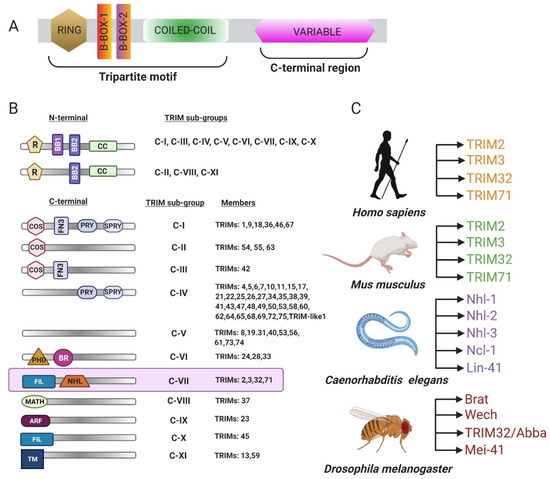
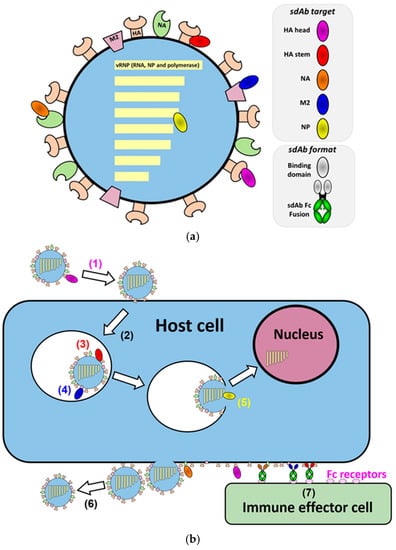
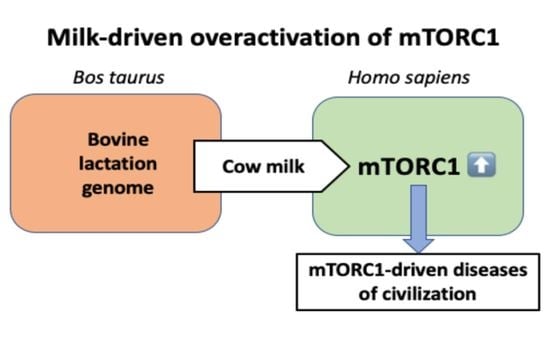
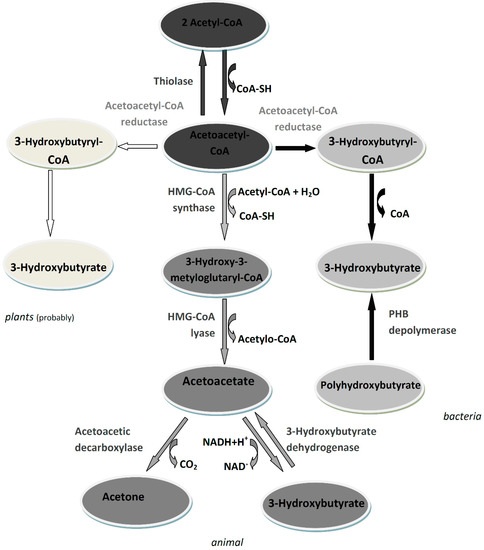
Biomolecules 2021, 11(3), 409; https://doi.org/10.3390/biom11030409 - 10 Mar 2021
Cited by 19 | Viewed by 5879
Abstract
Testosterone’s role in female depression is not well understood, with studies reporting conflicting results. Here, we use meta-analytical and Mendelian randomization techniques to determine whether serum testosterone levels differ between depressed and healthy women and whether such a relationship is casual. Our meta-analysis
[...] Read more.
Testosterone’s role in female depression is not well understood, with studies reporting conflicting results. Here, we use meta-analytical and Mendelian randomization techniques to determine whether serum testosterone levels differ between depressed and healthy women and whether such a relationship is casual. Our meta-analysis shows a significant association between absolute serum testosterone levels and female depression, which remains true for the premenopausal group while achieving borderline significance in the postmenopausal group. The results from our Mendelian randomization analysis failed to show any causal relationship between testosterone and depression. Our results show that women with depression do indeed display significantly different serum levels of testosterone. However, the directions of the effect of this relationship are conflicting and may be due to menopausal status. Since our Mendelian randomization analysis was insignificant, the difference in testosterone levels between healthy and depressed women is most likely a manifestation of the disease itself. Further studies could be carried out to leverage this newfound insight into better diagnostic capabilities culminating in early intervention in female depression.
Full article
(This article belongs to the Special Issue Androgen Receptors in Health and Diseases)
►
Show Figures