Differential Expression of miRNAs Involved in Response to Candidatus Liberibacter asiaticus Infection in Mexican Lime at Early and Late Stages of Huanglongbing Disease
Abstract
:1. Introduction
2. Results
2.1. Data Analysis of Small RNA Sequencing
2.2. Identification of Known and Novel miRNAs in Mexican Lime
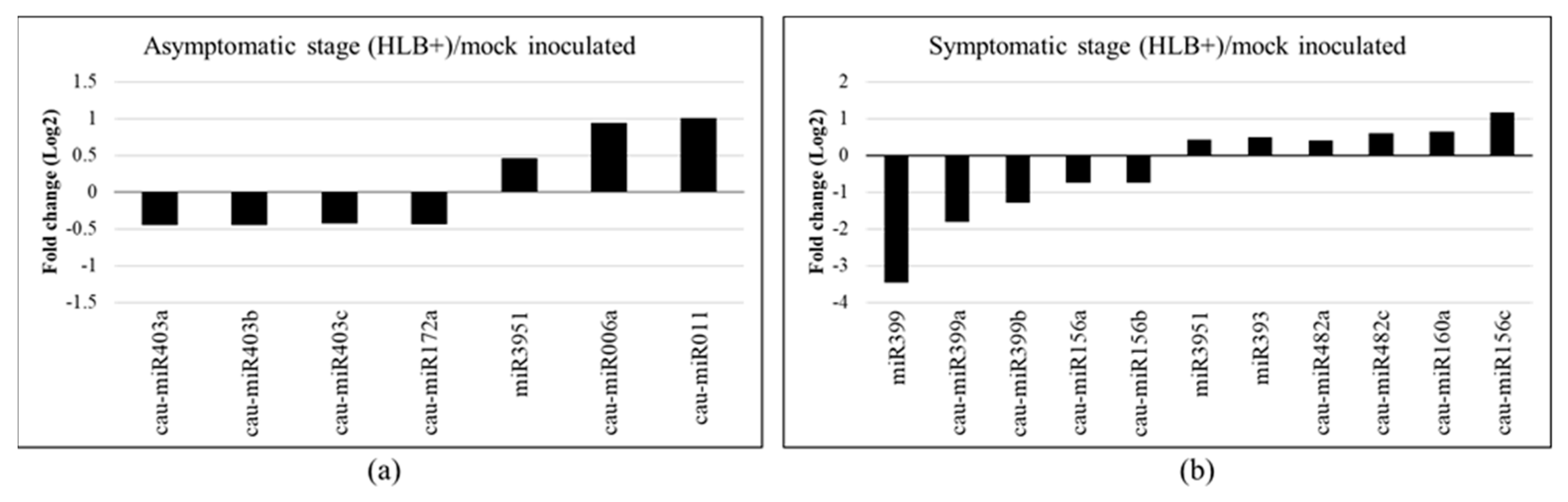
2.3. Differential Expression of miRNAs in Response to CLas in Citrus aurantifolia
2.4. Prediction of Potential Target Genes of the Differentially Expressed miRNAs
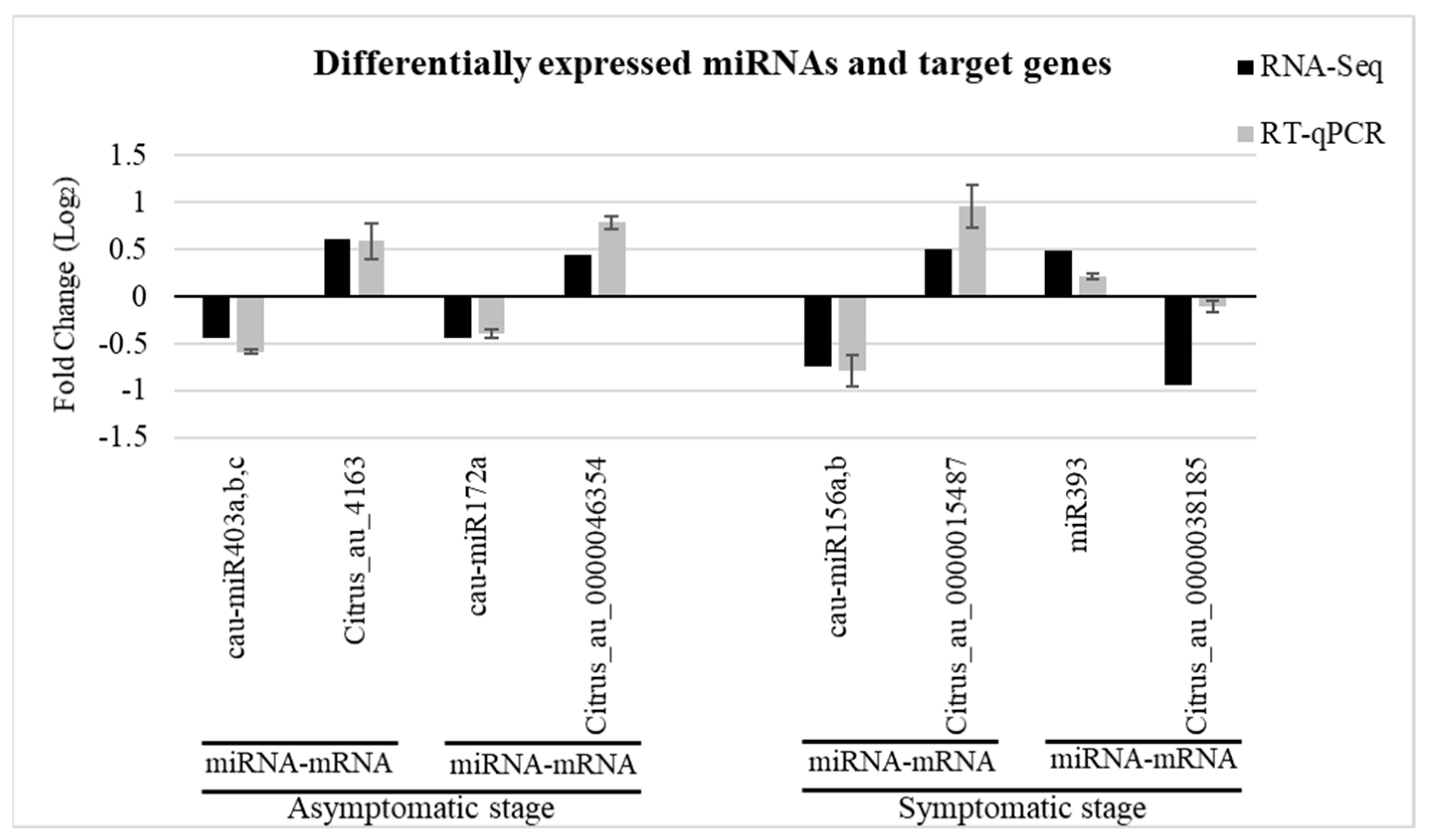
2.5. Validation of miRNAs and Target Genes
3. Discussion
4. Materials and Methods
4.1. Plant Material and Experimental Design
4.2. sRNA Library Construction and Sequencing
4.3. Identification of Known and Novel miRNAs in Mexican Lime
4.4. Differential Expression Analysis of miRNAs
4.5. Prediction and Annotation of Target Genes
4.6. Validation of miRNAs and Target Genes Expression by RT-qPCR
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Coletta-Filho, H.D.; Takita, M.A.; Targon, M.L.P.N.; Machado, M.A. Analysis of 16S rDNA sequences from citrus huanglongbing bacteria reveal a different “Ca. Liberibacter” strain associated with citrus disease in Sao Paulo. Plant Dis. 2005, 89, 848–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bové, J.M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 2006, 88, 7–37. [Google Scholar]
- Gottwald, T.R. Current Epidemiological Understanding of Citrus Huanglongbing. Annu. Rev. Phytopathol. 2010, 48, 119–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Servicio Nacional de Sanidad, Inocuidad y Calidad Agroalimentaria (SENASICA). Vigilancia del Huanglongbing. 2018. Available online: https://prod.senasica.gob.mx/SIRVEF/HLBV2.aspx (accessed on 2 October 2022).
- Albrecht, U.; Bowman, K.D. Gene expression in Citrus sinensis (L.) Osbeck following infection with the bacterial pathogen Candidatus Liberibacter asiaticus causing Huanglongbing in Florida. Plant Sci. 2008, 175, 291–306. [Google Scholar]
- Massenti, R.; Lo Bianco, R.; Sandhu, A.K.; Gu, L.; Sims, C. Huanglongbing modifies quality components and flavonoid content of ‘Valencia’ oranges. J. Sci. Food Agric. 2016, 96, 73–78. [Google Scholar] [CrossRef]
- Fan, J.; Chen, C.; Yu, Q.; Brlansky, R.H.; Li, Z.G.; Gmitter, F.G., Jr. Comparative iTRAQ proteome and transcriptome analyses of sweet orange infected by “Candidatus Liberibacter asiaticus”. Physiol. Plant. 2011, 143, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, F.; Uratsu, S.L.; Albrecht, U.; Reagan, R.L.; Leicht, E.; D’Souza, R.; Bowman, K.D.; Dandekar, A.M. Deep Transcriptome Profiling of Citrus Fruit in Response to Huanglongbing Disease. PLoS ONE 2012, 7, e38039. [Google Scholar] [CrossRef]
- Aritua, V.; Achor, D.; Gmitter, F.G.; Albrigo, G.; Wang, N. Transcriptional and microscopic analyses of citrus stem and root responses to Candidatus Liberibacter asiaticus infection. PLoS ONE 2013, 8, e73742. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Li, Y.; Zheng, Z.; Dai, Z.; Tao, Y.; Deng, X. Transcriptional analyses of mandarins seriously infected by ‘Candidatus Liberibacter asiaticus’. PLoS ONE 2015, 10, e0133652. [Google Scholar] [CrossRef]
