Glycation Interferes with the Activity of the Bi-Functional UDP-N-Acetylglucosamine 2-Epimerase/N-Acetyl-mannosamine Kinase (GNE)
Abstract
:1. Introduction
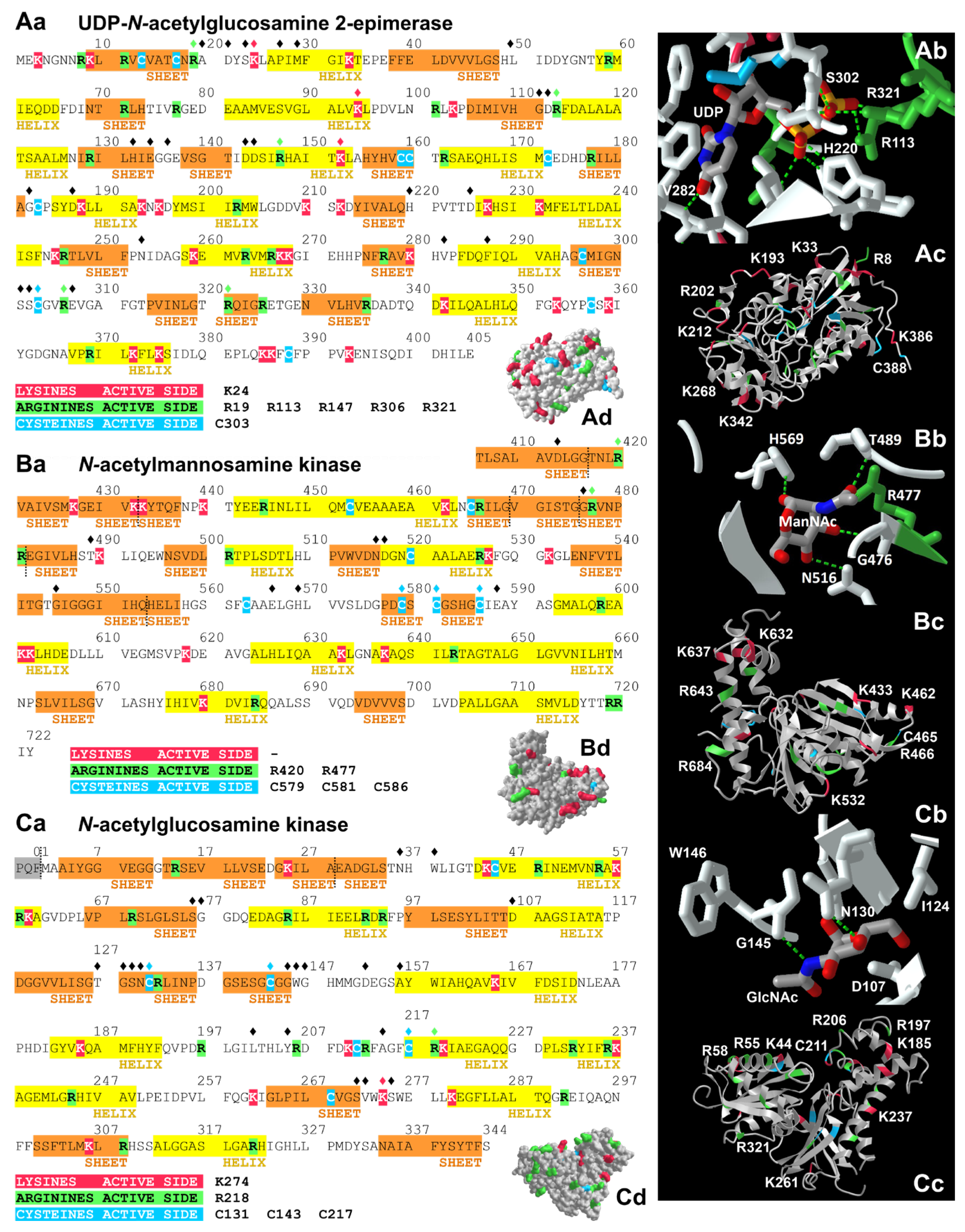
| Residues | Function | So-Far Known Mutations in Patients | In Vitro—Loss of Activity Due to Mutations (Compared to WT) |
| UDP-N-acetylglucosamine 2-epimerase (hGNE1, mRNA variant 2) | |||
| K24 | Involved in forming the vicinity of the active site [52] | n.d.a. | n.d.a. |
| R19 | UDP binding [52] | n.d.a. | n.d.a. |
| R113 | Involved in forming the vicinity of the active site [52] | n.d.a. | R113A: completely inactive [53] |
| R147 | Involved in forming the vicinity of the active site [52] | n.d.a. | n.d.a. |
| R306 | Involved in forming the vicinity of the active site [50] | R306Q [102] | n.d.a. |
| R321 | Interaction with the phosphate of UDP [53] | R321C [103] | n.d.a. |
| C303 | Hydrophobic interactions, but probably no specific function in the enzymatic reaction [50] | C303V [104] C303X [9] | C303V: epimerase 80%, kinase 60% C303X: epimerase 0%, kinase 0% [50] |
| N-acetylmannosamine kinase (hGNE1, mRNA variant 2) | |||
| Residues | Function | So-Far Known Mutations in Patients | In Vitro—Loss of Activity Due to Mutations (Compared to WT) |
| R420 | Interaction with the phosphate oxygens [51,52] | R420X [105] R420Q [106] → patient not yet suffering from myopathy | R420M: epimerase activity comparable to WT; kinase activity drastically reduced [51] |
| R477 | ManNAc binding [54] | n.d.a. | n.d.a. |
| C579 | Zinc binding [54] | C579Y [12] | n.d.a. |
| C581 | Zinc binding [54] | C581R [7] | n.d.a. |
| C586 | Zinc binding [54] | C586X [7] | n.d.a. |
| N-acetylglucosamine kinase | |||
| Residues | Function | In Vitro—Loss of Activity Due to Mutations (Compared to WT) | |
| K274 | Nucleotide binding [56] | n.d.a. | |
| R218 | Nucleotide binding [56] | n.d.a. | |
| C131 | Phosphate transfer from ATP to the hydroxyl groups of GlcNAc [55] | C131S: overall activity reduced (approx.. 10% activity remaining [55]) | |
| C143 | Phosphate transfer from ATP to the hydroxyl groups of GlcNAc [55] | C143S: overall activity reduced (approx.. 20% activity remaining [55]); 75% of WT activity [107] | |
| C217 | Nucleotide binding [56] | C217S: overall activity reduced (approx.. 30% activity remaining [55]) | |

2. Materials and Methods
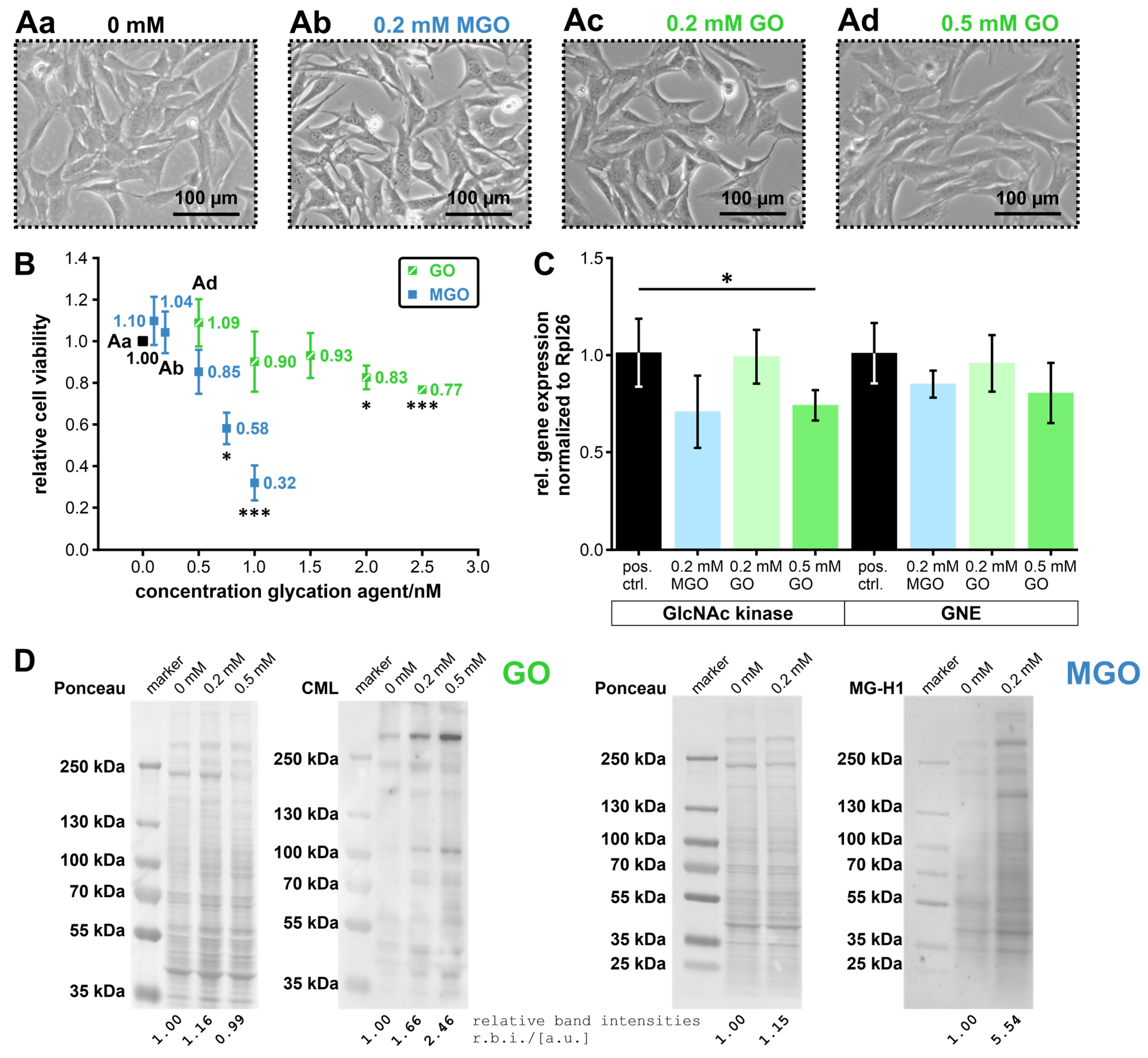
2.1. Cell Culture and MGO/GO-Treatment
2.2. RNA Extraction and RT-qPCR
| Gene Name | Direction | Sequence |
| RPL26 | forward | GGTCTATGCCCATTCGGAAGG |
| RPL26 | reverse | TCGTTCGATGTAGATGACGTACT |
| GAPDH | forward | CCTGGAGAAACCTGCCAAGTATG |
| GAPDH | reverse | AGAGTGGGAGTTGCTGTTGAAGTC |
| Isoform 1 (short) | forward | GGCGTCCGGGTTCTACGCA |
| Isoform 2 (long) | forward | GGAAACACACGCGCATCTCCAC |
| Isoform 1 and 2 | reverse | AATGGTCCCGCTGACCTCGC |
