Changes in a Protein Profile Can Account for the Altered Phenotype of the Yeast Saccharomyces cerevisiae Mutant Lacking the Copper-Zinc Superoxide Dismutase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Growth Conditions
2.2. Protein Extraction
2.3. Two-Dimensional Gel Electrophoresis (2-DE)
2.4. Image Analysis and Statistics
2.5. Mass Spectrometry
2.6. Protein Identification
2.7. Western Blot
2.8. RNA Samples
2.9. Real-Time PCR
3. Results
3.1. Comparative Proteome Analysis
3.2. Identification of Proteins
3.3. Functional Classes of the Identified Proteins
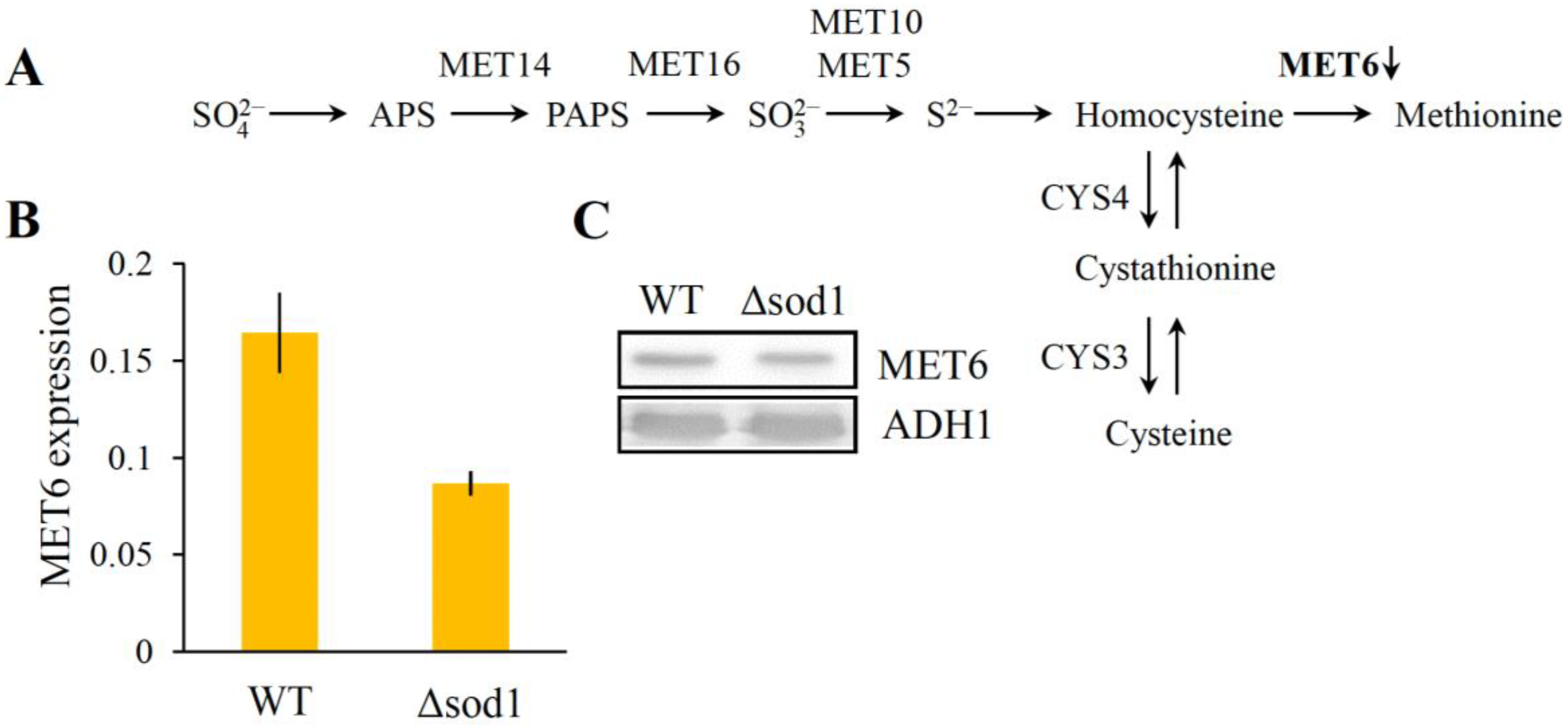
3.4. Decrease of Methionine Synthase Expression Causes Methionine Auxotrophy in the Δsod1 Mutant
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Culotta, V.C.; Yang, M.; O’Halloran, T.V. Activation of superoxide dismutases: Putting the metal to the pedal. Biochim. Biophys. Acta 2006, 1763, 747–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sturtz, L.A.; Diekert, K.; Jensen, L.T.; Lill, R.; Culotta, V.C. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J. Biol. Chem. 2001, 276, 38084–38089. [Google Scholar] [CrossRef] [PubMed]
- Herrero, E.; Ros, J.; Belli, G.; Cabiscol, E. Redox control and oxidative stress in yeast cells. Biochim. Biophys. Acta 2008, 1780, 1217–1235. [Google Scholar] [CrossRef]
- Kloppel, C.; Michels, C.; Zimmer, J.; Herrmann, J.M.; Riemer, J. In yeast redistribution of Sod1 to the mitochondrial intermembrane space provides protection against respiration derived oxidative stress. Biochem. Biophys. Res. Commun. 2010, 403, 114–119. [Google Scholar] [CrossRef]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montllor-Albalate, C.; Colin, A.E.; Chandrasekharan, B.; Bolaji, N.; Andersen, J.L.; Wayne Outten, F.; Reddi, A.R. Extra-mitochondrial Cu/Zn superoxide dismutase (Sod1) is dispensable for protection against oxidative stress but mediates peroxide signaling in Saccharomyces cerevisiae. Redox Biol. 2019, 21, 101064. [Google Scholar] [CrossRef]
- Baron, J.A.; Chen, J.S.; Culotta, V.C. Cu/Zn superoxide dismutase and the proton ATPase Pma1p of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2015, 462, 251–256. [Google Scholar] [CrossRef] [Green Version]
- Reddi, A.R.; Culotta, V.C. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell 2013, 152, 224–235. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Thornton, J.; Spírek, M.; Butow, R.A. Activation of the SPS amino acid-sensing pathway in Saccharomyces cerevisiae correlates with the phosphorylation state of a sensor component, Ptr3. Mol. Cell. Biol. 2008, 28, 551–563. [Google Scholar] [CrossRef] [Green Version]
- Montllor-Albalate, C.; Kim, H.; Thompson, A.E.; Jonke, A.P.; Torres, M.P.; Reddi, A.R. Sod1 integrates oxygen availability to redox regulate NADPH production and the thiol redoxome. Proc. Natl. Acad. Sci. USA 2022, 119, e2023328119. [Google Scholar] [CrossRef]