- Zhong, Y.; Cheng, C.Z.; Jiang, N.H.; Jiang, B.; Zhang, Y.Y.; Wu, B.; Hu, M.I.; Zeng, J.W.; Yan, H.X.; Yi, G.J.; et al. Comparative transcriptome and iTRAQ proteome analyses of citrus root responses to Candidatus Liberibacter asiaticus infection. PLoS ONE 2015, 10, e0126973. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Zhong, X.; Liu, X.; Lou, B.; Zhou, C.; Wang, X. Comparative transcriptome analysis unveils the tolerance mechanisms of Citrus hystrix in response to ‘Candidatus Liberibacter asiaticus’ infection. PLoS ONE 2017, 12, e0189229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, E.L.; Ramsey, J.; Saha, S.; Mishchuk, D.; Chavez, J.; Howe, K.; Zhong, X.; Flores-Gonzalez, M.; Mitrovic, E.; Polek, M.; et al. Multi-omics comparison reveals landscape of Citrus limon and Citrus sinensis response to ‘Candidatus Liberibacter asiaticus’. Phytofrontiers 2021, 1, 76–84. [Google Scholar] [CrossRef]
- Yuning, L.; Xianmei, Y.; Jingjing, Z.; Jinghua, D.; Luyang, L.; Jintian, L.; Benshui, S. Transcriptome analyses reveal the potential mechanisms for color changes of a sweet orange peel induced by Candidatus Liberibacter asiaticus. Gene 2022, 839, 146736. [Google Scholar] [CrossRef]
- Arce-Leal, Á.P.; Bautista, R.; Rodríguez-Negrete, E.A.; Manzanilla-Ramírez, M.Á.; Velázquez-Monreal, J.J.; Santos-Cervantes, M.E.; Méndez-Lozano, J.; Beuzón, C.R.; Bejarano, E.R.; Castillo, A.G.; et al. Gene expression profile of mexican lime (Citrus aurantifolia) trees in response to huanglongbing disease caused by Candidatus liberibacter asiaticus. Microorganisms 2020, 8, 528. [Google Scholar] [CrossRef] [Green Version]
- Mallory, A.C.; Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006, 38, S31–S36. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Chinnusamy, V.; Zhu, J.; Zhu, J.K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007, 12, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Mei, J.; Ren, G. Plant microRNAs: Biogenesis, homeostasis, and degradation. Front. Plant Sci. 2019, 10, 360. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. Plant microRNA: A small regulatory molecule with big impact. Dev. Biol. 2006, 289, 3–16. [Google Scholar] [CrossRef]
- Mishra, R.; Mohapatra, R.; Mahanty, B.; Joshi, R.K. Analysis of microRNAs and their targets from onion (Allium cepa) using genome survey sequences (GSS) and expressed sequence tags (ESTs). Bioinformation 2019, 15, 907. [Google Scholar] [CrossRef] [Green Version]
- Axtell, M.J. ShortStack: Comprehensive annotation and quantification of small RNA genes. RNA 2013, 19, 740–751. [Google Scholar] [CrossRef] [Green Version]
- Johnson, N.R.; Yeoh, J.M.; Coruh, C.; Axtell, M.J. Improved placement of multi-mapping small RNAs. G3 Genes Genomes Genet. 2016, 6, 2103–2111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Axtell, M.J.; Meyers, B.C. Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell 2018, 30, 272–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, J.; Sun, Y. miR-PREFeR: An accurate, fast and easy-to-use plant miRNA prediction tool using small RNA-Seq data. Bioinformatics 2014, 30, 2837–2839. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Gao, S.; Zhou, X.; Chellappan, P.; Chen, Z.; Zhou, X.; Zhang, X.; Fromuth, N.; Coutino, G.; Coffey, M.; et al. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol. Biol. 2011, 75, 93–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Zhu, H.; Li, N.; Batley, J.; Wang, Y. The miR393-Target Module Regulates Plant Development and Responses to Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2022, 23, 9477. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Mutka, A.M.; Fawley, S.; Tsao, T.; Kunkel, B.N. Auxin promotes susceptibility to Pseudomonas syringae via a mechanism independent of suppression of salicylic acid-mediated defenses. Plant J. 2013, 74, 746–754. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Zhang, J.; Wu, L.; Qi, Y.; Zhou, J.M. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010, 152, 2222–2231. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Sun, R.; Albrecht, U.; Padmanabhan, C.; Wang, A.; Coffey, M.D.; Girke, T.; Wang, Z.; Close, T.J.; Roose, M.; et al. Small RNA profiling reveals phosphorus deficiency as a contributing factor in symptom expression for citrus huanglongbing disease. Mol. Plant 2013, 6, 301–310. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Y.; Cheng, C.; Moniruzzaman, M.; Jiang, B.; Jiang, N.; Zhong, G. Expression of miRNAs and their target genes in roots of ‘Sanhu’ tangerine (Citrus reticulata blanco cv. ‘Sanhu’) in response to Candidatus Liberibacter asiaticus infection. J. Plant Dis. Prot. 2021, 128, 407–420. [Google Scholar] [CrossRef]
- Louzada, E.S.; Vazquez, O.E.; Braswell, W.E.; Yanev, G.; Devanaboina, M.; Kunta, M. Distribution of ‘Candidatus Liberibacter asiaticus’ above and below ground in Texas citrus. Phytopathology 2016, 106, 702–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balan, B.; Ibáñez, A.M.; Dandekar, A.M.; Caruso, T.; Martinelli, F. Identifying host molecular features strongly linked with responses to huanglongbing disease in citrus leaves. Front. Plant Sci. 2018, 9, 277. [Google Scholar] [CrossRef] [PubMed]
- Wang, N. The citrus huanglongbing crisis and potential solutions. Mol. Plant 2019, 12, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Schilbert, H.M.; Rempel, A.; Pucker, B. Comparison of read mapping and variant calling tools for the analysis of plant NGS data. Plants 2020, 9, 439. [Google Scholar] [CrossRef] [Green Version]
- Chávez Montes, R.; Rosas-Cárdenas, d.; De Paoli, E.; Accerbi, M.; Rymarquis, L.A.; Mahalingam, G.; Marsch-Martínez, N.; Meyers, B.C.; Green, P.J.; de Folter, S. Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat. Commun. 2014, 5, 3722. [Google Scholar] [CrossRef] [Green Version]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.B.; Qi, Y.P.; Yang, L.T.; Guo, P.; Li, Y.; Chen, L.S. Boron-deficiency-responsive microRNAs and their targets in Citrus sinensis leaves. BMC Plant Biol. 2015, 15, 271. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.B.; Yang, L.T.; Qi, Y.P.; Li, Y.; Li, Z.; Chen, Y.B.; Huang, Z.R.; Chen, L.S. Identification of boron-deficiency-responsive microRNAs in Citrus sinensis roots by Illumina sequencing. BMC Plant Biol. 2014, 14, 123. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.W.; Huang, J.H.; Li, C.P.; Yang, L.T.; Ye, X.; Lin, D.; Chen, L.S. MicroRNA-mediated responses to long-term magnesium-deficiency in Citrus sinensis roots revealed by Illumina sequencing. BMC Genom. 2017, 18, 657. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Liu, Y.; Zhu, A.; Wu, X.; Ye, J.; Yu, K.; Guo, W.; Deng, X. Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genom. 2010, 11, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, C.; Wang, C.; Zhang, C.; Korir, N.K.; Yu, H.; Ma, Z.; Fang, J. Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genom. 2010, 11, 431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, R.; Zhang, J.; Ma, Y.; Pan, X.; Dong, C.; Pang, S.; He, S.; Deng, L.; Yi, S.; Zheng, Y.; et al. Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots. Sci. Rep. 2017, 7, 42094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz, J.P.A.; Leblanc, O.; Rohr, C.; Grisolia, M.; Siena, L.A.; Podio, M.; Colono, C.; Azzaro, C.; Pessino, S.C. Small RNA-seq reveals novel regulatory components for apomixis in Paspalum notatum. BMC Genom. 2019, 20, 487. [Google Scholar] [CrossRef] [Green Version]
