USP18 Is Associated with PD-L1 Antitumor Immunity and Improved Prognosis in Colorectal Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. USP18 Expression Analysis
2.2. Kaplan-Meier (KM) Plotter Database Analysis
2.3. Cell Culture
2.4. Cell Transfection
2.5. qRT-PCR
2.6. Western Blotting
2.7. Cell Proliferation Assay
2.8. Data Acquisition and Analysis
2.9. Gene Set Enrichment Analysis (GSEA)
2.10. Functional Enrichment Analysis of the Immune Microenvironment
2.11. Single-Cell Sequencing Analysis
2.12. Protein–Protein Docking
2.13. Enzyme-Linked Immunosorbent Assay (ELISA)
2.14. Statistical Analysis
3. Results
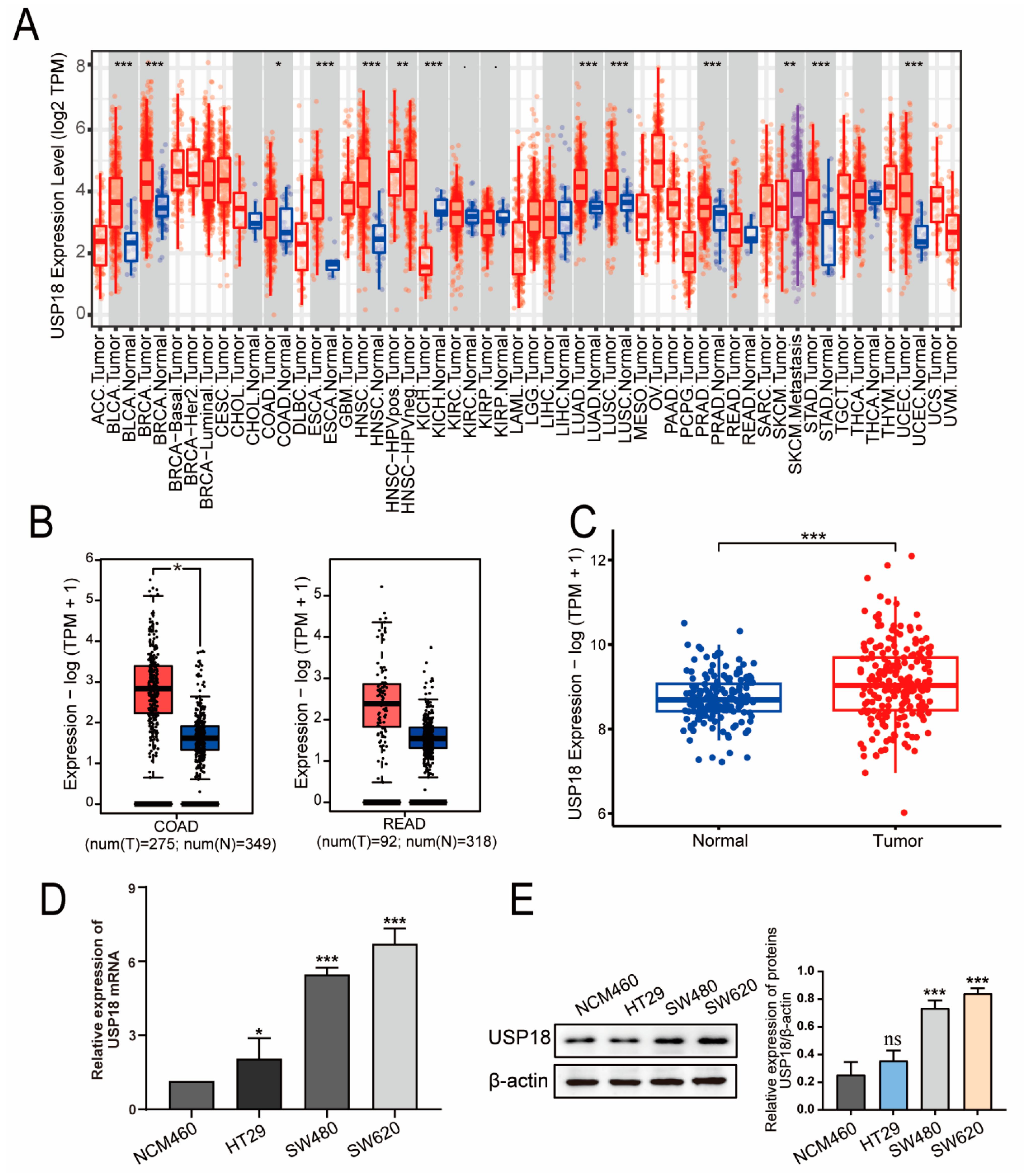
3.1. USP18 Is Highly Expressed in CRC Patients
3.2. Associations of USP18 Expression with Clinicopathological Features and the Prognosis of CRC Patients
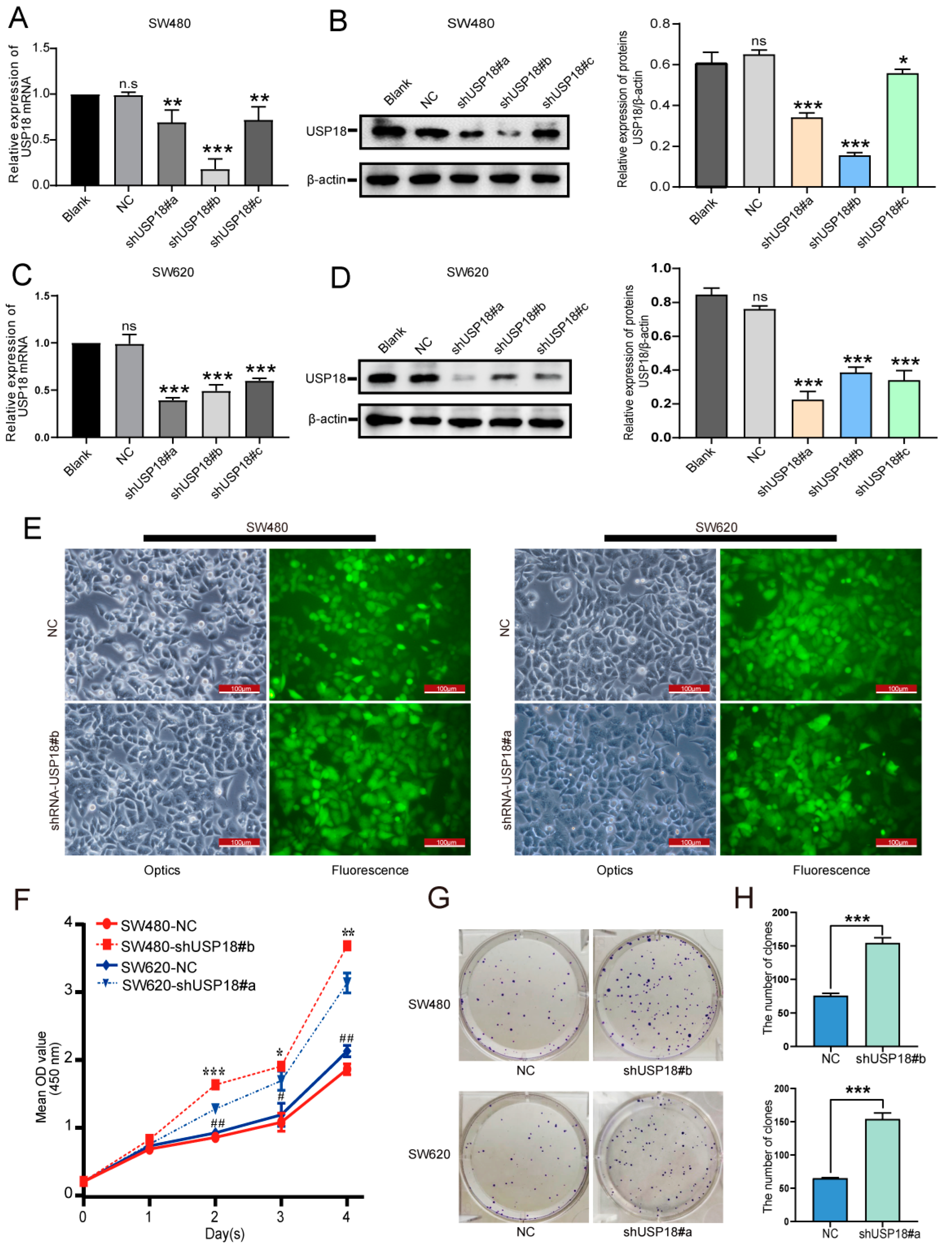
3.3. Silencing USP18 Promotes CRC Cell Proliferation and Colony Formation
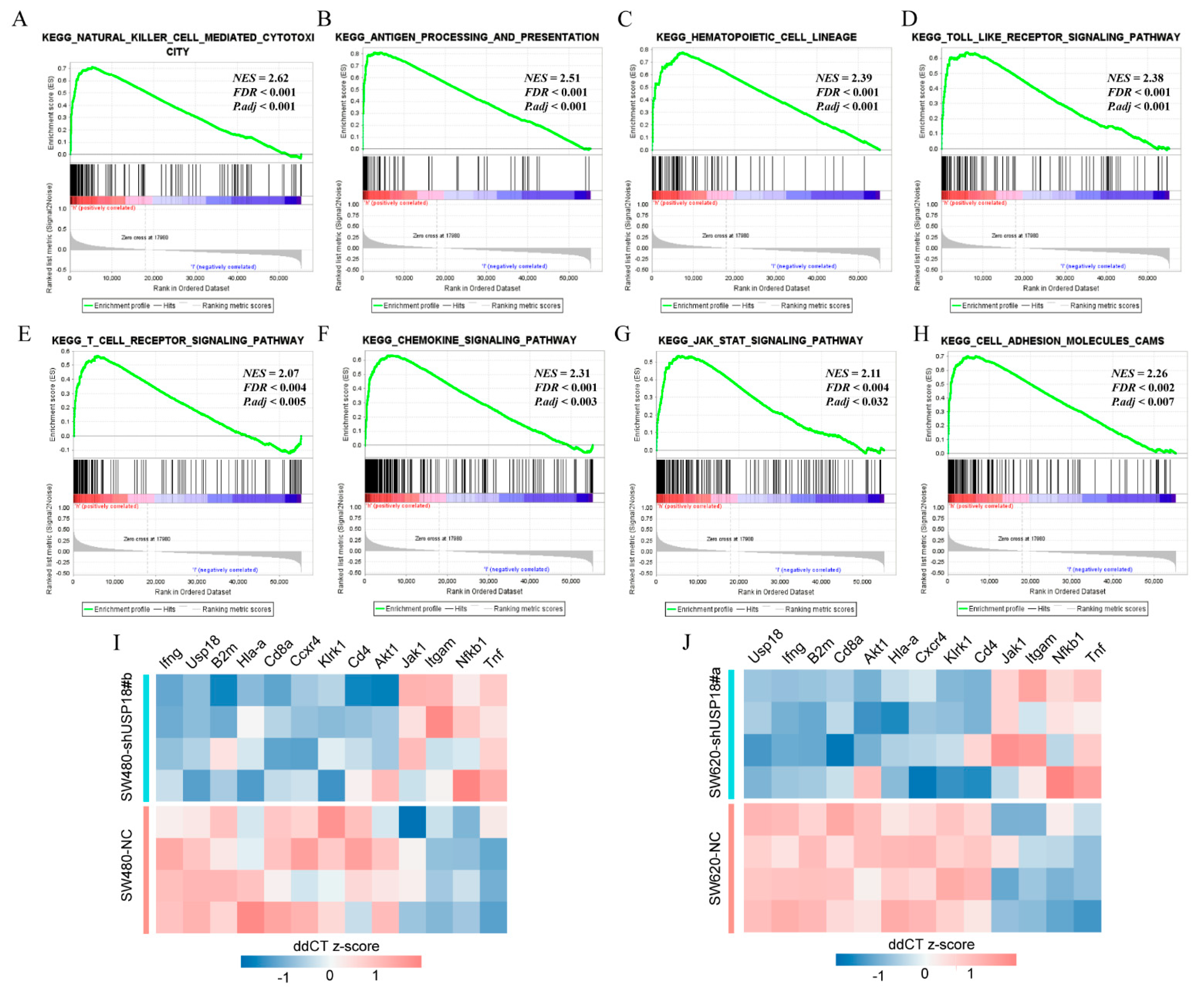
3.4. Potential Biological Functions and Pathways of USP18 in CRC
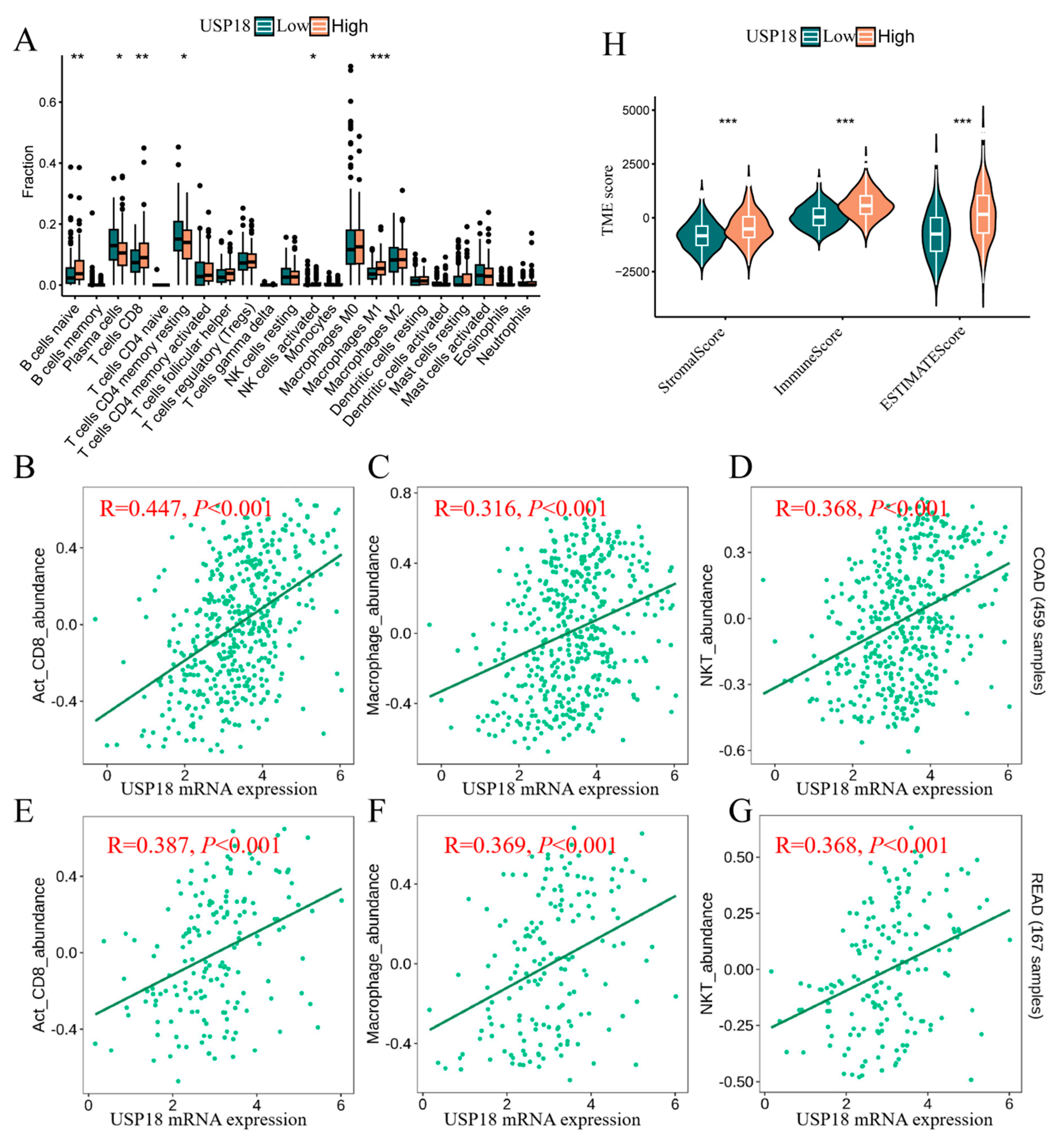
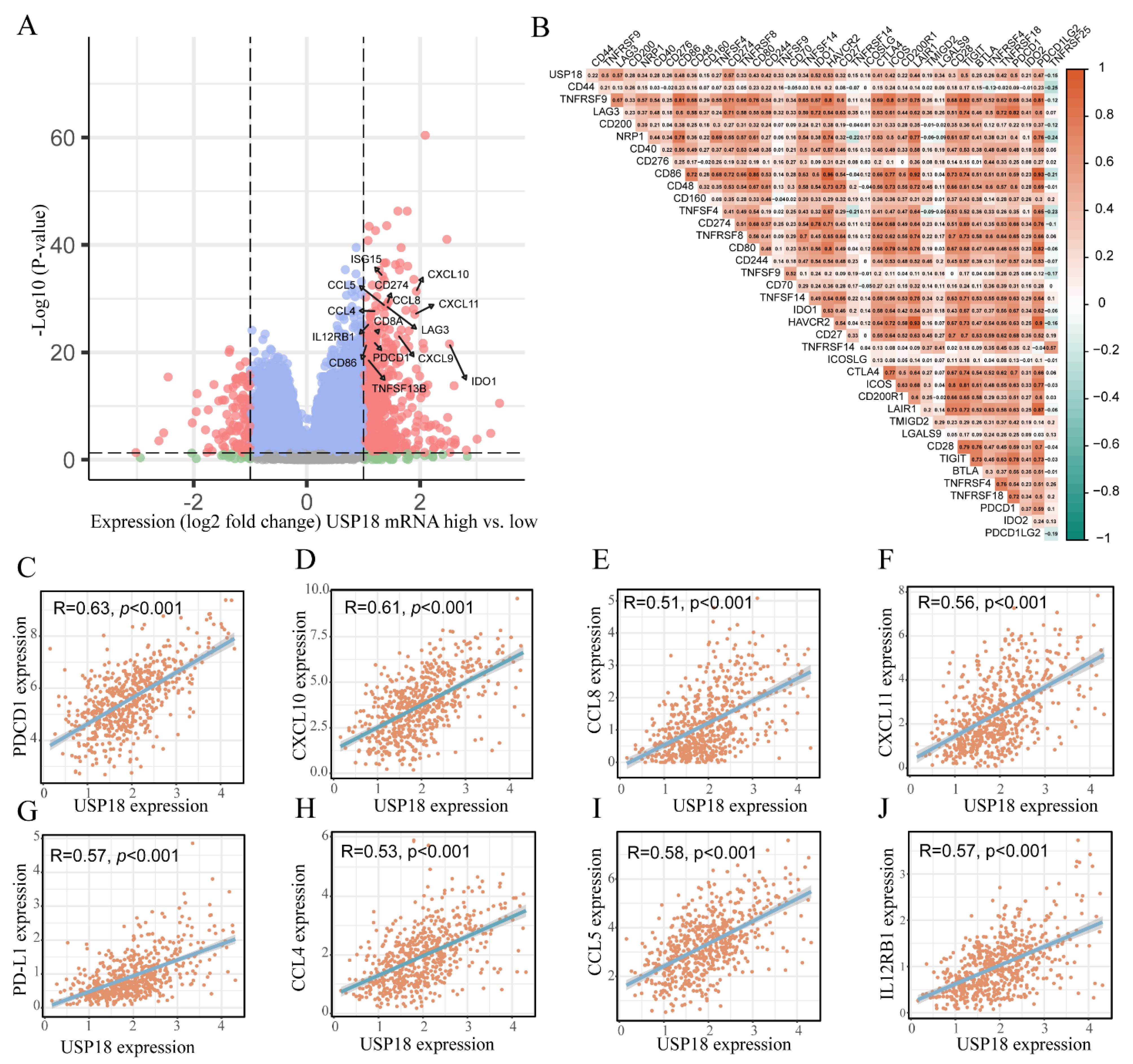
3.5. USP18 and Immune Cell Infiltration
3.6. Relationships of USP18 with Immune Checkpoint Genes in CRC
3.7. Regulation of PD-L1 Expression by USP18 Enhances Targeted Immunotherapy in CRC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- GBD 2019 Colorectal Cancer Collaborators. Global, regional, and national burden of colorectal cancer and its risk factors, 1990-2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet Gastroenterol. Hepatol. 2022, 7, 627–647. [Google Scholar] [CrossRef] [PubMed]
- Biller, L.H.; Schrag, D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA 2021, 325, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A., Jr. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Nikolouzakis, T.K.; Chrysos, E.; Docea, A.O.; Fragkiadaki, P.; Souglakos, J.; Tsiaoussis, J.; Tsatsakis, A. Current and Future Trends of Colorectal Cancer Treatment: Exploring Advances in Immunotherapy. Cancers 2024, 16, 1995. [Google Scholar] [CrossRef]