- Karachitos, A.; Galganska, H.; Wojtkowska, M.; Budzinska, M.; Stobienia, O.; Bartosz, G.; Kmita, H. Cu,Zn-superoxide dismutase is necessary for proper function of VDAC in Saccharomyces cerevisiae cells. FEBS Lett. 2009, 583, 449–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magrì, A.; Di Rosa, M.C.; Tomasello, M.F.; Guarino, F.; Reina, S.; Messina, A.; De Pinto, V. Overexpression of human SOD1 in VDAC1-less yeast restores mitochondrial functionality modulating beta-barrel outer membrane protein genes. Biochim. Biophys. Acta 2016, 1857, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Tsang, C.K.; Liu, Y.; Thomas, J.; Zhang, Y.; Zheng, X.F. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 2014, 5, 3446. [Google Scholar] [CrossRef] [Green Version]
- Reddi, A.R.; Culotta, V.C. Regulation of manganese antioxidants by nutrient sensing pathways in Saccharomyces cerevisiae. Genetics 2011, 189, 1261–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilinski, T.; Krawiec, Z.; Liczmanski, A.; Litwinska, J. Is hydroxyl radical generated by the Fenton reaction in vivo? Biochem. Biophys. Res. Commun. 1985, 130, 533–539. [Google Scholar] [CrossRef]
- Srinivasan, C.; Liba, A.; Imlay, J.A.; Valentine, J.S.; Gralla, E.B. Yeast lacking superoxide dismutase(s) show elevated levels of “free iron” as measured by whole cell electron paramagnetic resonance. J. Biol. Chem. 2000, 275, 29187–29192. [Google Scholar] [CrossRef] [Green Version]
- Wallace, M.A.; Liou, L.L.; Martins, J.; Clement, M.H.; Bailey, S.; Longo, V.D.; Valentine, J.S.; Gralla, E.B. Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis. Cross-compartment protection by CuZn-superoxide dismutase. J. Biol. Chem. 2004, 279, 32055–32062. [Google Scholar] [CrossRef] [Green Version]
- Bilinski, T.; Litwinska, J.; Blaszczynski, M. Selective killing of respiratory sufficient yeast-cells by paraquat. Acta Microbiol. Pol. 1985, 34, 15–17. [Google Scholar]
- Gralla, E.B.; Valentine, J.S. Null mutants of Saccharomyces cerevisiae Cu,Zn superoxide dismutase: Characterization and spontaneous mutation rates. J. Bacteriol. 1991, 173, 5918–5920. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Mirabal, H.R.; Thorsen, M.; Kielland-Brandt, M.C.; Toledano, M.B.; Winther, J.R. Cytoplasmic glutathione redox status determines survival upon exposure to the thiol-oxidant 4,4’-dipyridyl disulfide. FEMS Yeast Res. 2007, 7, 391–403. [Google Scholar] [CrossRef] [Green Version]
- Kwolek-Mirek, M.; Bednarska, S.; Bartosz, G.; Bilinski, T. Acrolein toxicity involves oxidative stress caused by glutathione depletion in the yeast Saccharomyces cerevisiae. Cell Biol. Toxicol. 2009, 25, 363–378. [Google Scholar] [CrossRef] [PubMed]
- Drakulic, T.; Temple, M.D.; Guido, R.; Jarolim, S.; Breitenbach, M.; Attfield, P.V.; Dawes, I.W. Involvement of oxidative stress response genes in redox homeostasis, the level of reactive oxygen species, and ageing in Saccharomyces cerevisiae. FEMS Yeast Res. 2005, 5, 1215–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maslanka, R.; Zadrag-Tecza, R.; Kwolek-Mirek, M. Linkage between Carbon Metabolism, Redox Status and Cellular Physiology in the Yeast Saccharomyces cerevisiae Devoid of SOD1 or SOD2 Gene. Genes 2020, 11, 780. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.M.; Dirmeier, R.; Engle, M.; Poyton, R.O. Mitochondrial protein oxidation in yeast mutants lacking manganese-(MnSOD) or copper- and zinc-containing superoxide dismutase (CuZnSOD): Evidence that MnSOD and CuZnSOD have both unique and overlapping functions in protecting mitochondrial proteins from oxidative damage. J. Biol. Chem. 2004, 279, 51817–51827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doronina, V.A.; Staniforth, G.L.; Speldewinde, S.H.; Tuite, M.F.; Grant, C.M. Oxidative stress conditions increase the frequency of de novo formation of the yeast [PSI+] prion. Mol. Microbiol. 2015, 96, 163–174. [Google Scholar] [CrossRef] [Green Version]
- Muid, K.A.; Karakaya, H.C.; Koc, A. Absence of superoxide dismutase activity causes nuclear DNA fragmentation during the aging process. Biochem. Biophys. Res. Commun. 2014, 444, 260–263. [Google Scholar] [CrossRef] [Green Version]
- Corson, L.B.; Folmer, J.; Strain, J.J.; Culotta, V.C.; Cleveland, D.W. Oxidative Stress and Iron Are Implicated in Fragmenting Vacuoles of Saccharomyces cerevisiae Lacking Cu,Zn-Superoxide Dismutase*. J. Biol. Chem. 1999, 274, 27590–27596. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhang, X.; Zhang, Z. Cu,Zn-superoxide dismutase is required for cell wall structure and for tolerance to cell wall-perturbing agents in Saccharomyces cerevisiae. FEBS Lett. 2010, 584, 1245–1250. [Google Scholar] [CrossRef] [Green Version]
- Barker, M.G.; Brimage, L.J.; Smart, K.A. Effect of Cu,Zn superoxide dismutase disruption mutation on replicative senescence in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 1999, 177, 199–204. [Google Scholar] [CrossRef]
- Longo, V.D.; Gralla, E.B.; Valentine, J.S. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J. Biol. Chem. 1996, 271, 12275–12280. [Google Scholar] [CrossRef] [Green Version]
- Unlu, E.S.; Koc, A. Effects of deleting mitochondrial antioxidant genes on life span. Ann. N. Y. Acad. Sci. 2007, 1100, 505–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wawryn, J.; Krzepilko, A.; Myszka, A.; Bilinski, T. Deficiency in superoxide dismutases shortens life span of yeast cells. Acta Biochim. Pol. 1999, 46, 249–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilinski, T.; Lukaszkiewicz, J.; Sledziewski, A. Demonstration of anaerobic catalase synthesis in the cz1 mutant of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1978, 83, 1225–1233. [Google Scholar] [CrossRef]
- Koziol, S.; Zagulski, M.; Bilinski, T.; Bartosz, G. Antioxidants protect the yeast Saccharomyces cerevisiae against hypertonic stress. Free Radic. Res. 2005, 39, 365–371. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Jungblut, P.R.; Seifert, R. Analysis by high-resolution two-dimensional electrophoresis of differentiation-dependent alterations in cytosolic protein pattern of HL-60 leukemic cells. J. Biochem. Biophys. Methods 1990, 21, 47–58. [Google Scholar] [CrossRef]
- Kwolek-Mirek, M.; Bednarska, S.; Dubicka-Lisowska, A.; Maslanka, R.; Zadrag-Tecza, R.; Kaszycki, P. Unbalance between Pyridine Nucleotide Cofactors in The SOD1 Deficient Yeast Saccharomyces cerevisiae Causes Hypersensitivity to Alcohols and Aldehydes. Int. J. Mol. Sci. 2023, 24, 659. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005, 39, 309–338. [Google Scholar] [CrossRef] [Green Version]