- Gramzow, L.; Klupsch, K.; Fernández-Pozo, N.; Hölzer, M.; Marz, M.; Rensing, S.A.; Theißen, G. Comparative transcriptomics identifies candidate genes involved in the evolutionary transition from dehiscent to indehiscent fruits in Lepidium (Brassicaceae). BMC Plant Biol. 2022, 22, 1–22. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W. Conservation and divergence in plant microRNAs. Plant Mol. Biol. 2012, 80, 3–16. [Google Scholar] [CrossRef]
- You, C.; Cui, J.; Wang, H.; Qi, X.; Kuo, L.Y.; Ma, H.; Gao, L.; Mo, B.; Chen, X. Conservation and divergence of small RNA pathways and microRNAs in land plants. Genome Biol. 2017, 18, 158. [Google Scholar] [CrossRef]
- Wu, X.M.; Kou, S.J.; Liu, Y.L.; Fang, Y.N.; Xu, Q.; Guo, W.W. Genomewide analysis of small RNA s in nonembryogenic and embryogenic tissues of citrus: Micro RNA-and si RNA-mediated transcript cleavage involved in somatic embryogenesis. Plant Biotechnol. J. 2015, 13, 383–394. [Google Scholar] [CrossRef]
- Fu, X.Z.; Zhang, X.Y.; Qiu, J.Y.; Zhou, X.; Yuan, M.; He, Y.Z.; Chang, P.C.; Cao, L.; Ling, L.L.; Peng, L.Z. Whole-transcriptome RNA sequencing reveals the global molecular responses and ceRNA regulatory network of mRNAs, lncRNAs, miRNAs and circRNAs in response to copper toxicity in Ziyang Xiangcheng (Citrus junos Sieb. Ex Tanaka). BMC Plant Biol. 2019, 19, 509. [Google Scholar] [CrossRef] [Green Version]
- Yin, F.; Qin, C.; Gao, J.; Liu, M.; Luo, X.; Zhang, W. Genome-wide identification and analysis of drought-responsive genes and microRNAs in tobacco. Int. J. Mol. Sci. 2015, 16, 5714–5740. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, L.; Tang, S.; Liu, J.; Zhang, H.; Zhi, H.; Jia, G.; Diao, X. Combined small RNA and degradome sequencing to identify miRNAs and their targets in response to drought in foxtail millet. BMC Genet. 2016, 17, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, N.; Ye, W.W.; Wu, X.L.; Shen, D.Y.; Wang, Y.C.; Xing, H.; Dou, D.L. Microarray profiling reveals microRNAs involving soybean resistance to Phytophthora sojae. Genome 2011, 54, 954–958. [Google Scholar] [CrossRef] [PubMed]
- Harvey, J.J.; Lewsey, M.G.; Patel, K.; Westwood, J.; Heimstädt, S.; Carr, J.P.; Baulcombe, D.C. An antiviral defense role of AGO2 in plants. PLoS ONE 2011, 6, e14639. [Google Scholar] [CrossRef] [PubMed]
- Incarbone, M.; Dunoyer, P. RNA silencing and its suppression: Novel insights from in planta analyses. Trends Plant Sci. 2013, 18, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Jagadeeswaran, G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 2008, 8, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.Z.; Xia, H.; Frazier, T.P.; Yao, Y.Y.; Bi, Y.P.; Li, A.Q.; Li, M.J.; Li, C.S.; Zhang, B.H.; Wang, X.J. Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.). BMC Plant Biol. 2010, 10, 3. [Google Scholar] [CrossRef] [Green Version]
- Shao, F.; Lu, Q.; Wilson, I.W.; Qiu, D. Genome-wide identification and characterization of the SPL gene family in Ziziphus jujuba. Gene 2017, 627, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Zhang, C.L.; Wang, G.L.; Wang, Y.X.; Qi, C.H.; Zhao, Q.; You, C.X.; Li, Y.Y.; Hao, Y.J. The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol. 2019, 19, 362. [Google Scholar] [CrossRef] [Green Version]
- Kishi-Kaboshi, M.; Seo, S.; Takahashi, A.; Hirochika, H. The MAMP-responsive MYB transcription factors MYB30, MYB55 and MYB110 activate the HCAA synthesis pathway and enhance immunity in rice. Plant Cell Physiol. 2018, 59, 903–915. [Google Scholar] [CrossRef]
- Sadka, A.; Shlizerman, L.; Kamara, I.; Blumwald, E. Primary metabolism in citrus fruit as affected by its unique structure. Front. Plant Sci. 2019, 10, 1167. [Google Scholar] [CrossRef]