| GNE1 | forward | GGTGGACAATGACGGCAACTGT |
| GNE1 | reverse | CAGTTCGTGCTGGTGGATGATC |
| GlcNAc kinase | forward | GGTAGTATGGCCGCGCTTTA |
| GlcNAc kinase | reverse | GGTGTGCAATCCAGTAGGCT |
2.3. Protein Isolation from Cell Culture
2.4. MTT Assay
2.5. Protein Expression in E.coli and Purification
2.6. Glycation of the Proteins (Protein-Ecoli)
2.7. Western Blot Analysis (Protein-CC and Protein-Ecoli)
2.8. Epimerase Activity Assay (Protein-Ecoli)
2.9. Kinase Activity Assay (Protein-Ecoli)
2.10. Structural Comparison
2.11. Statistical Analysis
3. Results
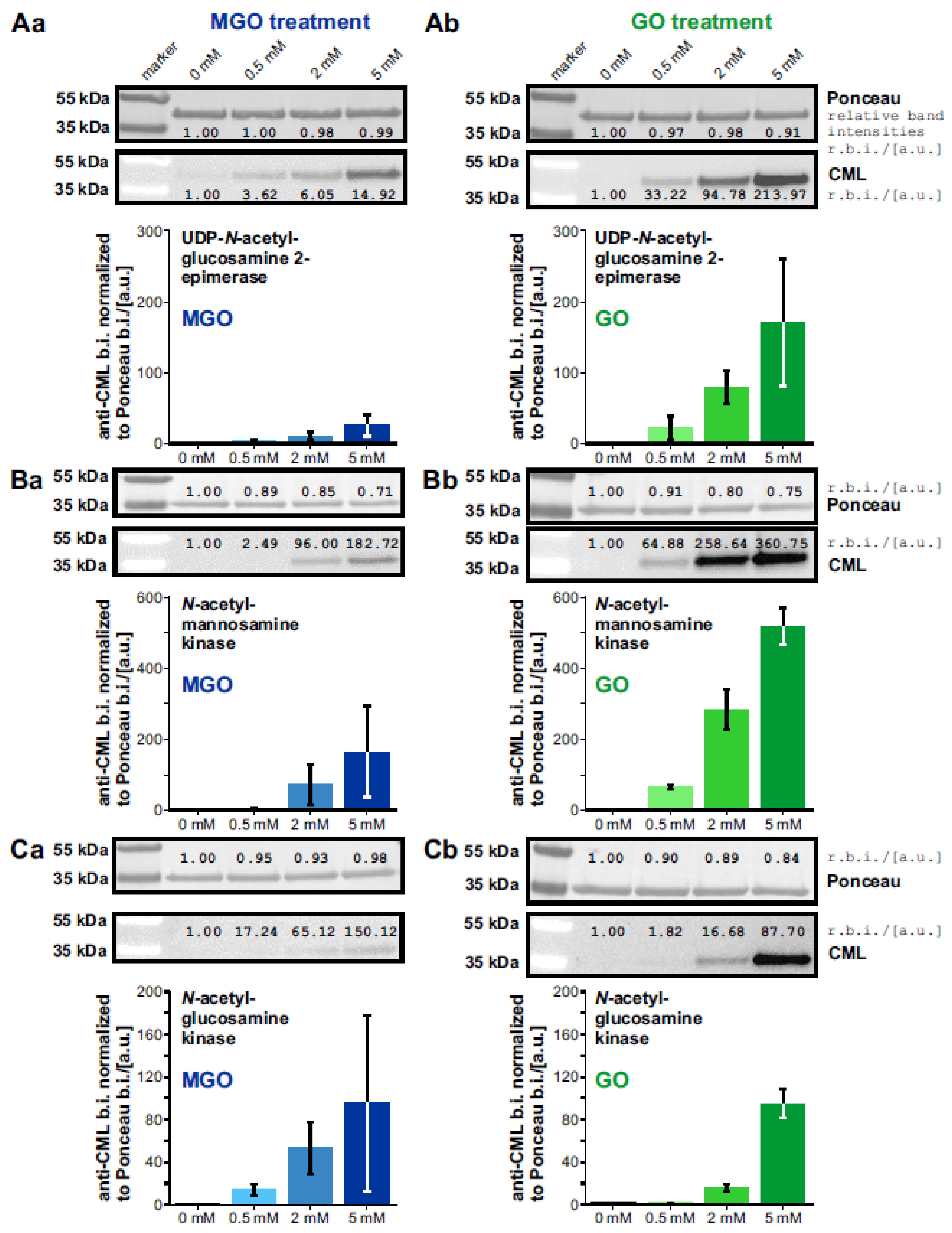
3.1. Expression and Glycation of the UDP-N-Acetylglucosamine 2-Epimerase, the N-Acetylmannosamine kinase, and the N-Acetylglucosamine Kinase
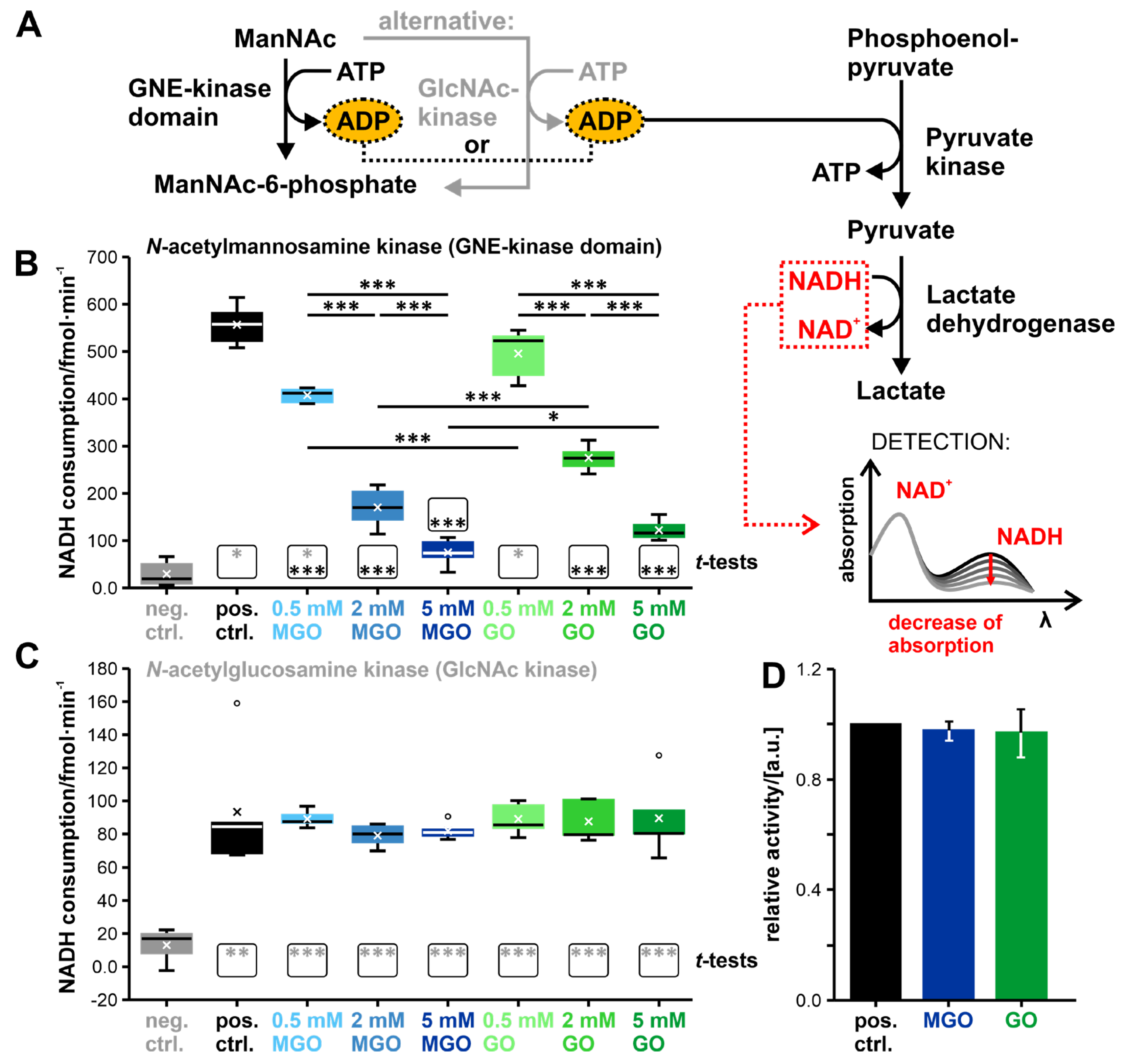
3.2. Glycation of the GNE-Domains Interferes with Their Enzymatic Activity—Activity of the GlcNAc Kinase Is Not Affected by Glycation
3.3. Expression of the UDP-N-Acetylglucosamine 2-Epimerase/N-Acetylmannosamine Kinase, and the N-Acetylglucosamine Kinase in MGO/GO-Treated Undifferentiated C2C12 Cells
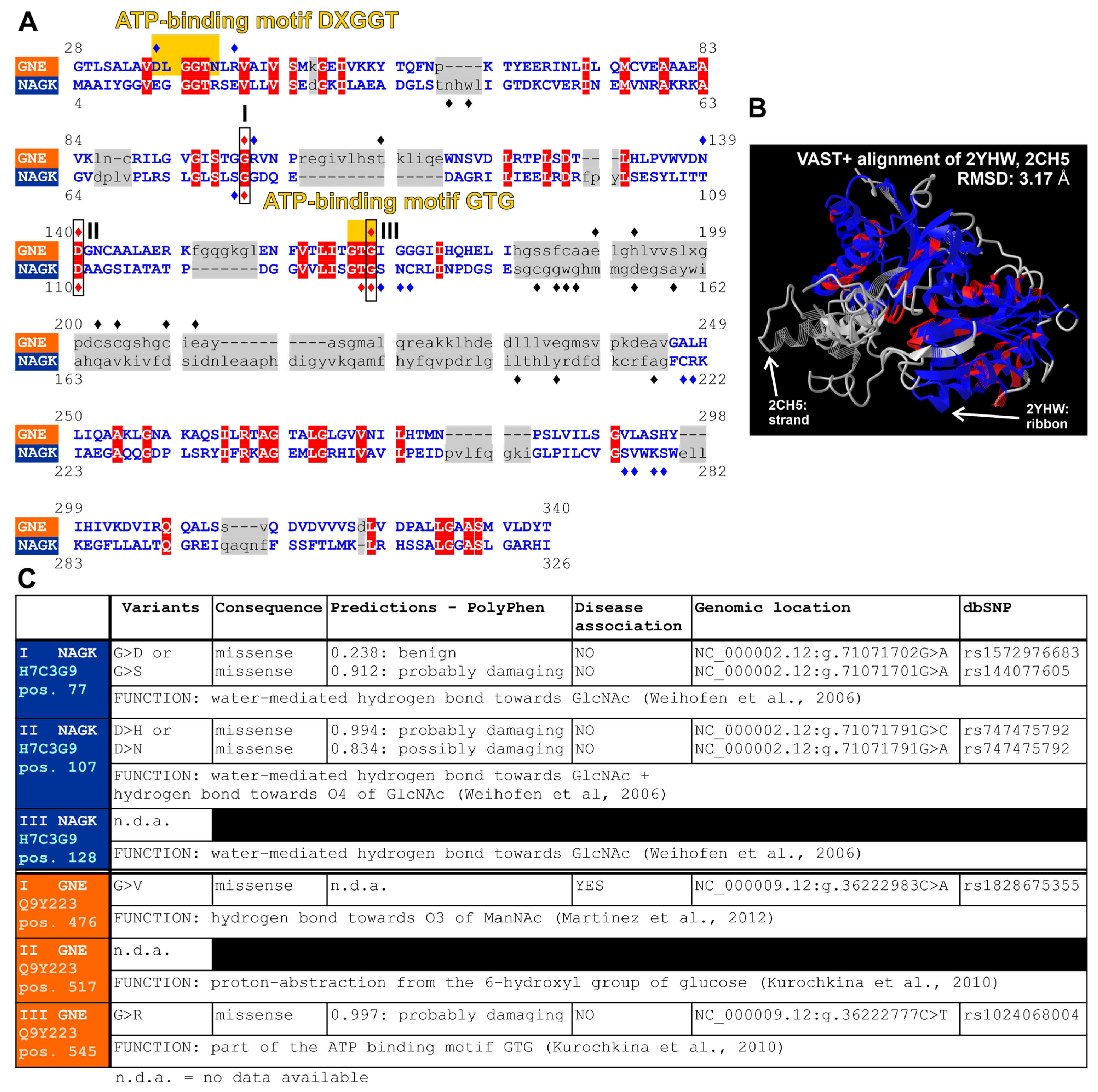
3.4. Structural Comparison of the UDP-N-Acetylglucosamine 2-Epimerase/N-Acetyl-mannosamine kinase and the N-Acetylglucosamine Kinase
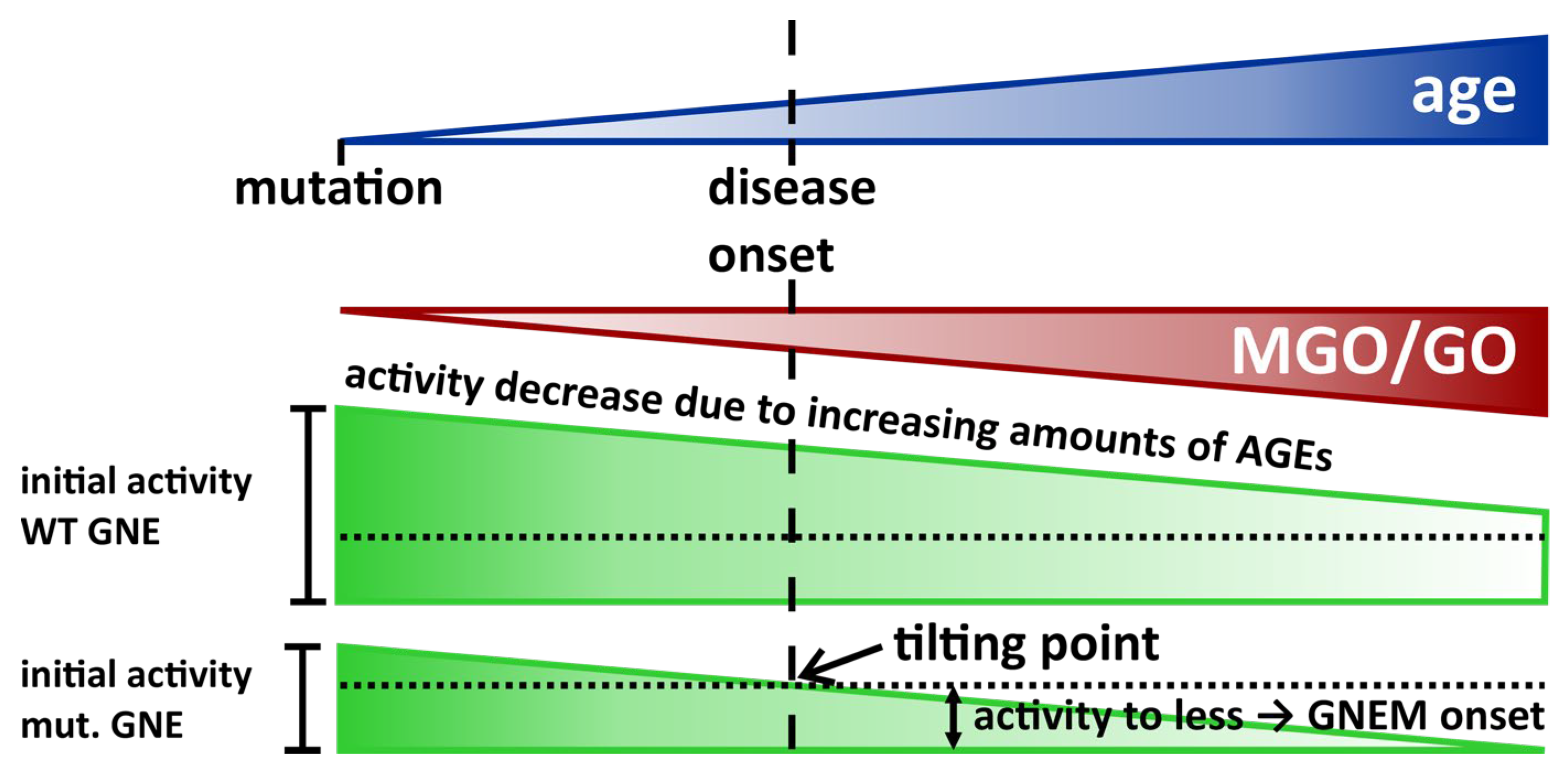
4. Discussion and Outlook
- Mutation introduces a new potential glycation site,
- Mutation has no effect on the number of potential glycation sites (both amino acids lead to a potential glycation site),
- Mutation deletes a potential glycation site.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keppler, O.T.; Hinderlich, S.; Langner, J.; Schwartz-Albiez, R.; Reutter, W.; Pawlita, M. UDP-GlcNAc 2-Epimerase: A Regulator of Cell Surface Sialylation. Science 1999, 284, 1372–1376. [Google Scholar] [CrossRef]
- Carrillo, N.; Malicdan, M.C.; Huizing, M. GNE Myopathy. In GeneReviews®; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2020. [Google Scholar]
- Traving, C.; Schauer, R. Structure, function and metabolism of sialic acids. Cell. Mol. Life Sci. 1998, 54, 1330–1349. [Google Scholar] [CrossRef] [PubMed]
- Nagasundaram, M.; Horstkorte, R.; Gnanapragassam, V.S. Sialic Acid Metabolic Engineering of Breast Cancer Cells Interferes with Adhesion and Migration. Molecules 2020, 25, 2632. [Google Scholar] [CrossRef] [PubMed]
- Bork, K.; Reutter, W.; Gerardy-Schahn, R.; Horstkorte, R. The intracellular concentration of sialic acid regulates the polysialylation of the neural cell adhesion molecule. FEBS Lett. 2005, 579, 5079–5083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassagañas, S.; Pérez-Garay, M.; Peracaula, R. Cell Surface Sialic Acid Modulates Extracellular Matrix Adhesion and Migration in Pancreatic Adenocarcinoma Cells. Pancreas 2014, 43, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Celeste, F.V.; Vilboux, T.; Ciccone, C.; de Dios, J.K.; Malicdan, M.C.V.; Leoyklang, P.; McKew, J.C.; Gahl, W.A.; Carrillo-Carrasco, N.; Huizing, M. Mutation Update for GNE Gene Variants Associated with GNE Myopathy. Hum. Mutat. 2014, 35, 915–926. [Google Scholar] [CrossRef] [Green Version]