- Maslanka, R.; Zadrag-Tecza, R. Duplication of DNA—A mechanism for the development of new functionality of genes. Postep. Biochem. 2015, 61, 388–397. [Google Scholar]
- McCord, J.M.; Fridovich, I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055. [Google Scholar] [CrossRef]
- Cai, Z.; Yan, L.J. Protein Oxidative Modifications: Beneficial Roles in Disease and Health. J. Biochem. Pharmacol. Res. 2013, 1, 15–26. [Google Scholar] [PubMed]
- Dahl, J.U.; Gray, M.J.; Jakob, U. Protein quality control under oxidative stress conditions. J. Mol. Biol. 2015, 427, 1549–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, A.B.; Sadowska-Bartosz, I.; Königstorfer, A.; Kettle, A.J.; Winterbourn, C.C. Superoxide dismutase protects ribonucleotide reductase from inactivation in yeast. Free Radic. Biol. Med. 2018, 116, 114–122. [Google Scholar] [CrossRef]
- Sehati, S.; Clement, M.H.; Martins, J.; Xu, L.; Longo, V.D.; Valentine, J.S.; Gralla, E.B. Metabolic alterations in yeast lacking copper-zinc superoxide dismutase. Free Radic. Biol. Med. 2011, 50, 1591–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado, M.L.; O’Connor, J.E.; Azorin, I.; Renau-Piqueras, J.; Gil, M.L.; Gozalbo, D. The glyceraldehyde-3-phosphate dehydrogenase polypeptides encoded by the Saccharomyces cerevisiae TDH1, TDH2 and TDH3 genes are also cell wall proteins. Microbiology 2001, 147, 411–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholls, C.; Li, H.; Liu, J.P. GAPDH: A common enzyme with uncommon functions. Clin. Exp. Pharmacol. Physiol. 2012, 39, 674–679. [Google Scholar] [CrossRef]
- Ringel, A.E.; Ryznar, R.; Picariello, H.; Huang, K.L.; Lazarus, A.G.; Holmes, S.G. Yeast Tdh3 (glyceraldehyde 3-phosphate dehydrogenase) is a Sir2-interacting factor that regulates transcriptional silencing and rDNA recombination. PLoS Genet. 2013, 9, e1003871. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, L.F.; Kennedy, B.K.; Harlow, E. A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc. Natl. Acad. Sci. USA 2001, 98, 3946–3951. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Romeo, A.; Kosman, D.J. Transcriptional remodeling and G1 arrest in dioxygen stress in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 24885–24893. [Google Scholar] [CrossRef] [Green Version]
- Chang, E.C.; Kosman, D.J. O2-dependent methionine auxotrophy in Cu,Zn superoxide dismutase-deficient mutants of Saccharomyces cerevisiae. J. Bacteriol. 1990, 172, 1840–1845. [Google Scholar] [CrossRef] [Green Version]
- Sahu, U.; Rajendra, V.K.H.; Kapnoor, S.S.; Bhagavat, R.; Chandra, N.; Rangarajan, P.N. Methionine synthase is localized to the nucleus in Pichia pastoris and Candida albicans and to the cytoplasm in Saccharomyces cerevisiae. J. Biol. Chem. 2017, 292, 14730–14746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, D.; Surdin-Kerjan, Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1997, 61, 503–532. [Google Scholar] [PubMed]