- Ishida, Y.; Agata, Y.; Shibahara, K.; Honjo, T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992, 11, 3887–3895. [Google Scholar] [CrossRef]
- Pauken, K.E.; Torchia, J.A.; Chaudhri, A.; Sharpe, A.H.; Freeman, G.J. Emerging concepts in PD-1 checkpoint biology. Semin. Immunol. 2021, 52, 101480. [Google Scholar] [CrossRef]
- Cai, J.; Wang, D.; Zhang, G.; Guo, X. The Role Of PD-1/PD-L1 Axis in Treg Development and Function: Implications for Cancer Immunotherapy. OncoTargets Ther. 2019, 12, 8437–8445. [Google Scholar] [CrossRef]
- Gao, M.; Shi, J.; Xiao, X.; Yao, Y.; Chen, X.; Wang, B.; Zhang, J. PD-1 regulation in immune homeostasis and immunotherapy. Cancer Lett. 2024, 588, 216726. [Google Scholar] [CrossRef]
- Doroshow, D.B.; Bhalla, S.; Beasley, M.B.; Sholl, L.M.; Kerr, K.M.; Gnjatic, S.; Wistuba, I.I.; Rimm, D.L.; Tsao, M.S.; Hirsch, F.R. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2021, 18, 345–362. [Google Scholar] [CrossRef]
- Wu, M.; Huang, Q.; Xie, Y.; Wu, X.; Ma, H.; Zhang, Y.; Xia, Y. Improvement of the anticancer efficacy of PD-1/PD-L1 blockade via combination therapy and PD-L1 regulation. J. Hematol. Oncol. 2022, 15, 24. [Google Scholar] [CrossRef]
- Hong, Y.; Chen, Q.; Wang, Z.; Zhang, Y.; Li, B.; Guo, H.; Huang, C.; Kong, X.; Mo, P.; Xiao, N.; et al. Targeting Nuclear Receptor Coactivator SRC-1 Prevents Colorectal Cancer Immune Escape by Reducing Transcription and Protein Stability of PD-L1. Adv. Sci. 2024, 11, e2310037. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.X.; Istl, A.C.; Quan, D.; Skaro, A.; Tang, E.; Zheng, X. PD-1 and PD-L1 inhibitors in cold colorectal cancer: Challenges and strategies. Cancer Immunol. Immunother. 2023, 72, 3875–3893. [Google Scholar] [CrossRef] [PubMed]