- Carrillo, N.; Malicdan, M.C.; Huizing, M. GNE Myopathy: Etiology, Diagnosis, and Therapeutic Challenges. Neurotherapeutics 2018, 15, 900–914. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg, I.; Avidan, N.; Potikha, T.; Hochner, H.; Chen, M.; Olender, T.; Barash, M.; Shemesh, M.; Sadeh, M.; Gil Grabov-Nardini, G.; et al. The UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase gene is mutated in recessive hereditary inclusion body myopathy. Nat. Genet. 2001, 29, 83–87. [Google Scholar] [CrossRef]
- Huizing, M.; Carrillo-Carrasco, N.; Malicdan, M.C.V.; Noguchi, S.; Gahl, W.A.; Mitrani-Rosenbaum, S.; Argov, Z.; Nishino, I. GNE myopathy: New name and new mutation nomenclature. Neuromuscul. Disord. 2014, 24, 387–389. [Google Scholar] [CrossRef] [Green Version]
- Arai, A.; Tanaka, K.; Ikeuchi, T.; Igarashi, S.; Kobayashi, H.; Asaka, T.; Ms, H.D.; Saito, M.; Tanaka, H.; Kawasaki, S.; et al. A novel mutation in the GNE gene and a linkage disequilibrium in Japanese pedigrees. Ann. Neurol. 2002, 52, 516–519. [Google Scholar] [CrossRef]
- Cho, A.; Hayashi, Y.K.; Monma, K.; Oya, Y.; Noguchi, S.; Nonaka, I.; Nishino, I. Mutation profile of the GNE gene in Japanese patients with distal myopathy with rimmed vacuoles (GNE myopathy). J. Neurol. Neurosurg. Psychiatry 2014, 85, 914–917. [Google Scholar] [CrossRef] [PubMed]
- Weiss, P.; Tietze, F.; Gahl, W.A.; Seppala, R.; Ashwell, G. Identification of the metabolic defect in sialuria. J. Biol. Chem. 1989, 264, 17635–17636. [Google Scholar] [CrossRef] [PubMed]
- Seppala, R.; Lehto, V.-P.; Gahl, W.A. Mutations in the Human UDP-N-Acetylglucosamine 2-Epimerase Gene Define the Disease Sialuria and the Allosteric Site of the Enzyme. Am. J. Hum. Genet. 1999, 64, 1563–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enns, G.M.; Seppala, R.; Musci, T.J.; Weisiger, K.; Ferrell, L.D.; Wenger, D.A.; Gahl, W.A.; Packman, S. Clinical course and biochemistry of sialuria. J. Inherit. Metab. Dis. 2001, 24, 328–336. [Google Scholar] [CrossRef]
- Noguchi, S.; Keira, Y.; Murayama, K.; Ogawa, M.; Fujita, M.; Kawahara, G.; Oya, Y.; Imazawa, M.; Goto, Y.-I.; Hayashi, Y.K.; et al. Reduction of UDP-N-acetylglucosamine 2-Epimerase/N-Acetylmannosamine Kinase Activity and Sialylation in Distal Myopathy with Rimmed Vacuoles. J. Biol. Chem. 2004, 279, 11402–11407. [Google Scholar] [CrossRef] [Green Version]