- Lecoq, K.; Belloc, I.; Desgranges, C.; Daignan-Fornier, B. Role of adenosine kinase in Saccharomyces cerevisiae: Identification of the ADO1 gene and study of the mutant phenotypes. Yeast 2001, 18, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Alifano, P.; Fani, R.; Lio, P.; Lazcano, A.; Bazzicalupo, M.; Carlomagno, M.S.; Bruni, C.B. Histidine biosynthetic pathway and genes: Structure, regulation, and evolution. Microbiol. Rev. 1996, 60, 44–69. [Google Scholar] [CrossRef]
- Pearce, D.A.; Sherman, F. Toxicity of copper, cobalt, and nickel salts is dependent on histidine metabolism in the yeast Saccharomyces cerevisiae. J. Bacteriol. 1999, 181, 4774–4779. [Google Scholar] [CrossRef] [Green Version]
- Tibbetts, A.S.; Appling, D.R. Saccharomyces cerevisiae expresses two genes encoding isozymes of 5-aminoimidazole-4-carboxamide ribonucleotide transformylase. Arch. Biochem. Biophys. 1997, 340, 195–200. [Google Scholar] [CrossRef]
- Tibbetts, A.S.; Appling, D.R. Characterization of two 5-aminoimidazole-4-carboxamide ribonucleotide transformylase/inosine monophosphate cyclohydrolase isozymes from Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 20920–20927. [Google Scholar] [CrossRef] [Green Version]
- Garay, E.; Campos, S.E.; Gonzalez de la Cruz, J.; Gaspar, A.P.; Jinich, A.; Deluna, A. High-resolution profiling of stationary-phase survival reveals yeast longevity factors and their genetic interactions. PLoS Genet. 2014, 10, e1004168. [Google Scholar] [CrossRef] [Green Version]
- Lecoq, K.; Konrad, M.; Daignan-Fornier, B. Yeast GMP kinase mutants constitutively express AMP biosynthesis genes by phenocopying a hypoxanthine-guanine phosphoribosyltransferase defect. Genetics 2000, 156, 953–961. [Google Scholar] [CrossRef]
- Kang, H.A.; Hershey, J.W. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J. Biol. Chem. 1994, 269, 3934–3940. [Google Scholar] [CrossRef]
- Thompson, G.M.; Cano, V.S.; Valentini, S.R. Mapping eIF5A binding sites for Dys1 and Lia1: In Vivo evidence for regulation of eIF5A hypusination. FEBS Lett. 2003, 555, 464–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, K.; Abid, M.R.; Miyazaki, M. Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett. 1996, 384, 151–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krokowski, D.; Tchorzewski, M.; Boguszewska, A.; Grankowski, N. Acquisition of a stable structure by yeast ribosomal P0 protein requires binding of P1A-P2B complex: In vitro formation of the stalk structure. Biochim. Biophys. Acta 2005, 1724, 59–70. [Google Scholar] [CrossRef]
- Bautista-Santos, A.; Zinker, S. The P1/P2 protein heterodimers assemble to the ribosomal stalk at the moment when the ribosome is committed to translation but not to the native 60S ribosomal subunit in Saccharomyces cerevisiae. Biochemistry 2014, 53, 4105–4112. [Google Scholar] [CrossRef] [PubMed]
- Abramczyk, O.; Zien, P.; Zielinski, R.; Pilecki, M.; Hellman, U.; Szyszka, R. The protein kinase 60S is a free catalytic CK2alpha’ subunit and forms an inactive complex with superoxide dismutase SOD1. Biochem. Biophys. Res. Commun. 2003, 307, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Zielinski, R.; Pilecki, M.; Kubinski, K.; Zien, P.; Hellman, U.; Szyszka, R. Inhibition of yeast ribosomal stalk phosphorylation by Cu-Zn superoxide dismutase. Biochem. Biophys. Res. Commun. 2002, 296, 1310–1316. [Google Scholar] [CrossRef]