- Hussain, S.B.; Guo, L.X.; Shi, C.Y.; Khan, M.A.; Bai, Y.X.; Du, W.; Liu, Y.Z. Assessment of sugar and sugar accumulation-related gene expression profiles reveal new insight into the formation of low sugar accumulation trait in a sweet orange (Citrus sinensis) bud mutant. Mol. Biol. Rep. 2020, 47, 2781–2791. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Chen, C.; Brlansky, R.H.; Gmitter, F.G., Jr.; Li, Z.G. Changes in carbohydrate metabolism in Citrus sinensis infected with ‘Candidatus Liberibacter asiaticus’. Plant Pathol. 2010, 59, 1037–1043. [Google Scholar] [CrossRef]
- Martinelli, F.; Ibanez, A.M.; Reagan, R.L.; Davino, S.; Dandekar, A.M. Stress responses in citrus peel: Comparative analysis of host responses to Huanglongbing disease and puffing disorder. Sci. Hortic. 2015, 192, 409–420. [Google Scholar] [CrossRef]
- Smeekens, S. Sugar-induced signal transduction in plants. Annu. Rev. Plant Biol. 2000, 51, 49–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pu, T.; Mo, Z.; Su, L.; Yang, J.; Wan, K.; Wang, L.; Liu, R.; Liu, Y. Genome-wide identification and expression analysis of the ftsH protein family and its response to abiotic stress in Nicotiana tabacum L. BMC Genom. 2022, 23, 503. [Google Scholar] [CrossRef]
- Kadirjan-Kalbach, D.K.; Yoder, D.W.; Ruckle, M.E.; Larkin, R.M.; Osteryoung, K.W. FtsHi1/ARC1 is an essential gene in Arabidopsis that links chloroplast biogenesis and division. Plant J. 2012, 72, 856–867. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, D.; Li, S.; Su, Y.; Liang, Q.; Meng, H.; Shen, S.; Fan, Y.; Liu, C.; Zhang, C. FtsHi4 is essential for embryogenesis due to its influence on chloroplast development in Arabidopsis. PLoS ONE 2014, 9, e99741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Li, S.; Chen, D.; Xi, Y.; Xu, X.; Ye, N.; Zhang, J.; Peng, X.; Zhu, G. Impairment of FtsHi5 function affects cellular redox balance and photorespiratory metabolism in Arabidopsis. Plant Cell Physiol. 2018, 59, 2526–2535. [Google Scholar] [CrossRef]
- Li, R.; Chen, D.; Wang, T.; Wan, Y.; Li, R.; Fang, R.; Wang, Y.; Hu, F.; Zhou, H.; Li, L.; et al. High throughput deep degradome sequencing reveals microRNAs and their targets in response to drought stress in mulberry (Morus alba). PLoS ONE 2017, 12, e0172883. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.Q.; Xue, H.W. The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ. 2019, 42, 2931–2944. [Google Scholar] [CrossRef] [Green Version]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhou, Z.; Shen, M.; Ge, L.; Liu, F. Ubiquitin-Conjugating Enzyme E2 E Inhibits the Accumulation of Rice Stripe Virus in Laodelphax striatellus (Fallén). Viruses 2020, 12, 908. [Google Scholar] [CrossRef]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W.R. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008, 53, 731–738. [Google Scholar] [CrossRef] [Green Version]
- Ehya, F.; Monavarfeshani, A.; Fard, E.M.; Farsad, L.K.; Nekouei, M.K.; Mardi, M.; Salekdeh, G.H. Phytoplasma-responsive microRNAs modulate hormonal, nutritional, and stress signalling pathways in Mexican lime trees. PLoS ONE 2013, 8, e66372. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Fang, Y.N.; Wu, X.M.; Qing, M.; Li, C.C.; Xie, K.D.; Deng, X.X.; Guo, W.W. The miR399-CsUBC24 module regulates reproductive development and male fertility in citrus. Plant Physiol. 2020, 183, 1681–1695. [Google Scholar] [CrossRef]
- Kim, E.J.; Lee, S.H.; Park, C.H.; Kim, S.H.; Hsu, C.C.; Xu, S.; Wang, Z.Y.; Kim, S.K.; Kim, T.W. Plant U-Box40 mediates degradation of the brassinosteroid-responsive transcription factor BZR1 in Arabidopsis roots. Plant Cell 2019, 31, 791–808. [Google Scholar] [CrossRef]
