 73, 3077–3083. [Google Scholar] [CrossRef]
- Laybourn-Parry, J.; Pearce, D.A. The biodiversity and ecology of Antarctic lakes: Models for evolution. Philos. Trans. Roy. Soc. B 2007, 362, 2273–2289. [Google Scholar] [CrossRef]
- Mosier, A.C.; Murray, A.E.; Fritsen, C.H. Microbiota within the perennial ice cover of Lake Vida, Antarctica. FEMS Microbiol. Ecol. 2007, 59, 274–288. [Google Scholar] [CrossRef]
- Kalyuzhnaya, M.G.; lsapidus, A.; Ivanova, N.; Copeland, A.C.; McHardy, A.C.; Szeto, E.; Salamov, A.; Grigoriev, I.V.; Suciu, D.; Levine, S.R. High-resolution metagenomics targets specific functional types in complex microbial communities. Nat. Biotechnol. 2008, 26, 1029–1034. [Google Scholar] [CrossRef]
- Kemp, P.F.; Aller, J.Y. Estimating prokaryotic diversity: When are 16S rDNA libraries large enough? Limnol. Oceanogr. 2004, 2, 114–125. [Google Scholar] [CrossRef]
- Newton, R.J.; Jones, S.E.; Eiler, A.; McMahon, K.D.; Bertlisson, S. A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. 2011, 75, 14–49. [Google Scholar] [CrossRef]
- Ravenschlag, K.; Sahm, K.; Pernthaler, J.; Amann, R. High Bacterial Diversity in Permanently Cold Marine Sediments. Appl. Environ. Microb. 1999, 65, 3982–3989. [Google Scholar]
- Rogers, S.O.; Ma, L.J.; Zhao, Y.; Catranis, C.M.; Starmer, W.T.; Castello, J.D. Recommendations for elimination of contaminants and authentication of isolates in ancient ice cores. In Life in Ancient Ice; Castello, J.D., Rogers, S.O., Eds.; Princeton University Press: Princeton, NJ, USA, 2005; pp. 5–21. [Google Scholar]
- Rogers, S.O.; Theraisnathan, V.; Ma, L.J.; Zhao, Y.; Zhang, G.; Shin, S.-G.; Castello, J.; Starmer, W. Comparisons of protocols to decontaminate environmental ice samples for biological and molecular examinations. Appl. Environ. Microbiol. 2005, 70, 2540–2544. [Google Scholar]
- Chevreux, B.; Wetter, T.; Suhai, S. Genome sequence assembly using trace signals and additional sequence information, Computer Science and Biology. In Proceedings of the German Conference on Bioinformatics (GCB), Hannover, Germany, 4–6 October 1999; pp. 45–56.
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A. The Metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinforma. 2008, 9, 386. [Google Scholar] [CrossRef] [Green Version]
- Goecks, J.; Nekrutenko, A.; Taylor, J. The Galaxy Team. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010, 11, R86. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, K. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar]
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Rogers, S.O.; Shtarkman, Y.M.; Koçer, Z.A.; Edgar, R.; Veerapaneni, R.; D'Elia, T. Ecology of Subglacial Lake Vostok (Antarctica), Based on Metagenomic/Metatranscriptomic Analyses of Accretion Ice. Biology 2013, 2, 629-650. https://doi.org/10.3390/biology2020629
Rogers SO, Shtarkman YM, Koçer ZA, Edgar R, Veerapaneni R, D'Elia T. Ecology of Subglacial Lake Vostok (Antarctica), Based on Metagenomic/Metatranscriptomic Analyses of Accretion Ice. Biology. 2013; 2(2):629-650. https://doi.org/10.3390/biology2020629
Chicago/Turabian StyleRogers, Scott O., Yury M. Shtarkman, Zeynep A. Koçer, Robyn Edgar, Ram Veerapaneni, and Tom D'Elia. 2013. "Ecology of Subglacial Lake Vostok (Antarctica), Based on Metagenomic/Metatranscriptomic Analyses of Accretion Ice" Biology 2, no. 2: 629-650. https://doi.org/10.3390/biology2020629