- Szyszka, R. Protein kinases phosphorylating acidic ribosomal proteins from yeast cells. Folia Microbiol. 1999, 44, 142–152. [Google Scholar] [CrossRef]
- Zadrag, R.; Kwolek-Mirek, M.; Bartosz, G.; Bilinski, T. Relationship between the replicative age and cell volume in Saccharomyces cerevisiae. Acta Biochim. Pol. 2006, 53, 747–751. [Google Scholar] [CrossRef] [Green Version]
- Zadrag-Tecza, R.; Kwolek-Mirek, M.; Bartosz, G.; Bilinski, T. Cell volume as a factor limiting the replicative lifespan of the yeast Saccharomyces cerevisiae. Biogerontology 2009, 10, 481–488. [Google Scholar] [CrossRef]
- Justice, M.C.; Hsu, M.J.; Tse, B.; Ku, T.; Balkovec, J.; Schmatz, D.; Nielsen, J. Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J. Biol. Chem. 1998, 273, 3148–3151. [Google Scholar] [CrossRef] [Green Version]
- Perentesis, J.P.; Phan, L.D.; Gleason, W.B.; LaPorte, D.C.; Livingston, D.M.; Bodley, J.W. Saccharomyces cerevisiae elongation factor 2. Genetic cloning, characterization of expression, and G-domain modeling. J. Biol. Chem. 1992, 267, 1190–1197. [Google Scholar] [CrossRef] [PubMed]
- Carr-Schmid, A.; Durko, N.; Cavallius, J.; Merrick, W.C.; Kinzy, T.G. Mutations in a GTP-binding motif of eukaryotic elongation factor 1A reduce both translational fidelity and the requirement for nucleotide exchange. J. Biol. Chem. 1999, 274, 30297–30302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munshi, R.; Kandl, K.A.; Carr-Schmid, A.; Whitacre, J.L.; Adams, A.E.; Kinzy, T.G. Overexpression of translation elongation factor 1A affects the organization and function of the actin cytoskeleton in yeast. Genetics 2001, 157, 1425–1436. [Google Scholar]
- Park, S.G.; Cha, M.K.; Jeong, W.; Kim, I.H. Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 5723–5732. [Google Scholar] [CrossRef] [Green Version]
- MacDiarmid, C.W.; Taggart, J.; Kerdsomboon, K.; Kubisiak, M.; Panascharoen, S.; Schelble, K.; Eide, D.J. Peroxiredoxin chaperone activity is critical for protein homeostasis in zinc-deficient yeast. J. Biol. Chem. 2013, 288, 31313–31327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weids, A.J.; Grant, C.M. The yeast peroxiredoxin Tsa1 protects against protein-aggregate-induced oxidative stress. J. Cell Sci. 2014, 127, 1327–1335. [Google Scholar] [CrossRef] [Green Version]
- Vogel, C.; Silva, G.M.; Marcotte, E.M. Protein expression regulation under oxidative stress. Mol. Cell. Proteom. 2011, 10, M111.009217. [Google Scholar] [CrossRef] [Green Version]


| Spot | Accession Number (ORF) | Protein Name | Description | Molecular Mass [kDa] | pI b | MOWSE Score | Number of Peptides Matched | Sequence Coverage [%] | emPAI | |
|---|---|---|---|---|---|---|---|---|---|---|
| Calculated a | Theoretical b | |||||||||
| 1. | YOR133W YDR385W | EFT1 EFT2 p | Elongation factor 2 Elongation factor 2 | 93.69 93.69 | 93.28 93.28 | 6.23 6.23 | 3559 3559 | 72 72 | 39 39 | 5.85 5.85 |
| 2. | YER091C | MET6 | Cobalamin-independent methionine synthase | 85.98 | 85.85 | 6.41 | 3703 | 64 | 35 | 5.02 |
| 3. | YMR120C YLR028C | ADE17 ADE16 p | Enzyme of ‘de novo’ purine biosynthesis Enzyme of ‘de novo’ purine biosynthesis | 65.56 65.64 | 65.26 65.28 | 6.52 6.52 | 2280 1075 | 41 18 | 26 14 | 4.02 1.19 |