- Payandeh, Z.; Khalili, S.; Somi, M.H.; Mard-Soltani, M.; Baghbanzadeh, A.; Hajiasgharzadeh, K.; Samadi, N.; Baradaran, B. PD-1/PD-L1-dependent immune response in colorectal cancer. J. Cell Physiol. 2020, 235, 5461–5475. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, R.; Wang, B.; Lian, J.; Yao, Y.; Sun, H.; Zhang, C.; Fang, L.; Guan, X.; Shi, J.; et al. Blocking IL-17A enhances tumor response to anti-PD-1 immunotherapy in microsatellite stable colorectal cancer. J. Immunother. Cancer 2021, 9, e001895. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Bi, D.; Xie, R.; Li, M.; Guo, J.; Liu, H.; Guo, X.; Fang, J.; Ding, T.; Zhu, H.; et al. Fusobacterium nucleatum enhances the efficacy of PD-L1 blockade in colorectal cancer. Signal Transduct. Target. Ther. 2021, 6, 398. [Google Scholar] [CrossRef]
- Ren, D.; Hua, Y.; Yu, B.; Ye, X.; He, Z.; Li, C.; Wang, J.; Mo, Y.; Wei, X.; Chen, Y.; et al. Predictive biomarkers and mechanisms underlying resistance to PD1/PD-L1 blockade cancer immunotherapy. Mol. Cancer 2020, 19, 19. [Google Scholar] [CrossRef]
- Zhu, D.; Xu, R.; Huang, X.; Tang, Z.; Tian, Y.; Zhang, J.; Zheng, X. Deubiquitinating enzyme OTUB1 promotes cancer cell immunosuppression via preventing ER-associated degradation of immune checkpoint protein PD-L1. Cell Death Differ. 2021, 28, 1773–1789. [Google Scholar] [CrossRef]
- Xiao, X.; Shi, J.; He, C.; Bu, X.; Sun, Y.; Gao, M.; Xiang, B.; Xiong, W.; Dai, P.; Mao, Q.; et al. ERK and USP5 govern PD-1 homeostasis via deubiquitination to modulate tumor immunotherapy. Nat. Commun. 2023, 14, 2859. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, X.; Lao, M.; Sun, K.; He, L.; Xu, J.; Duan, Y.; Chen, Y.; Ying, H.; Li, M.; et al. Targeting ubiquitin-specific protease 8 sensitizes anti-programmed death-ligand 1 immunotherapy of pancreatic cancer. Cell Death Differ. 2023, 30, 560–575. [Google Scholar] [CrossRef]