- Saito, F.; Tomimitsu, H.; Arai, K.; Nakai, S.; Kanda, T.; Shimizu, T.; Mizusawa, H.; Matsumura, K. A Japanese patient with distal myopathy with rimmed vacuoles: Missense mutations in the epimerase domain of the UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE) gene accompanied by hyposialylation of skeletal muscle glycoproteins. Neuromuscul. Disord. 2004, 14, 158–161. [Google Scholar] [CrossRef]
- Gagiannis, D.; Orthmann, A.; Danssmann, I.; Schwarzkopf, M.; Weidemann, W.; Horstkorte, R. Reduced sialylation status in UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase (GNE)-deficient mice. Glycoconj. J. 2007, 24, 125–130. [Google Scholar] [CrossRef]
- Hinderlich, S.; Salama, I.; Eisenberg, I.; Potikha, T.; Mantey, L.R.; Yarema, K.J.; Horstkorte, R.; Argov, Z.; Sadeh, M.; Reutter, W.; et al. The homozygous M712T mutation of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase results in reduced enzyme activities but not in altered overall cellular sialylation in hereditary inclusion body myopathy. FEBS Lett. 2004, 566, 105–109. [Google Scholar] [CrossRef]
- Sela, I.; Goss, V.; Becker-Cohen, M.; Dell, A.; Haslam, S.M.; Mitrani-Rosenbaum, S. The glycomic sialylation profile of GNE Myopathy muscle cells does not point to consistent hyposialylation of individual glycoconjugates. Neuromuscul. Disord. 2020, 30, 621–630. [Google Scholar] [CrossRef]
- Watts, G.D.; Thorne, M.; Kovach, M.; Pestronk, A.; E Kimonis, V. Clinical and genetic heterogeneity in chromosome 9p associated hereditary inclusion body myopathy: Exclusion of GNE and three other candidate genes. Neuromuscul. Disord. 2003, 13, 559–567. [Google Scholar] [CrossRef]
- National Library of Medicine. Data Tables—NCBI Datasets. Available online: https://www.ncbi.nlm.nih.gov/datasets/tables/genes/?table_type=transcripts&key=5c64e7feb14b55b61e8dcb1678dbaf67 (accessed on 22 August 2022).
- Yardeni, T.; Choekyi, T.; Jacobs, K.; Ciccone, C.; Patzel, K.; Anikster, Y.; Gahl, W.A.; Kurochkina, N.; Huizing, M. Identification, Tissue Distribution, and Molecular Modeling of Novel Human Isoforms of the Key Enzyme in Sialic Acid Synthesis, UDP-GlcNAc 2-Epimerase/ManNAc Kinase. Biochemistry 2011, 50, 8914–8925. [Google Scholar] [CrossRef] [Green Version]
- Awasthi, K.; Bhattacharya, S.; Bhattacharya, A. Tissue-specific isoform expression of GNE gene in human tissues. J. Muscle Res. Cell Motil. 2022, 43, 49–61. [Google Scholar] [CrossRef]
- Lucka, L.; Krause, M.; Danker, K.; Reutter, W.; Horstkorte, R. Primary structure and expression analysis of human UDP-N-acetyl-glucosamine-2-epimerase/N-acetylmannosamine kinase, the bifunctional enzyme in neuraminic acid biosynthesis1. FEBS Lett. 1999, 454, 341–344. [Google Scholar] [CrossRef] [Green Version]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the Human Tissue-specific Expression by Genome-wide Integration of Transcriptomics and Antibody-based Proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [Green Version]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 22 August 2022).