| 4. | YLR044C | PDC1 | Major of three pyruvate decarboxylase isoenzymes | 61.68 | 61.49 | 6.11 | 2856 | 54 | 19 | 4.54 |
| 5. | YBR248C | HIS7 | Imidazole glycerol phosphate synthase | 61.54 | 61.05 | 5.15 | 1219 | 22 | 18 | 1.83 |
| 6. | YPR080W YBR118W | TEF1 TEF2 p | Translational elongation factor 1 alpha Translational elongation factor 1 alpha | 50.40 50.40 | 50.04 50.04 | 9.58 9.58 | 684 684 | 16 16 | 10 10 | 1.01 1.01 |
| 7. | YHR174W | ENO2 | Enolase II, a phosphopyruvate hydratase | 46.94 | 46.91 | 5.85 | 9716 | 150 | 35 | 43.84 |
| 8. | YHR068W | DYS1 | Deoxyhypusine synthase | 43.26 | 42.88 | 5.49 | 643 | 11 | 9 | 1.09 |
| 9. | YCL018W | LEU2 | Beta-isopropylmalate dehydrogenase | 39.04 | 38.95 | 5.48 | 872 | 16 | 15 | 2.67 |
| 10. | YOL086C | ADH1 | Alcohol dehydrogenase I | 37.28 | 36.84 | 6.67 | 4286 | 89 | 20 | 9.83 |
| 11. | YKL060C | FBA1 | Fructose 1,6-bisphosphate aldolase | 39.88 | 39.61 | 5.59 | 1043 | 18 | 11 | 2.05 |
| 12. | YJR105W | ADO1 | Adenosine kinase | 36.52 | 36.36 | 4.77 | 1047 | 15 | 10 | 1.84 |
| 13. | YGR192C | TDH3 | Glyceraldehyde-3-phosphate dehydrogenase | 35.84 | 35.74 | 6.96 | 1978 | 39 | 21 | 7.35 |
| 14. | YKL152C | GPM1 | Tetrameric phosphoglycerate mutase | 27.59 | 27.61 | 9.34 | 706 | 13 | 10 | 2.92 |
| 15. | YML028W | TSA1 | Thioredoxin peroxidase | 21.69 | 21.58 | 4.77 | 957 | 20 | 9 | 3.88 |
| 16. | YDR454C | GUK1 | Guanylate kinase | 20.68 | 20.64 | 7.34 | 789 | 15 | 7 | 2.88 |
| 17. | YLR109W | AHP1 | Thiol-specific peroxiredoxin | 19.27 | 19.11 | 4.78 | 831 | 13 | 9 | 3.99 |
| 18. | YJR104C | SOD1 | Cytosolic copper-zinc superoxide dismutase | 15.96 | 15.85 | 5.91 | 963 | 16 | 10 | 9.06 |
| 19. | YDR382W | RPP2B | Ribosomal protein P2 beta | 11.04 | 11.03 | 3.80 | 793 | 11 | 3 | 1.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwolek-Mirek, M.; Dubicka-Lisowska, A.; Bednarska, S.; Zadrag-Tecza, R.; Kaszycki, P. Changes in a Protein Profile Can Account for the Altered Phenotype of the Yeast Saccharomyces cerevisiae Mutant Lacking the Copper-Zinc Superoxide Dismutase. Metabolites 2023, 13, 459. https://doi.org/10.3390/metabo13030459
Kwolek-Mirek M, Dubicka-Lisowska A, Bednarska S, Zadrag-Tecza R, Kaszycki P. Changes in a Protein Profile Can Account for the Altered Phenotype of the Yeast Saccharomyces cerevisiae Mutant Lacking the Copper-Zinc Superoxide Dismutase. Metabolites. 2023; 13(3):459. https://doi.org/10.3390/metabo13030459
Chicago/Turabian StyleKwolek-Mirek, Magdalena, Aleksandra Dubicka-Lisowska, Sabina Bednarska, Renata Zadrag-Tecza, and Pawel Kaszycki. 2023. "Changes in a Protein Profile Can Account for the Altered Phenotype of the Yeast Saccharomyces cerevisiae Mutant Lacking the Copper-Zinc Superoxide Dismutase" Metabolites 13, no. 3: 459. https://doi.org/10.3390/metabo13030459
APA StyleKwolek-Mirek, M., Dubicka-Lisowska, A., Bednarska, S., Zadrag-Tecza, R., & Kaszycki, P. (2023). Changes in a Protein Profile Can Account for the Altered Phenotype of the Yeast Saccharomyces cerevisiae Mutant Lacking the Copper-Zinc Superoxide Dismutase. Metabolites, 13(3), 459. https://doi.org/10.3390/metabo13030459