- Yang, Z.; Xu, G.; Wang, B.; Liu, Y.; Zhang, L.; Jing, T.; Tang, M.; Xu, X.; Jiao, K.; Xiang, L.; et al. USP12 downregulation orchestrates a protumourigenic microenvironment and enhances lung tumour resistance to PD-1 blockade. Nat. Commun. 2021, 12, 4852. [Google Scholar] [CrossRef]
- Malakhov, M.P.; Malakhova, O.A.; Kim, K.I.; Ritchie, K.J.; Zhang, D.E. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 2002, 277, 9976–9981. [Google Scholar] [CrossRef] [PubMed]
- Qu, T.; Zhang, W.; Yan, C.; Ren, D.; Wang, Y.; Guo, Y.; Guo, Q.; Wang, J.; Liu, L.; Han, L.; et al. ISG15 targets glycosylated PD-L1 and promotes its degradation to enhance antitumor immune effects in lung adenocarcinoma. J. Transl. Med. 2023, 21, 341. [Google Scholar] [CrossRef] [PubMed]
- Oyoshi, H.; Du, J.; Sakai, S.A.; Yamashita, R.; Okumura, M.; Motegi, A.; Hojo, H.; Nakamura, M.; Hirata, H.; Sunakawa, H.; et al. Comprehensive single-cell analysis demonstrates radiotherapy-induced infiltration of macrophages expressing immunosuppressive genes into tumor in esophageal squamous cell carcinoma. Sci. Adv. 2023, 9, eadh9069. [Google Scholar] [CrossRef]
- Basters, A.; Knobeloch, K.P.; Fritz, G. USP18—A multifunctional component in the interferon response. Biosci. Rep. 2018, 38, BSR20180250. [Google Scholar] [CrossRef]
- Aricò, E.; Castiello, L.; Capone, I.; Gabriele, L.; Belardelli, F. Type I Interferons and Cancer: An Evolving Story Demanding Novel Clinical Applications. Cancers 2019, 11, 1943. [Google Scholar] [CrossRef]
- Ding, N.; You, A.B.; Yang, H.; Hu, G.S.; Lai, C.P.; Liu, W.; Ye, F. A Tumor-suppressive Molecular Axis EP300/circRERE/miR-6837-3p/MAVS Activates Type I IFN Pathway and Antitumor Immunity to Suppress Colorectal Cancer. Clin. Cancer Res. 2023, 29, 2095–2109. [Google Scholar] [CrossRef] [PubMed]