- Cardini, C.; Leloir, L.F. Enzymatic formation of acetylgalactosamine. J. Biol. Chem. 1957, 225, 317–324. [Google Scholar] [CrossRef]
- Comb, D.; Roseman, S. Enzymic synthesis of N-acetyl-D-mannosamine. Biochim. Biophys. Acta 1958, 29, 653–654. [Google Scholar] [CrossRef] [Green Version]
- Glaser, L. On the mechanism of N-acetylmannosamine formation. Biochim. Biophys. Acta 1960, 41, 534–536. [Google Scholar] [CrossRef]
- Warren, L.; Felsenfeld, H. The biosynthesis of N-acetylneuraminic acid. Biochem. Biophys. Res. Commun. 1961, 4, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Warren, L.; Felsenfeld, H. N-acetylmannosamine-6-phosphate and N-acetylneuraminic acid-9-phosphate as intermediates in sialic acid biosynthesis. Biochem. Biophys. Res. Commun. 1961, 5, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Roseman, S. Enzymatic phosphorylation of N-acetyl-D-mannosamine. Proc. Natl. Acad. Sci. USA 1961, 47, 955–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinderlich, S.; Stäsche, R.; Zeitler, R.; Reutter, W. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver. Purification and characterization of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. J. Biol. Chem. 1997, 272, 24313–24318. [Google Scholar] [CrossRef] [Green Version]
- Stäsche, R.; Hinderlich, S.; Weise, C.; Effertz, K.; Lucka, L.; Moormann, P.; Reutter, W. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver. Molecular cloning and functional expression of UDP-N-acetyl-glucosamine 2-epimerase/N-acetylmannosamine kinase. J. Biol. Chem. 1997, 272, 24319–24324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blume, A.; Weidemann, W.; Stelzl, U.; Wanker, E.E.; Lucka, L.; Donner, P.; Reutter, W.; Horstkorte, R.; Hinderlich, S. Domain-specific characteristics of the bifunctional key enzyme of sialic acid biosynthesis, UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. Biochem. J. 2004, 384, 599–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohls, D.; Sulea, T.; O Purisima, E.; E MacKenzie, R.; Vrielink, A. The crystal structure of the formiminotransferase domain of formiminotransferase-cyclodeaminase: Implications for substrate channeling in a bifunctional enzyme. Structure 2000, 8, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Knighton, D.R.; Kan, C.-C.; Howland, E.F.; Janson, C.A.; Hostomska, Z.; Welsh, K.M.; Matthews, D.A. Structure of and kinetic channelling in bifunctional dihydrofolate reductase–thymidylate synthase. Nat. Struct. Biol. 1994, 1, 186–194. [Google Scholar] [CrossRef]
- Nagradova, N. Interdomain Communications in Bifunctional Enzymes: How Are Different Activities Coordinated? IUBMB Life 2003, 55, 459–466. [Google Scholar] [CrossRef]
- Allen, M.B.; Walker, D.G. Kinetic characterization of N-acetyl-D-glucosamine kinase from rat liver and kidney. Biochem. J. 1980, 185, 577–582. [Google Scholar] [CrossRef] [Green Version]
- Hinderlich, S.; Berger, M.; Keppler, O.T.; Pawlita, M.; Reutter, W. Biosynthesis of N-Acetylneuraminic Acid in Cells Lacking UDP-N-Acetylglucosamine 2-Epimerase/N-Acetylmannosamine Kinase. Biol. Chem. 2001, 382, 291–297. [Google Scholar] [CrossRef]
- Hinderlich, S.; Nohring, S.; Weise, C.; Franke, P.; Stasche, R.; Reutter, W. Purification and characterization of N-acetylglucosamine kinase from rat liver. Comparison with UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. Eur. J. Biochem. 1998, 252, 133–139. [Google Scholar] [CrossRef]
- Hinderlich, S.; Weidemann, W.; Yardeni, T.; Horstkorte, R.; Huizing, M. UDP-GlcNAc 2-Epimerase/ManNAc Kinase (GNE): A Master Regulator of Sialic Acid Synthesis. Top. Curr. Chem. 2015, 366, 97–137. [Google Scholar] [CrossRef] [Green Version]
- Roseman, S. Enzymatic synthesis of cytidine 5′-monophospho-sialic acids. Proc. Natl. Acad. Sci. USA 1962, 48, 437–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, L.; Blacklow, R.S. The Biosynthesis of Cytidine 5′-Monophospho-N-acetylneuraminic Acid by an Enzyme from Neisseria meningitidis. J. Biol. Chem. 1962, 237, 3527–3534. [Google Scholar] [CrossRef] [PubMed]
- Harduin-Lepers, A.; Recchi, M.-A.; Delannoy, P. 1994, the year of sialyltransferases. Glycobiology 1995, 5, 741–758. [Google Scholar] [CrossRef] [PubMed]
- Bork, K.; Weidemann, W.; Berneck, B.; Kuchta, M.; Bennmann, D.; Thate, A.; Huber, O.; Gnanapragassam, V.S.; Horstkorte, R. The expression of sialyltransferases is regulated by the bioavailability and biosynthesis of sialic acids. Gene Expr. Patterns 2017, 23–24, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Kornfeld, S.; Kornfeld, R.; Neufeld, E.F.; O’Brien, P.J. The feedback control of sugar nucleotide biosynthesis in liver. Proc. Natl. Acad. Sci. USA 1964, 52, 371–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarema, K.J.; Goon, S.; Bertozzi, C.R. Metabolic selection of glycosylation defects in human cells. Nat. Biotechnol. 2001, 19, 553–558. [Google Scholar] [CrossRef]
- Penner, J.; Mantey, L.R.; Elgavish, S.; Ghaderi, D.; Cirak, S.; Berger, M.; Krause, S.; Lucka, L.; Voit, T.; Mitrani-Rosenbaum, S.; et al. Influence of UDP-GlcNAc 2-Epimerase/ManNAc Kinase Mutant Proteins on Hereditary Inclusion Body Myopathy. Biochemistry 2006, 45, 2968–2977. [Google Scholar] [CrossRef]
- Effertz, K.; Hinderlich, S.; Reutter, W. Selective Loss of either the Epimerase or Kinase Activity of UDP-N-acetylglucosamine 2-Epimerase/N-Acetylmannosamine Kinase due to Site-directed Mutagenesis Based on Sequence Alignments. J. Biol. Chem. 1999, 274, 28771–28778. [Google Scholar] [CrossRef] [Green Version]
- Kurochkina, N.; Yardeni, T.; Huizing, M. Molecular modeling of the bifunctional enzyme UDP-GlcNAc 2-epimerase/ManNAc kinase and predictions of structural effects of mutations associated with HIBM and sialuria. Glycobiology 2010, 20, 322–337. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.-C.; Huang, C.-H.; Lai, S.-J.; Yang, C.S.; Hsiao, T.-H.; Lin, C.-H.; Fu, P.-K.; Ko, T.-P.; Chen, Y. Mechanism and inhibition of human UDP-GlcNAc 2-epimerase, the key enzyme in sialic acid biosynthesis. Sci. Rep. 2016, 6, 23274. [Google Scholar] [CrossRef] [Green Version]
- Martinez, J.; Nguyen, L.D.; Hinderlich, S.; Zimmer, R.; Tauberger, E.; Reutter, W.; Saenger, W.; Fan, H.; Moniot, S. Crystal Structures of N-Acetylmannosamine Kinase Provide Insights into Enzyme Activity and Inhibition. J. Biol. Chem. 2012, 287, 13656–13665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, M.; Chen, H.; Reutter, W.; Hinderlich, S. Structure and function of N-acetylglucosamine kinase: Identification of two active site cysteines. Eur. J. Biochem. 2002, 269, 4212–4218. [Google Scholar] [CrossRef] [PubMed]
- Weihofen, W.A.; Berger, M.; Chen, H.; Saenger, W.; Hinderlich, S. Structures of Human N-Acetylglucosamine Kinase in Two Complexes with N-Acetylglucosamine and with ADP/Glucose: Insights into Substrate Specificity and Regulation. J. Mol. Biol. 2006, 364, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Tasca, G.; Ricci, E.; Monforte, M.; Laschena, F.; Ottaviani, P.; Rodolico, C.; Barca, E.; Silvestri, G.; Iannaccone, E.; Mirabella, M.; et al. Muscle imaging findings in GNE myopathy. J. Neurol. 2012, 259, 1358–1365. [Google Scholar] [CrossRef]
- Liu, C.-Y.; Yao, J.; Kovacs, W.C.; Shrader, J.A.; Joe, G.; Ouwerkerk, R.; Mankodi, A.K.; Gahl, W.A.; Summers, R.M.; Carrillo, N. Skeletal Muscle Magnetic Resonance Biomarkers in GNE Myopathy. Neurology 2021, 96, e798–e808. [Google Scholar] [CrossRef]
- Nonaka, I.; Sunohara, N.; Satoyoshi, E.; Teresawa, K.; Yonemoto, K. Autosomal recessive distal muscular dystrophy: A comparative study with distal myopathy with rimmed vacoule formation. Ann. Neurol. 1985, 17, 51–59. [Google Scholar] [CrossRef]
- Argov, Z.; Yarom, R. “Rimmed vacuole myopathy” sparing the quadriceps: A unique disorder in Iranian Jews. J. Neurol. Sci. 1984, 64, 33–43. [Google Scholar] [CrossRef]
- Mori-Yoshimura, M.; Hayashi, Y.K.; Yonemoto, N.; Nakamura, H.; Murata, M.; Takeda, S.; Nishino, I.; Kimura, E. Nationwide patient registry for GNE myopathy in Japan. Orphanet J. Rare Dis. 2014, 9, 150. [Google Scholar] [CrossRef]
- Pogoryelova, O.; Cammish, P.; Mansbach, H.; Argov, Z.; Nishino, I.; Skrinar, A.; Chan, Y.; Nafissi, S.; Shamshiri, H.; Kakkis, E.; et al. Phenotypic stratification and genotype–phenotype correlation in a heterogeneous, international cohort of GNE myopathy patients: First report from the GNE myopathy Disease Monitoring Program, registry portion. Neuromuscul. Disord. 2018, 28, 158–168. [Google Scholar] [CrossRef] [Green Version]