- Liu, W.; Tang, X.; Qi, X.; Fu, X.; Ghimire, S.; Ma, R.; Li, S.; Zhang, N.; Si, H. The Ubiquitin Conjugating Enzyme: An Important Ubiquitin Transfer Platform in Ubiquitin-Proteasome System. Int. J. Mol. Sci. 2020, 21, 2894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, C.; Zhang, X.; Liu, H.; Xu, J.R. Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLoS Pathog. 2018, 14, e1006875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, J.; Jeong, D.H.; De Paoli, E.; Park, S.; Rosen, B.D.; Li, Y.; González, A.J.; Yan, Z.; Kitto, S.L.; Grusak, M.A.; et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011, 25, 2540–2553. [Google Scholar] [CrossRef] [Green Version]
- Shivaprasad, P.V.; Chen, H.M.; Patel, K.; Bond, D.M.; Santos, B.A.; Baulcombe, D.C. A microRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, H.; Hong, G.; Li, L.; Zhang, X.; Kong, Y.; Sun, Z.; Li, J.; Chen, J.; He, Y. miR156/SPL9 regulates reactive oxygen species accumulation and immune response in Arabidopsis thaliana. Phytopathology 2019, 109, 632–642. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.L.; Huang, Y.Y.; Zheng, Y.P.; Liu, X.X.; Zhou, S.X.; Yang, X.M.; Liu, S.L.; Li, Y.; Li, J.L.; Zhao, S.L.; et al. Osa-miR535 targets SQUAMOSA promoter binding protein-like 4 to regulate blast disease resistance in rice. Plant J. 2022, 110, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Didion, J.P.; Martin, M.; Collins, F.S. Atropos: Specific, sensitive, and speedy trimming of sequencing reads. PeerJ 2017, 5, e3720. [Google Scholar] [CrossRef] [Green Version]
- Arce-Leal, Á.P.; Bautista, R.; Rodríguez-Negrete, E.A.; Manzanilla-Ramírez, M.Á.; Velázquez-Monreal, J.J.; Méndez-Lozano, J.; Bejarano, E.R.; Castillo, A.G.; Claros, M.G.; Leyva-López, N.E. De novo assembly and functional annotation of Citrus aurantifolia transcriptome from Candidatus Liberibacter asiaticus infected and non-infected trees. Data Brief 2020, 29, 105198. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Lunardon, A.; Johnson, N.R.; Hagerott, E.; Phifer, T.; Polydore, S.; Coruh, C.; Axtell, M.J. Integrated annotations and analyses of small RNA–producing loci from 47 diverse plants. Genome Res. 2020, 30, 497–513. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [Green Version]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [Green Version]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007, 3, 12. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hartung, J.S.; Levy, L. Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with citrus huanglongbing. J. Microbiol. Methods 2006, 66, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Kou, S.J.; Wu, X.M.; Liu, Z.; Liu, Y.L.; Xu, Q.; Guo, W.W. Selection and validation of suitable reference genes for miRNA expression normalization by quantitative RT-PCR in citrus somatic embryogenic and adult tissues. Plant Cell Rep. 2012, 31, 2151–2163. [Google Scholar] [CrossRef] [PubMed]
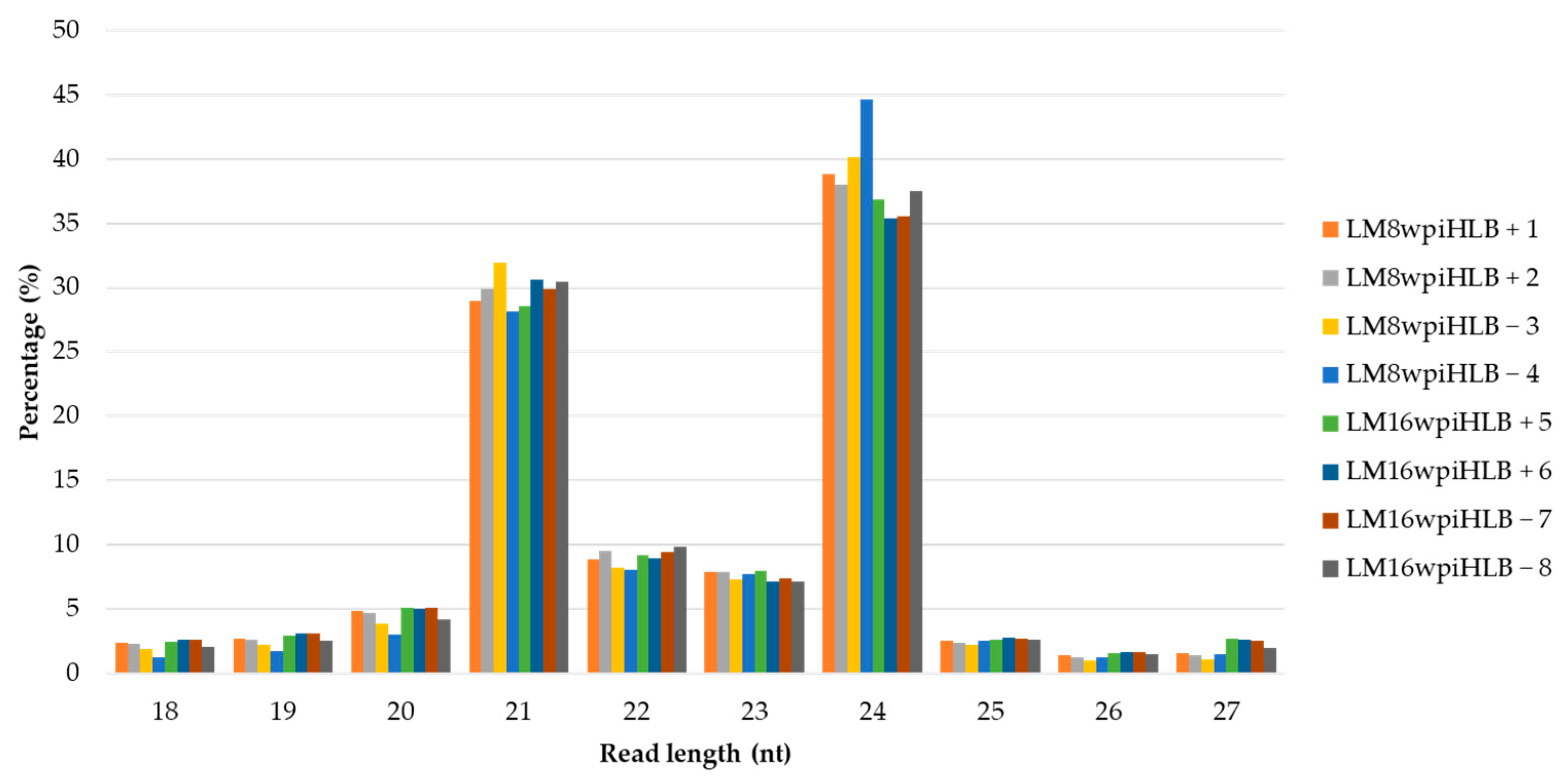
| Condition | Libraries | Raw Reads | Clean Reads | Reads Mapped to Transcriptome | miRNAs Identified | Total miRNAs |
|---|---|---|---|---|---|---|
| Asymptomatic (early) stage | LM8wpiHLB + 1 | 38,662,026 | 23,579,913 | 12,691,904 (53.82%) | 24 | 46 |
| LM8wpiHLB + 2 | 48,578,991 | 29,405,251 | 16,123,718 (54.83%) | 28 | ||
| Mock inoculated | LM8wpiHLB − 3 | 34,095,266 | 22,307,669 | 11,744,853 (52.64%) | 29 | |
| LM8wpiHLB − 4 | 54,610,772 | 37,857,718 | 18,498,221 (48.86%) | 27 | ||