- Krug, J.; Rodrian, G.; Petter, K.; Yang, H.; Khoziainova, S.; Guo, W.; Bénard, A.; Merkel, S.; Gellert, S.; Maschauer, S.; et al. N-glycosylation Regulates Intrinsic IFN-γ Resistance in Colorectal Cancer: Implications for Immunotherapy. Gastroenterology 2023, 164, 392–406.e395. [Google Scholar] [CrossRef]
- Győrffy, B.; Surowiak, P.; Budczies, J.; Lánczky, A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 2013, 8, e82241. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef]
- Taylor, J.P.; Cash, M.N.; Santostefano, K.E.; Nakanishi, M.; Terada, N.; Wallet, M.A. CRISPR/Cas9 knockout of USP18 enhances type I IFN responsiveness and restricts HIV-1 infection in macrophages. J. Leukoc. Biol. 2018, 103, 1225–1240. [Google Scholar] [CrossRef]
- Cong, X.L.; Lo, M.C.; Reuter, B.A.; Yan, M.; Fan, J.B.; Zhang, D.E. Usp18 promotes conventional CD11b+ dendritic cell development. J. Immunol. 2012, 188, 4776–4781. [Google Scholar] [CrossRef]
- Miyauchi, S.; Arimoto, K.I.; Liu, M.; Zhang, Y.; Zhang, D.E. Reprogramming of tumor-associated macrophages via NEDD4-mediated CSF1R degradation by targeting USP18. Cell Rep. 2023, 42, 113560. [Google Scholar] [CrossRef]
- Kang, J.A.; Jeon, Y.J. Emerging Roles of USP18: From Biology to Pathophysiology. Int. J. Mol. Sci. 2020, 21, 6825. [Google Scholar] [CrossRef]
- Liu, X.; Lu, Y.; Chen, Z.; Liu, X.; Hu, W.; Zheng, L.; Chen, Y.; Kurie, J.M.; Shi, M.; Mustachio, L.M.; et al. The Ubiquitin-Specific Peptidase USP18 Promotes Lipolysis, Fatty Acid Oxidation, and Lung Cancer Growth. Mol. Cancer Res. 2021, 19, 667–677. [Google Scholar] [CrossRef]
- Song, W.; Yang, K.; Luo, J.; Gao, Z.; Gao, Y. Dysregulation of USP18/FTO/PYCR1 signaling network promotes bladder cancer development and progression. Aging 2021, 13, 3909–3925. [Google Scholar] [CrossRef]