- Nonaka, I.; Sunohara, N.; Ishiura, S.; Satoyoshi, E. Familial distal myopathy with rimmed vacuole and lamellar (myeloid) body formation. J. Neurol. Sci. 1981, 51, 141–155. [Google Scholar] [CrossRef]
- Mori-Yoshimura, M.; Oya, Y.; Yajima, H.; Yonemoto, N.; Kobayashi, Y.; Hayashi, Y.K.; Noguchi, S.; Nishino, I.; Murata, M. GNE myopathy: A prospective natural history study of disease progression. Neuromuscul. Disord. 2014, 24, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Quintana, M.; Shrader, J.; Slota, C.; Joe, G.; McKew, J.; Fitzgerald, M.; Gahl, W.; Berry, S.; Carrillo, N. Bayesian model of disease progression in GNE myopathy. Stat. Med. 2019, 38, 1459–1474. [Google Scholar] [CrossRef] [PubMed]
- Nishino, I.; Carrillo-Carrasco, N.; Argov, Z. GNE myopathy: Current update and future therapy. J. Neurol. Neurosurg. Psychiatry 2015, 86, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Krause, S.; Schlotter-Weigel, B.; Walter, M.C.; Najmabadi, H.; Wiendl, H.; Müller-Höcker, J.; Müller-Felber, W.; Pongratz, D.; Lochmüller, H. A novel homozygous missense mutation in the GNE gene of a patient with quadriceps-sparing hereditary inclusion body myopathy associated with muscle inflammation. Neuromuscul. Disord. 2003, 13, 830–834. [Google Scholar] [CrossRef] [PubMed]
- Kannan, M.A.; Challa, S.; Urtizberea, A.J.; Krahn, M.; Jabeen, A.S.; Borgohain, R. Distal myopathy with rimmed vacuoles and inflammation: A genetically proven case. Neurol. India 2012, 60, 631–634. [Google Scholar] [CrossRef] [PubMed]
- Tanboon, J.; Rongsa, K.; Pithukpakorn, M.; Boonyapisit, K.; Limwongse, C.; Sangruchi, T. A Novel Mutation of the GNE Gene in Distal Myopathy with Rimmed Vacuoles: A Case with Inflammation. Case Rep. Neurol. 2014, 6, 55–59. [Google Scholar] [CrossRef]
- Malicdan, M.C.V.; Noguchi, S.; Hayashi, Y.K.; Nonaka, I.; Nishino, I. Prophylactic treatment with sialic acid metabolites precludes the development of the myopathic phenotype in the DMRV-hIBM mouse model. Nat. Med. 2009, 15, 690–695. [Google Scholar] [CrossRef]
- Krentsis, I.M.; Sela, I.; Eiges, R.; Blanchard, V.; Berger, M.; Cohen, M.B.; Mitrani-Rosenbaum, S. GNE Is Involved in the Early Development of Skeletal and Cardiac Muscle. PLoS ONE 2011, 6, e21389. [Google Scholar] [CrossRef] [Green Version]
- Mori-Yoshimura, M.; Kimura, A.; Tsuru, A.; Yajima, H.; Segawa, K.; Mizuno, K.; Oya, Y.; Noguchi, S.; Nishino, I.; Takahashi, Y. Assessment of thrombocytopenia, sleep apnea, and cardiac involvement in GNE myopathy patients. Muscle Nerve 2022, 65, 284–290. [Google Scholar] [CrossRef]
- Galeano, B.; Klootwijk, R.; Manoli, I.; Sun, M.; Ciccone, C.; Darvish, D.; Starost, M.F.; Zerfas, P.M.; Hoffmann, V.J.; Hoogstraten-Miller, S.; et al. Mutation in the key enzyme of sialic acid biosynthesis causes severe glomerular proteinuria and is rescued by N-acetylmannosamine. J. Clin. Investig. 2007, 117, 1585–1594. [Google Scholar] [CrossRef] [Green Version]
- Niethamer, T.K.; Yardeni, T.; Leoyklang, P.; Ciccone, C.; Astiz-Martinez, A.; Jacobs, K.; Dorward, H.M.; Zerfas, P.M.; Gahl, W.A.; Huizing, M. Oral monosaccharide therapies to reverse renal and muscle hyposialylation in a mouse model of GNE myopathy. Mol. Genet. Metab. 2012, 107, 748–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malicdan, M.C.V.; Noguchi, S.; Tokutomi, T.; Goto, Y.-I.; Nonaka, I.; Hayashi, Y.K.; Nishino, I. Peracetylated N-Acetylmannosamine, a Synthetic Sugar Molecule, Efficiently Rescues Muscle Phenotype and Biochemical Defects in Mouse Model of Sialic Acid-deficient Myopathy. J. Biol. Chem. 2012, 287, 2689–2705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakani, S.; Yardeni, T.; Poling, J.; Ciccone, C.; Niethamer, T.; Klootwijk, E.D.; Manoli, I.; Darvish, D.; Hoogstraten-Miller, S.; Zerfas, P.; et al. The Gne M712T Mouse as a Model for Human Glomerulopathy. Am. J. Pathol. 2012, 180, 1431–1440. [Google Scholar] [CrossRef] [Green Version]
- Yonekawa, T.; Malicdan, M.C.V.; Cho, A.; Hayashi, Y.K.; Nonaka, I.; Mine, T.; Yamamoto, T.; Nishino, I.; Noguchi, S. Sialyllactose ameliorates myopathic phenotypes in symptomatic GNE myopathy model mice. Brain 2014, 137, 2670–2679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pogoryelova, O.; Coraspe, J.A.G.; Nikolenko, N.; Lochmüller, H.; Roos, A. GNE myopathy: From clinics and genetics to pathology and research strategies. Orphanet J. Rare Dis. 2018, 13, 70. [Google Scholar] [CrossRef]
- Wang, J.; Youkharibache, P.; Zhang, D.; Lanczycki, C.J.; Geer, R.C.; Madej, T.; Phan, L.; Ward, M.; Lu, S.; Marchler, G.H.; et al. iCn3D, a web-based 3D viewer for sharing 1D/2D/3D representations of biomolecular structures. Bioinformatics 2020, 36, 131–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Youkharibache, P.; Marchler-Bauer, A.; Lanczycki, C.; Zhang, D.; Lu, S.; Madej, T.; Marchler, G.H.; Cheng, T.; Chong, L.C.; et al. iCn3D: From Web-Based 3D Viewer to Structural Analysis Tool in Batch Mode. Front. Mol. Biosci. 2022, 9, 831740. [Google Scholar] [CrossRef]
- Mercuri, E.; Bönnemann, C.G.; Muntoni, F. Muscular dystrophies. Lancet 2019, 394, 2025–2038. [Google Scholar] [CrossRef]
- Monnier, V.M.; Cerami, A. Nonenzymatic Browning in Vivo: Possible Process for Aging of Long-Lived Proteins. Science 1981, 211, 491–493. [Google Scholar] [CrossRef]
- Zeng, J.; Davies, M.J. Evidence for the Formation of Adducts and S-(Carboxymethyl)cysteine on Reaction of α-Dicarbonyl Compounds with Thiol Groups on Amino Acids, Peptides, and Proteins. Chem. Res. Toxicol. 2005, 18, 1232–1241. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, M.W.; Hedegaard, R.V.; Andersen, J.M.; de Courten, B.; Bügel, S.; Nielsen, J.; Skibsted, L.H.; Dragsted, L.O. Advanced glycation endproducts in food and their effects on health. Food Chem. Toxicol. 2013, 60, 10–37. [Google Scholar] [CrossRef] [PubMed]
- Horstkorte, R.; Nöhring, S.; Danker, K.; Effertz, K.; Reutter, W.; Lucka, L. Protein kinase C phosphorylates and regulates UDP-N -acetylglucosamine-2-epimerase/N -acetylmannosamine kinase. FEBS Lett. 2000, 470, 315–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennmann, D.; Horstkorte, R.; Hofmann, B.; Jacobs, K.; Navarrete-Santos, A.; Simm, A.; Bork, K.; Gnanapragassam, V.S. Advanced Glycation Endproducts Interfere with Adhesion and Neurite Outgrowth. PLoS ONE 2014, 9, e112115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledl, F.; Schleicher, R.N.H.E. New Aspects of the Maillard Reaction in Foods and in the Human Body. Angew. Chem. Int. Ed. 1990, 29, 565–594. [Google Scholar] [CrossRef]
- Schwimmer, S.; Olcott, H.S. Reaction between Glycine and the Hexose Phosphates. J. Am. Chem. Soc. 1953, 75, 4855–4856. [Google Scholar] [CrossRef]
- Schalkwijk, C.; Stehouwer, C.D.A. Methylglyoxal, a Highly Reactive Dicarbonyl Compound, in Diabetes, Its Vascular Complications, and Other Age-Related Diseases. Physiol. Rev. 2020, 100, 407–461. [Google Scholar] [CrossRef] [PubMed]
- Eggen, M.D.; Glomb, M.A. Analysis of Glyoxal- and Methylglyoxal-Derived Advanced Glycation End Products during Grilling of Porcine Meat. J. Agric. Food Chem. 2021, 69, 15374–15383. [Google Scholar] [CrossRef]
- Scheffler, J.; Bork, K.; Bezold, V.; Rosenstock, P.; Gnanapragassam, V.S.; Horstkorte, R. Ascorbic acid leads to glycation and interferes with neurite outgrowth. Exp. Gerontol. 2019, 117, 25–30. [Google Scholar] [CrossRef]
- Sell, D.R. Ageing promotes the increase of early glycation Amadori product as assessed by ϵ-N-(2-furoylmethyl)-l-lysine (furosine) levels in rodent skin collagen: The relationship to dietary restriction and glycoxidation. Mech. Ageing Dev. 1997, 95, 81–99. [Google Scholar] [CrossRef]
- Hodge, J.E. Dehydrated Foods, Chemistry of Browning Reactions in Model Systems. J. Agric. Food Chem. 1953, 1, 928–943. [Google Scholar] [CrossRef]
- Twarda-Clapa, A.; Olczak, A.; Białkowska, A.M.; Koziołkiewicz, M. Advanced Glycation End-Products (AGEs): Formation, Chemistry, Classification, Receptors, and Diseases Related to AGEs. Cells 2022, 11, 1312. [Google Scholar] [CrossRef] [PubMed]