| Symptomatic (late) stage | LM16wpiHLB + 5 | 47,150,743 | 25,075,202 | 13,584,848 (54.17%) | 23 | |
| LM16wpiHLB + 6 | 41,940,661 | 22,189,688 | 12,312,553 (55.48%) | 24 | ||
| Mock inoculated | LM16wpiHLB − 7 | 51,599,735 | 26,326,495 | 14,677,662 (55.75%) | 23 | |
| LM16wpiHLB − 8 | 50,672,438 | 30,719,833 | 16,774,553 (54.60%) | 26 |
| miRNA 1 | Sequence (5′ to 3′) | Length | miRNA Location in C. aurantifolia Transcriptome | |
|---|---|---|---|---|
| Known miRNAs | cau-miR156a,b | UUGACGGAAGAUAGAGAGCAC | 21 | Citrus_au_5549, Citrus_au_5550 |
| cau-miR156c | AUGACAGAAGAGAGAGAGUAC | 21 | Citrus_au_0000117891 | |
| cau-miR159a | UUUGGAUUGAAGGGAGCUCUA | 21 | Citrus_au_0000113744 | |
| cau-miR160a | UGCCUGGCUCCCUGUAUGCCA | 21 | Citrus_au_3426 | |
| cau-miR162a | UCGAUAAACCUCUGCAUCCAG | 21 | Citrus_au_0000053652 | |
| cau-miR164a,b | UGGAGAAGCAGGGCACGUGCA | 21 | Citrus_au_0000038181, Citrus_au_4480 | |
| cau-miR164c,d | CAUGUGCCCUUCUUCCCCAUC | 21 | Citrus_au_4480, Citrus_au_0000038181 | |
| cau-miR166a | UCGGACCAGGCUUCAUUCCCU | 21 | Citrus_au_3052 | |
| cau-miR166b | UCGGACCAGGCUUCAUUCCCC | 21 | Citrus_au_7116 | |
| cau-miR167a | UGAAGCUGCCAGCAUGAUCUGA | 22 | Citrus_au_7532 | |
| cau-miR168a | UCGCUUGGUGCAGGUCGGGAA | 21 | Citrus_au_0000028247 | |
| cau-miR172a | GUAGCAUCAUCAAGAUUCAC | 20 | Citrus_au_1099 | |
| cau-miR391a | UGCAGGUGAGAUGAUACCGUCA | 22 | Citrus_au_9968 | |
| cau-miR391b,c | CCGGAAUCAUUUCUCCCGCGUG | 22 | Citrus_au_0000052759, Citrus_au_9968 | |
| cau-miR393a | UCCAAAGGGAUCGCAUUGAUCU | 22 | Citrus_au_0000052858 | |
| cau-miR396a | UUCCACAGCUUUCUUGAACUG | 21 | Citrus_au_0000051770 | |
| cau-miR399a | UGCCAAAGGAGAGUUGCCCUA | 21 | Citrus_au_6097 | |
| cau-miR399b | UGCCAAAGGAGAGUUGCCCUG | 21 | Citrus_au_0000054379 | |
| cau-miR403a,b,c | UUAGAUUCACGCACAAACUCG | 21 | Citrus_au_4163, Citrus_au_0000097776, Citrus_au_0000097777 | |
| cau-miR482a,b,c | UUUUUCCCACACCUCCCAUCCC | 22 | Citrus_au_4020, Citrus_au_0000120512, Citrus_au_0000054570 | |
| cau-miR482d | UCUUGCCCACCCCUCCCAUUCC | 22 | Citrus_au_7459 | |
| Novel miRNAs | cau-miR001 | AUGUUGCUUGAUGAUAUUUAGUGU | 24 | Citrus_au_0000073759 |
| cau-miR002 | CCGUUUCAUCUUGUCCUCCAG | 21 | Citrus_au_0000096565 | |
| cau-miR003 | CUGUAGAAGGCUCCUGUGACC | 21 | Citrus_au_5642 | |
| cau-miR004 | AAAGUUAGGGAUAAGUUAAAAGAC | 24 | Citrus_au_0000021657 | |
| cau-miR005 | UCUAAGAAAACUUCAAUAGCU | 21 | Citrus_au_8635 | |
| cau-miR006a,b | UAGCAGCUGGGCUGUGAUAGGCCA | 24 | Citrus_au_0000043483, Citrus_au_5236 | |
| cau-miR007 | AGUUUCAGGAUUGGUUUGGGAUUC | 24 | Citrus_au_0000074192 | |
| cau-miR008 | GUGUUGCUUGAUGAUAUUUAGUGU | 24 | Citrus_au_0000073759 | |
| cau-miR009 | UCGCAGGAGCUUUCUACGGUU | 21 | Citrus_au_5642 | |
| cau-miR010 | AAUGUUGAGCUAAGGUUUUGC | 21 | Citrus_au_3278 | |
| cau-miR011 | UGACUCGUGAGUUGGCCUUG | 20 | Citrus_au_2177 | |
| cau-miR012a,b,c | UUUUGUUGCAUGAUGCUGAUAA | 22 | Citrus_au_0000041993, Citrus_au_0000105499, Citrus_au_0000041992 | |
| cau-miR013 | AAAACCUCAACUCAGCACUGA | 21 | Citrus_au_3278 | |
| cau-miR014 | CUUUCAGCAGCCUUCGGCGUC | 21 | Citrus_au_0000116384 |
| Stage | miRNA 1 | miRNA Differential Expression (Log2 FC) | mRNA Differential Expression (Log2 FC) | Target Gene | Predicted Function | Inhibition |
|---|---|---|---|---|---|---|
| 8 wpi | cau-miR403a,b,c | −0.445 −0.449 −0.430 | 0.609 | Citrus_au_4163 | Putative ubiquitin-conjugating enzyme E2 38 | Cleavage |
| 0.714 | Citrus_au_0000009992 | Flavin-containing monooxygenase FMO GS-OX-like 2 | Translation | |||
| cau-miR006a | 0.939 | −0.603 | Citrus_au_0000110774 | FtsH extracellular protease family | Cleavage | |
| −1.248 | Citrus_au_0000046578 | U-box domain-containing protein 40 (PUB40) | Cleavage | |||