- Arimoto, K.I.; Miyauchi, S.; Troutman, T.D.; Zhang, Y.; Liu, M.; Stoner, S.A.; Davis, A.G.; Fan, J.B.; Huang, Y.J.; Yan, M.; et al. Expansion of interferon inducible gene pool via USP18 inhibition promotes cancer cell pyroptosis. Nat. Commun. 2023, 14, 251. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, T.; Xiao, J.; Zhu, K.; Wei, R.; Wu, Z.; Meng, H.; Li, Y.; Yuan, J. Modification of BECN1 by ISG15 plays a crucial role in autophagy regulation by type I IFN/interferon. Autophagy 2015, 11, 617–628. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, L.; Zhang, S.; Yang, J.; Zhu, A.; Sun, J.; Kalvakolanu, D.V.; Cong, X.; Zhang, J.; Tang, J.; et al. Taxifolin inhibits melanoma proliferation/migration impeding USP18/Rac1/JNK/β-catenin oncogenic signaling. Phytomedicine 2024, 123, 155199. [Google Scholar] [CrossRef]
- Potu, H.; Sgorbissa, A.; Brancolini, C. Identification of USP18 as an important regulator of the susceptibility to IFN-alpha and drug-induced apoptosis. Cancer Res. 2010, 70, 655–665. [Google Scholar] [CrossRef]
- Diao, W.; Guo, Q.; Zhu, C.; Song, Y.; Feng, H.; Cao, Y.; Du, M.; Chen, H. USP18 promotes cell proliferation and suppressed apoptosis in cervical cancer cells via activating AKT signaling pathway. BMC Cancer 2020, 20, 741. [Google Scholar] [CrossRef]
- Honke, N.; Shaabani, N.; Zhang, D.E.; Hardt, C.; Lang, K.S. Multiple functions of USP18. Cell Death Dis. 2016, 7, e2444. [Google Scholar] [CrossRef]
- Mustachio, L.M.; Lu, Y.; Kawakami, M.; Roszik, J.; Freemantle, S.J.; Liu, X.; Dmitrovsky, E. Evidence for the ISG15-Specific Deubiquitinase USP18 as an Antineoplastic Target. Cancer Res. 2018, 78, 587–592. [Google Scholar] [CrossRef]
- Pinto-Fernandez, A.; Salio, M.; Partridge, T.; Chen, J.; Vere, G.; Greenwood, H.; Olie, C.S.; Damianou, A.; Scott, H.C.; Pegg, H.J.; et al. Deletion of the deISGylating enzyme USP18 enhances tumour cell antigenicity and radiosensitivity. Br. J. Cancer 2021, 124, 817–830. [Google Scholar] [CrossRef]