- Vistoli, G.; De Maddis, D.; Cipak, A.; Zarkovic, N.; Carini, M.; Aldini, G. Advanced glycoxidation and lipoxidation end products (AGEs and ALEs): An overview of their mechanisms of formation. Free Radic. Res. 2013, 47, 3–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, R.; Matsumoto, K.; Ling, X.; Suzuki, H.; Araki, T.; Horiuchi, S. Glycolaldehyde, a reactive intermediate for advanced glycation end products, plays an important role in the generation of an active ligand for the macrophage scavenger receptor. Diabetes 2000, 49, 1714–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monnier, V.M.; Kohn, R.R.; Cerami, A. Accelerated age-related browning of human collagen in diabetes mellitus. Proc. Natl. Acad. Sci. USA 1984, 81, 583–587. [Google Scholar] [CrossRef] [Green Version]
- Verzijl, N.; DeGroot, J.; Oldehinkel, E.; Bank, R.A.; Thorpe, S.R.; Baynes, J.W.; Bayliss, M.T.; Bijlsma, J.W.; Lafeber, F.P.; Tekoppele, J.M. Age-related accumulation of Maillard reaction products in human articular cartilage collagen. Biochem. J. 2000, 350, 381–387. [Google Scholar] [CrossRef]
- Ahmed, N.; Thornalley, P.; Dawczynski, J.; Franke, S.; Strobel, J.; Stein, G.; Haik, G.M. Methylglyoxal-Derived Hydroimidazolone Advanced Glycation End-Products of Human Lens Proteins. Investig. Ophthalmol. Vis. Sci. 2003, 44, 5287–5292. [Google Scholar] [CrossRef]
- Lee, H.J.; Howell, S.K.; Sanford, R.J.; Beisswenger, P.J. Methylglyoxal Can Modify GAPDH Activity and Structure. Ann. N. Y. Acad. Sci. 2005, 1043, 135–145. [Google Scholar] [CrossRef]
- Morcos, M.; Du, X.; Pfisterer, F.; Hutter, H.; Sayed, A.A.R.; Thornalley, P.; Ahmed, N.; Baynes, J.; Thorpe, S.; Kukudov, G.; et al. Glyoxalase-1 prevents mitochondrial protein modification and enhances lifespan in Caenorhabditis elegans. Aging Cell 2008, 7, 260–269. [Google Scholar] [CrossRef]
- Nishino, I.; Noguchi, S.; Murayama, K.; Driss, A.; Sugie, K.; Oya, Y.; Nagata, T.; Chida, K.; Takahashi, T.; Takusa, Y.; et al. Distal myopathy with rimmed vacuoles is allelic to hereditary inclusion body myopathy. Neurology 2002, 59, 1689–1693. [Google Scholar] [CrossRef]
- Mori-Yoshimura, M.; Monma, K.; Suzuki, N.; Aoki, M.; Kumamoto, T.; Tanaka, K.; Tomimitsu, H.; Nakano, S.; Sonoo, M.; Shimizu, J.; et al. Heterozygous UDP-GlcNAc 2-epimerase and N-acetylmannosamine kinase domain mutations in the GNE gene result in a less severe GNE myopathy phenotype compared to homozygous N-acetylmannosamine kinase domain mutations. J. Neurol. Sci. 2012, 318, 100–105. [Google Scholar] [CrossRef]
- Tomimitsu, H.; Ishikawa, K.; Shimizu, J.; Ohkoshi, N.; Kanazawa, I.; Mizusawa, H. Distal myopathy with rimmed vacuoles: Novel mutations in the GNE gene. Neurology 2002, 59, 451–454. [Google Scholar] [CrossRef]
- Tomimitsu, H.; Shimizu, J.; Ishikawa, K.; Ohkoshi, N.; Kanazawa, I.; Mizusawa, H. Distal myopathy with rimmed vacuoles (DMRV): New GNE mutations and splice variant. Neurology 2004, 62, 1607–1610. [Google Scholar] [CrossRef] [PubMed]
- Zieger, B.; Boeckelmann, D.; Anani, W.; Falet, H.; Zhu, J.; Glonnegger, H.; Full, H.; Andresen, F.; Erlacher, M.; Lausch, E.; et al. Novel GNE Gene Variants Associated with Severe Congenital Thrombocytopenia and Platelet Sialylation Defect. Thromb. Haemost. 2022, 122, 1139–1146. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Dutta, S.; Moon, I.S. Upregulation of dendritic arborization by N-acetyl-D-glucosamine kinase is not dependent on its kinase activity. Mol Cells. 2014, 37, 322–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, M.U.; Thorpe, S.R.; Baynes, J.W. Identification of N epsilon-carboxymethyllysine as a degradation product of fructoselysine in glycated protein. J. Biol. Chem. 1986, 261, 4889–4894. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.U.; Frye, E.B.; Degenhardt, T.P.; Thorpe, S.R.; Baynes, J.W. N ε-(Carboxyethyl)lysine, a product of the chemical modification of proteins by methylglyoxal, increases with age in human lens proteins. Biochem. J. 1997, 324, 565–570. [Google Scholar] [CrossRef]
- Alt, N.; Schieberle, P. Identification of N 7 -(1-Carboxyethyl)-Arginine, a Novel Posttranslational Protein Modification of Arginine Formed at High Hydrostatic Pressure. Ann. N. Y. Acad. Sci. 2005, 1043, 55–58. [Google Scholar] [CrossRef]
- Alt, N.; Schieberle, P. Model Studies on the Influence of High Hydrostatic Pressure on the Formation of Glycated Arginine Modifications at Elevated Temperatures. J. Agric. Food Chem. 2005, 53, 5789–5797. [Google Scholar] [CrossRef]
- Iijima, K.; Murata, M.; Takahara, H.; Irie, S.; Fujimoto, D. Identification of N(omega)-carboxymethylarginine as a novel acid-labileadvanced glycation end product in collagen. Biochem. J. 2000, 347, 23–27. [Google Scholar] [CrossRef]
- Lo, T.W.; E Westwood, M.; McLellan, A.C.; Selwood, T.; Thornalley, P.J. Binding and modification of proteins by methylglyoxal under physiological conditions. A kinetic and mechanistic study with N alpha-acetylarginine, N alpha-acetylcysteine, and N alpha-acetyllysine, and bovine serum albumin. J. Biol. Chem. 1994, 269, 32299–32305. [Google Scholar] [CrossRef]
- Thornalley, P.J. Pharmacology of methylglyoxal: Formation, modification of proteins and nucleic acids, and enzymatic detoxification-A role in pathogenesis and antiproliferative chemotherapy. Gen. Pharmacol. Vasc. Syst. 1996, 27, 565–573. [Google Scholar] [CrossRef]
- Thornalley, P.J.; Langborg, A.; Minhas, H.S. Formation of glyoxal, methylglyoxal and 3-deoxyglucosone in the glycation of proteins by glucose. Biochem. J. 1999, 344, 109–116. [Google Scholar] [CrossRef]
- Allaman, I.; Bélanger, M.; Magistretti, P.J. Methylglyoxal, the dark side of glycolysis. Front. Neurosci. 2015, 9, 23. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, T.; Namki, M. Formation of Two-Carbon Sugar Fragment at an Early Stage of the Browning Reaction of Sugar with Amine. Agric. Biol. Chem. 1980, 44, 2575–2580. [Google Scholar] [CrossRef]
- Wolff, S.P.; Crabbe, M.J.C.; Thornalley, P.J. The autoxidation of glyceraldehyde and other simple monosaccharides. Experientia 1984, 40, 244–246. [Google Scholar] [CrossRef]
- Wolff, S.P.; Dean, R.T. Glucose autoxidation and protein modification. The potential role of ‘autoxidative glycosylation’ in diabetes. Biochem. J. 1987, 245, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Wells-Knecht, K.J.; Zyzak, D.V.; Litchfield, J.E.; Thorpe, S.R.; Baynes, J.W. Identification of Glyoxal and Arabinose as Intermediates in the Autoxidative Modification of Proteins by Glucose. Biochemistry 1995, 34, 3702–3709. [Google Scholar] [CrossRef] [PubMed]
- Nagao, M.; Fujita, Y.; Wakabayashi, K.; Nukaya, H.; Kosuge, T.; Sugimura, T. Mutagens in coffee and other beverages. Environ. Health Perspect. 1986, 67, 89–91. [Google Scholar] [CrossRef]
- Carr, A.; Rowe, S. Factors Affecting Vitamin C Status and Prevalence of Deficiency: A Global Health Perspective. Nutrients 2020, 12, 1963. [Google Scholar] [CrossRef]
- Glomb, M.A.; Monnier, V.M. Mechanism of Protein Modification by Glyoxal and Glycolaldehyde, Reactive Intermediates of the Maillard Reaction. J. Biol. Chem. 1995, 270, 10017–10026. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, A.E.N.; Freire, A.M.J.P.; Voit, E.O. A quantitative model of the generation of N∊-(carboxymethyl)lysine in the Maillard reaction between collagen and glucose. Biochem. J. 2003, 376, 109–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, S.; Bichler, J.; Wells-Knecht, K.J.; Thorpe, S.R.; Baynes, J.W. N.epsilon.-(Carboxymethyl)lysine Is a Dominant Advanced Glycation End Product (AGE) Antigen in Tissue Proteins. Biochemistry 1995, 34, 10872–10878. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.B.; Walker, D.G. The isolation and preliminary characterization of N-acetyl-D-glucosamine kinase from rat kidney and liver. Biochem. J. 1980, 185, 565–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibrat, J.-F.; Madej, T.; Bryant, S.H. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996, 6, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Madej, T.; Lanczycki, C.J.; Zhang, D.; Thiessen, P.A.; Geer, R.C.; Marchler-Bauer, A.; Bryant, S.H. MMDB and VAST+: Tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 2014, 42, D297–D303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madej, T.; Marchler-Bauer, A.; Lanczycki, C.; Zhang, D.; Bryant, S.H. Biological Assembly Comparison with VAST+. Struct. Bioinform. Methods Protoc. 2020, 2112, 175–186. [Google Scholar] [CrossRef] [Green Version]
- National Library of Medicine. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 20 October 2022).