| cau-miR011 | 1.010 | −1.177 | Citrus_au_0000014317 | Sucrose transport protein SUC3 | Cleavage | |
| −1.290 | Citrus_au_2600 | MAPK/ERK kinase kinase E493 (MAPKKK10) | Cleavage | |||
| cau-miR172a | −0.433 | 0.446 | Citrus_au_0000046354 | Myb domain protein 105 (MYB105) | Cleavage | |
| 0.491 | Citrus_au_0000042060 | Replication protein A 70 kDa DNA-binding subunit C (RPA1C) | Cleavage | |||
| miR3951 | 0.467 | −0.642 | Citrus_au_4749 | Leucine-rich repeat (LRR) family protein | Cleavage | |
| −0.762 | Citrus_au_0000103503 | Leucine-rich repeat (LRR) family protein | Cleavage | |||
| 16 wpi | cau-miR156a,b | −0.746 −0.748 | 0.883 | Citrus_au_0000112798 | Squamosa promoter-binding-like protein 3 | Cleavage |
| 0.504 | Citrus_au_0000015487 | Squamosa promoter-binding-like protein 4 | Cleavage | |||
| cau-miR156c | 1.160 | −1.247 | Citrus_au_0000049123 | Squamosa promoter-binding-like protein 4 | Cleavage | |
| −2.045 | Citrus_au_3104 | UDP-glycosyltransferase 73C3 | Cleavage | |||
| cau-miR160a | 0.649 | −1.109 | Citrus_au_0000004007 | Auxin response factor 17 (ARF17) | Cleavage | |
| −0.554 | Citrus_au_0000004009 | Leucine aminopeptidase 3, chloroplastic | Cleavage | |||
| cau-miR399a,b | −1.806 −1.291 | 1.277 | Citrus_au_0000032113 | Probable ubiquitin-conjugating enzyme E2 24 (UBC24) | Cleavage | |
| 1.371 | Citrus_au_0000046079 | Dicarboxylate transporter 1, chloroplastic (DIT1) | Translation | |||
| cau-miR482a,c | 0.500 | −0.444 | Citrus_au_10466 | Disease resistance protein (TIR-NBS-LRR class) family | Translation | |
| −0.509 | Citrus_au_5033 | hydrolases, acting on ester bonds | Cleavage | |||
| miR393 | 0.487 | −0.392 | Citrus_au_0000018820 | NADPH--cytochrome P450 reductase 1 (ATR1) | Cleavage | |
| −0.944 | Citrus_au_0000038185 | Amino acid kinase family protein | Cleavage | |||
| miR3951 | 0.418 | −0.878 | Citrus_au_4749 | Leucine-rich repeat (LRR) family protein | Cleavage | |
| −0.504 | Citrus_au_0000041265 | Calcium-dependent protein kinase 21 (CPK21) | Cleavage | |||
| miR399 | −3.455 | 1.277 | Citrus_au_0000032113 | Probable ubiquitin-conjugating enzyme E2 24 (UBC24) | Cleavage | |
| 1.073 | Citrus_au_0000042988 | C2H2-like zinc finger protein | Cleavage |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bojórquez-Orozco, A.M.; Arce-Leal, Á.P.; Montes, R.A.C.; Santos-Cervantes, M.E.; Cruz-Mendívil, A.; Méndez-Lozano, J.; Castillo, A.G.; Rodríguez-Negrete, E.A.; Leyva-López, N.E. Differential Expression of miRNAs Involved in Response to Candidatus Liberibacter asiaticus Infection in Mexican Lime at Early and Late Stages of Huanglongbing Disease. Plants 2023, 12, 1039. https://doi.org/10.3390/plants12051039
Bojórquez-Orozco AM, Arce-Leal ÁP, Montes RAC, Santos-Cervantes ME, Cruz-Mendívil A, Méndez-Lozano J, Castillo AG, Rodríguez-Negrete EA, Leyva-López NE. Differential Expression of miRNAs Involved in Response to Candidatus Liberibacter asiaticus Infection in Mexican Lime at Early and Late Stages of Huanglongbing Disease. Plants. 2023; 12(5):1039. https://doi.org/10.3390/plants12051039
Chicago/Turabian StyleBojórquez-Orozco, Ana Marlenne, Ángela Paulina Arce-Leal, Ricardo A. Chávez Montes, María Elena Santos-Cervantes, Abraham Cruz-Mendívil, Jesús Méndez-Lozano, Araceli G. Castillo, Edgar A. Rodríguez-Negrete, and Norma Elena Leyva-López. 2023. "Differential Expression of miRNAs Involved in Response to Candidatus Liberibacter asiaticus Infection in Mexican Lime at Early and Late Stages of Huanglongbing Disease" Plants 12, no. 5: 1039. https://doi.org/10.3390/plants12051039