- Hong, B.; Li, H.; Lu, Y.; Zhang, M.; Zheng, Y.; Qian, J.; Yi, Q. USP18 is crucial for IFN-γ-mediated inhibition of B16 melanoma tumorigenesis and antitumor immunity. Mol. Cancer 2014, 13, 132. [Google Scholar] [CrossRef]
- Malakhova, O.A.; Kim, K.I.; Luo, J.K.; Zou, W.; Kumar, K.G.; Fuchs, S.Y.; Shuai, K.; Zhang, D.E. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006, 25, 2358–2367. [Google Scholar] [CrossRef]
- Zhang, X.; Cheng, L.; Gao, C.; Chen, J.; Liao, S.; Zheng, Y.; Xu, L.; He, J.; Wang, D.; Fang, Z.; et al. Androgen Signaling Contributes to Sex Differences in Cancer by Inhibiting NF-κB Activation in T Cells and Suppressing Antitumor Immunity. Cancer Res. 2023, 83, 906–921. [Google Scholar] [CrossRef]
- Burkart, C.; Arimoto, K.; Tang, T.; Cong, X.; Xiao, N.; Liu, Y.C.; Kotenko, S.V.; Ellies, L.G.; Zhang, D.E. Usp18 deficient mammary epithelial cells create an antitumour environment driven by hypersensitivity to IFN-λ and elevated secretion of Cxcl10. EMBO Mol. Med. 2013, 5, 1035–1050. [Google Scholar] [CrossRef] [PubMed]
- Bule, P.; Aguiar, S.I.; Aires-Da-Silva, F.; Dias, J.N.R. Chemokine-Directed Tumor Microenvironment Modulation in Cancer Immunotherapy. Int. J. Mol. Sci. 2021, 22, 9804. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Grochans, S.; Gutowska, I.; Barczak, K.; Baranowska-Bosiacka, I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of Receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 Ligands. Int. J. Mol. Sci. 2020, 21, 7619. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, Q.; Chen, T.; Su, R.; Pan, C.; Qian, J.; Huang, H.; Yin, S.; Xie, H.; Zhou, L.; et al. Blocking CD47 promotes antitumour immunity through CD103(+) dendritic cell-NK cell axis in murine hepatocellular carcinoma model. J. Hepatol. 2022, 77, 467–478. [Google Scholar] [CrossRef]