- Nishimasu, H.; Fushinobu, S.; Shoun, H.; Wakagi, T. Crystal Structures of an ATP-dependent Hexokinase with Broad Substrate Specificity from the Hyperthermophilic Archaeon Sulfolobus tokodaii. J. Biol. Chem. 2007, 282, 9923–9931. [Google Scholar] [CrossRef] [Green Version]
- National Library of Medicine. Available online: https://www.ncbi.nlm.nih.gov/snp/ (accessed on 22 August 2022).
- UniProt. Available online: https://www.uniprot.org (accessed on 16 November 2022).
- Uchida, K.; Kawakishi, S. 2-Oxo-histidine as a novel biological marker for oxidatively modified proteins. FEBS Lett. 1993, 332, 208–210. [Google Scholar] [CrossRef] [Green Version]
- Coussons, P.J.; Jacoby, J.; McKay, A.; Kelly, S.; Price, N.C.; Hunt, J.V. Glucose Modification of Human Serum Albumin: A Structural Study. Free Radic. Biol. Med. 1997, 22, 1217–1227. [Google Scholar] [CrossRef]
- Fonda, I.; Kenig, M.; Gaberc-Porekar, V.; Pristovaek, P.; Menart, V. Attachment of Histidine Tags to Recombinant Tumor Necrosis Factor-Alpha Drastically Changes Its Properties. Sci. World J. 2002, 2, 1312–1325. [Google Scholar] [CrossRef] [Green Version]
- Freydank, A.-C.; Brandt, W.; Dräger, B. Protein structure modeling indicates hexahistidine-tag interference with enzyme activity. Proteins Struct. Funct. Bioinform. 2008, 72, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Kimple, M.E.; Brill, A.L.; Pasker, R.L. Overview of affinity tags for protein purification. Curr. Protoc. Protein Sci. 2013, 73, 9.9.1–9.9.23. [Google Scholar] [CrossRef] [Green Version]
- Thielges, M.C.; Chung, J.K.; Axup, J.Y.; Fayer, M.D. Influence of Histidine Tag Attachment on Picosecond Protein Dynamics. Biochemistry 2011, 50, 5799–5805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koito, W.; Araki, T.; Horiuchi, S.; Nagai, R. Conventional Antibody against Nε-(Carboxymethyl)Lysine (CML) Shows Cross-Reaction to Nε-(Carboxyethyl)Lysine (CEL): Immunochemical Quantification of CML with a Specific Antibody. J. Biochem. 2004, 136, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Rosenstock, P.; Bezold, V.; Bork, K.; Scheffler, J.; Horstkorte, R. Glycation interferes with natural killer cell function. Mech. Ageing Dev. 2019, 178, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Randell, E.; Vasdev, S.; Gill, V.; Gadag, V.; Newhook, L.A.; Grant, M.; Hagerty, D. Plasma methylglyoxal and glyoxal are elevated and related to early membrane alteration in young, complication-free patients with Type 1 diabetes. Mol. Cell. Biochem. 2007, 305, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Dhananjayan, K.; Irrgang, F.; Raju, R.; Harman, D.G.; Moran, C.; Srikanth, V.; Münch, G. Determination of glyoxal and methylglyoxal in serum by UHPLC coupled with fluorescence detection. Anal. Biochem. 2019, 573, 51–66. [Google Scholar] [CrossRef]
- Boyden, S.E.; Duncan, A.R.; Estrella, E.A.; Lidov, H.G.; Mahoney, L.J.; Katz, J.S.; Kunkel, L.M.; Kang, P.B. Molecular diagnosis of hereditary inclusion body myopathy by linkage analysis and identification of a novel splice site mutation in GNE. BMC Med. Genet. 2011, 12, 87. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Chanana, P.; Bharadwaj, R.; Bhattacharya, S.; Arya, R. Functional characterization of GNE mutations prevalent in Asian subjects with GNE myopathy, an ultra-rare neuromuscular disorder. Biochimie 2022, 199, 36–45. [Google Scholar] [CrossRef]
- Hart, G.W. Nutrient regulation of signaling and transcription. J. Biol. Chem. 2019, 294, 2211–2231. [Google Scholar] [CrossRef] [Green Version]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef] [PubMed]
- Glomb, M.A.; Pfahler, C. Amides Are Novel Protein Modifications Formed by Physiological Sugars. J. Biol. Chem. 2001, 276, 41638–41647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldensperger, T.; Glomb, M.A. Pathways of Non-enzymatic Lysine Acylation. Front. Cell Dev. Biol. 2021, 9, 664553. [Google Scholar] [CrossRef]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/search/GlcNAc (accessed on 15 November 2022).
- Hinderlich, S.; Berger, M.; Schwarzkopf, M.; Effertz, K.; Reutter, W. Molecular cloning and characterization of murine and human N-acetylglucosamine kinase. JBIC J. Biol. Inorg. Chem. 2000, 267, 3301–3308. [Google Scholar] [CrossRef] [Green Version]
- Campbell, S.; Mesaros, C.; Izzo, L.; Affronti, H.; Noji, M.; E Schaffer, B.; Tsang, T.; Sun, K.; Trefely, S.; Kruijning, S.; et al. Glutamine deprivation triggers NAGK-dependent hexosamine salvage. Elife 2021, 10, e62644. [Google Scholar] [CrossRef] [PubMed]
- Szwergold, B.S.; Howell, S.; Beisswenger, P.J. Human fructosamine-3-kinase: Purification, sequencing, substrate specificity, and evidence of activity in vivo. Diabetes 2001, 50, 2139–2147. [Google Scholar] [CrossRef] [Green Version]
- Van Schaftingen, E.; Collard, F.; Wiame, E.; Veiga-Da-Cunha, M. Enzymatic repair of Amadori products. Amino Acids 2012, 42, 1143–1150. [Google Scholar] [CrossRef]
- Tanhäuserová, V.; Kuricová, K.; Pácal, L.; Bartáková, V.; Řehořová, J.; Svojanovský, J.; Olšovský, J.; Bělobrádková, J.; Kaňková, K. Genetic variability in enzymes of metabolic pathways conferring protection against non-enzymatic glycation versus diabetes-related morbidity and mortality. Clin. Chem. Lab. Med. 2014, 52, 77–83. [Google Scholar] [CrossRef]
- Avemaria, F.; Carrera, P.; Lapolla, A.; Sartore, G.; Chilelli, N.C.; Paleari, R.; Ambrosi, A.; Ferrari, M.; Mosca, A. Possible role of fructosamine 3-kinase genotyping for the management of diabetic patients. Clin. Chem. Lab. Med. 2015, 53, 1315–1320. [Google Scholar] [CrossRef]
- De Bruyne, S.; van Schie, L.; Himpe, J.; De Somer, F.; Everaert, I.; Derave, W.; Broecke, C.V.D.; Huizing, M.; Bostan, N.; Speeckaert, M.; et al. A Potential Role for Fructosamine-3-Kinase in Cataract Treatment. Int. J. Mol. Sci. 2021, 22, 3841. [Google Scholar] [CrossRef]
- Thornalley, P.J. The glyoxalase system in health and disease. Mol. Asp. Med. 1993, 14, 287–371. [Google Scholar] [CrossRef] [PubMed]
- Mannervik, B. Molecular enzymology of the glyoxalase system. Drug Metab. Drug Interact. 2008, 23, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Hovatta, I.; Tennant, R.S.; Helton, R.; Marr, R.A.; Singer, O.; Redwine, J.M.; Ellison, J.A.; Schadt, E.E.; Verma, I.M.; Lockhart, D.J.; et al. Glyoxalase 1 and glutathione reductase 1 regulate anxiety in mice. Nature 2005, 438, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Distler, M.G.; Plant, L.D.; Sokoloff, G.; Hawk, A.J.; Aneas, I.; Wuenschell, G.E.; Termini, J.; Meredith, S.C.; Nobrega, M.A.; Palmer, A.A. Glyoxalase 1 increases anxiety by reducing GABAA receptor agonist methylglyoxal. J. Clin. Investig. 2012, 122, 2306–2315. [Google Scholar] [CrossRef] [Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hagenhaus, V.; Gorenflos López, J.L.; Rosenstengel, R.; Neu, C.; Hackenberger, C.P.R.; Celik, A.; Weinert, K.; Nguyen, M.-B.; Bork, K.; Horstkorte, R.; et al. Glycation Interferes with the Activity of the Bi-Functional UDP-N-Acetylglucosamine 2-Epimerase/N-Acetyl-mannosamine Kinase (GNE). Biomolecules 2023, 13, 422. https://doi.org/10.3390/biom13030422
Hagenhaus V, Gorenflos López JL, Rosenstengel R, Neu C, Hackenberger CPR, Celik A, Weinert K, Nguyen M-B, Bork K, Horstkorte R, et al. Glycation Interferes with the Activity of the Bi-Functional UDP-N-Acetylglucosamine 2-Epimerase/N-Acetyl-mannosamine Kinase (GNE). Biomolecules. 2023; 13(3):422. https://doi.org/10.3390/biom13030422
Chicago/Turabian StyleHagenhaus, Vanessa, Jacob L. Gorenflos López, Rebecca Rosenstengel, Carolin Neu, Christian P. R. Hackenberger, Arif Celik, Klara Weinert, Mai-Binh Nguyen, Kaya Bork, Rüdiger Horstkorte, and et al. 2023. "Glycation Interferes with the Activity of the Bi-Functional UDP-N-Acetylglucosamine 2-Epimerase/N-Acetyl-mannosamine Kinase (GNE)" Biomolecules 13, no. 3: 422. https://doi.org/10.3390/biom13030422