- Guia, S.; Cognet, C.; de Beaucoudrey, L.; Tessmer, M.S.; Jouanguy, E.; Berger, C.; Filipe-Santos, O.; Feinberg, J.; Camcioglu, Y.; Levy, J.; et al. A role for interleukin-12/23 in the maturation of human natural killer and CD56+ T cells in vivo. Blood 2008, 111, 5008–5016. [Google Scholar] [CrossRef]
- Wang, Z.; Kang, W.; Li, O.; Qi, F.; Wang, J.; You, Y.; He, P.; Suo, Z.; Zheng, Y.; Liu, H.M. Abrogation of USP7 is an alternative strategy to downregulate PD-L1 and sensitize gastric cancer cells to T cells killing. Acta Pharm. Sin. B 2021, 11, 694–707. [Google Scholar] [CrossRef]
- Zheng, R.; Gao, F.; Mao, Z.; Xiao, Y.; Yuan, L.; Huang, Z.; Lv, Q.; Qin, C.; Du, M.; Zhang, Z.; et al. LncRNA BCCE4 Genetically Enhances the PD-L1/PD-1 Interaction in Smoking-Related Bladder Cancer by Modulating miR-328-3p-USP18 Signaling. Adv. Sci. 2023, 10, e2303473. [Google Scholar] [CrossRef]


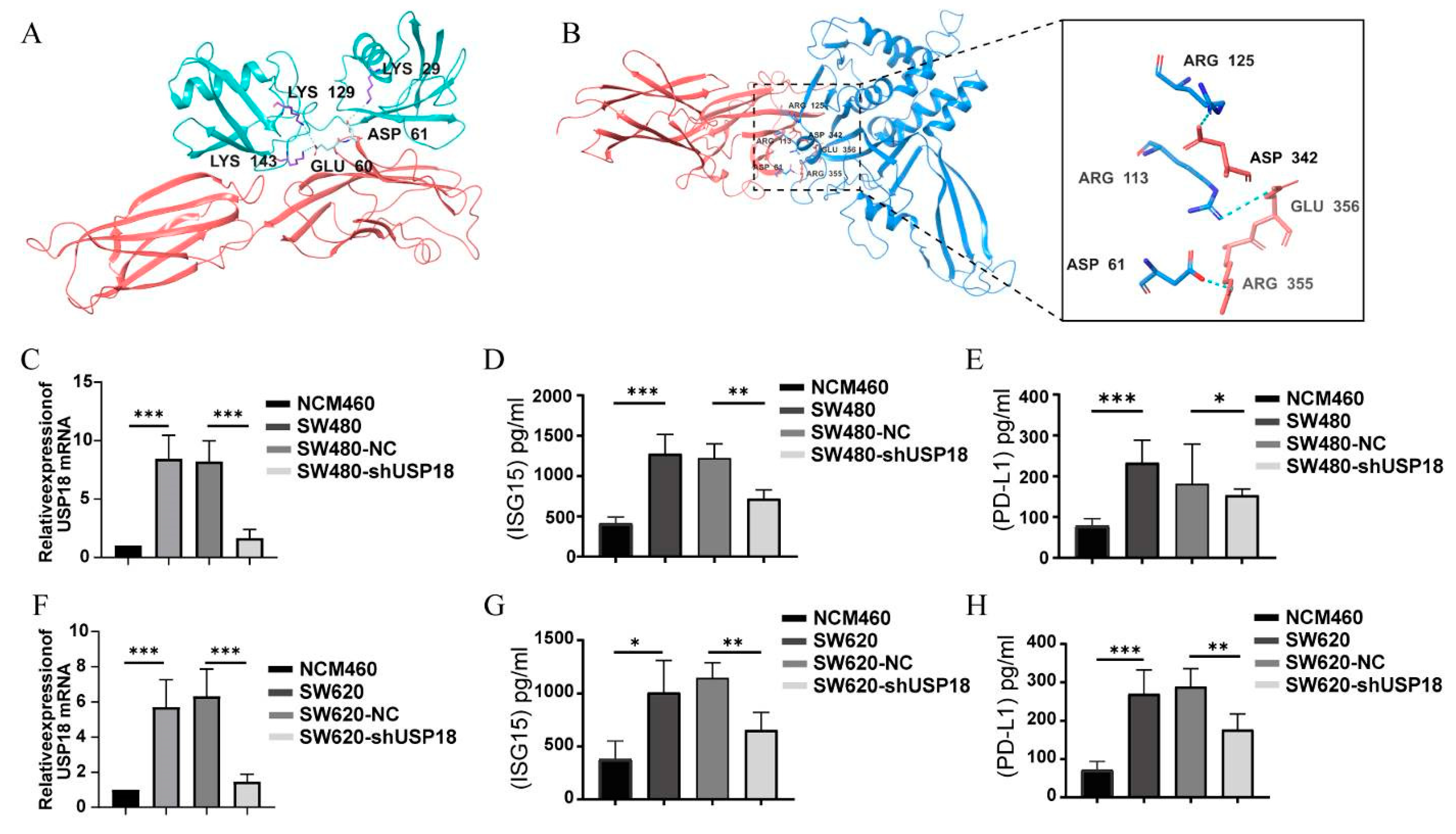
| Protein-Protein Complex | Docking Score | ΔG (kcal mol−1) | Kd (M) | ICs Charged–Charged | ICs Charged–Apolar |
|---|---|---|---|---|---|
| USP18-PD-L1 | −256.35 | −11.3 | 4.9 × 10−9 | 12 | 24 |
| ISG15-PD-L1 | −223.24 | −12.0 | 1.6 × 10−9 | 20 | 18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jifu, C.; Lu, L.; Ding, J.; Lv, M.; Xia, J.; Wang, J.; Wang, P. USP18 Is Associated with PD-L1 Antitumor Immunity and Improved Prognosis in Colorectal Cancer. Biomolecules 2024, 14, 1191. https://doi.org/10.3390/biom14091191
Jifu C, Lu L, Ding J, Lv M, Xia J, Wang J, Wang P. USP18 Is Associated with PD-L1 Antitumor Immunity and Improved Prognosis in Colorectal Cancer. Biomolecules. 2024; 14(9):1191. https://doi.org/10.3390/biom14091191
Chicago/Turabian StyleJifu, Cili, Linxia Lu, Jiaxin Ding, Mengjun Lv, Jun Xia, Jingtao Wang, and Peijun Wang. 2024. "USP18 Is Associated with PD-L1 Antitumor Immunity and Improved Prognosis in Colorectal Cancer" Biomolecules 14, no. 9: 1191. https://doi.org/10.3390/biom14091191





