Genome-Wide Identification and Characterization of the HMGR Gene Family in Taraxacum kok-saghyz Provide Insights into Its Regulation in Response to Ethylene and Methyl Jsamonate Treatments
Abstract
:1. Introduction
2. Results
2.1. Identification of TkHMGR Family Members in T. kok-saghyz
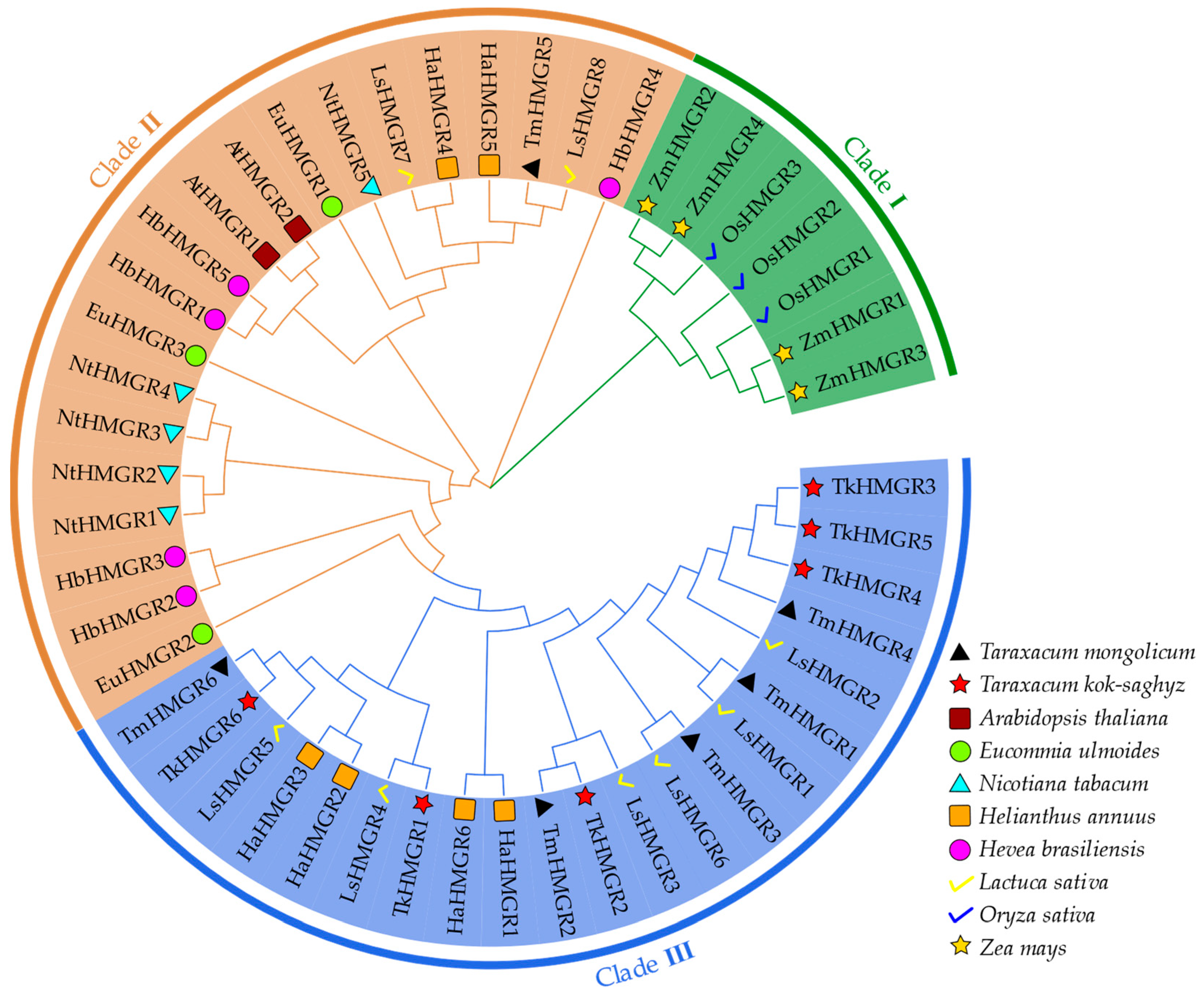
2.2. Phylogenetic Relationship, Conserved Structure, Motif Composition, and Gene Structures of TkHMGRs
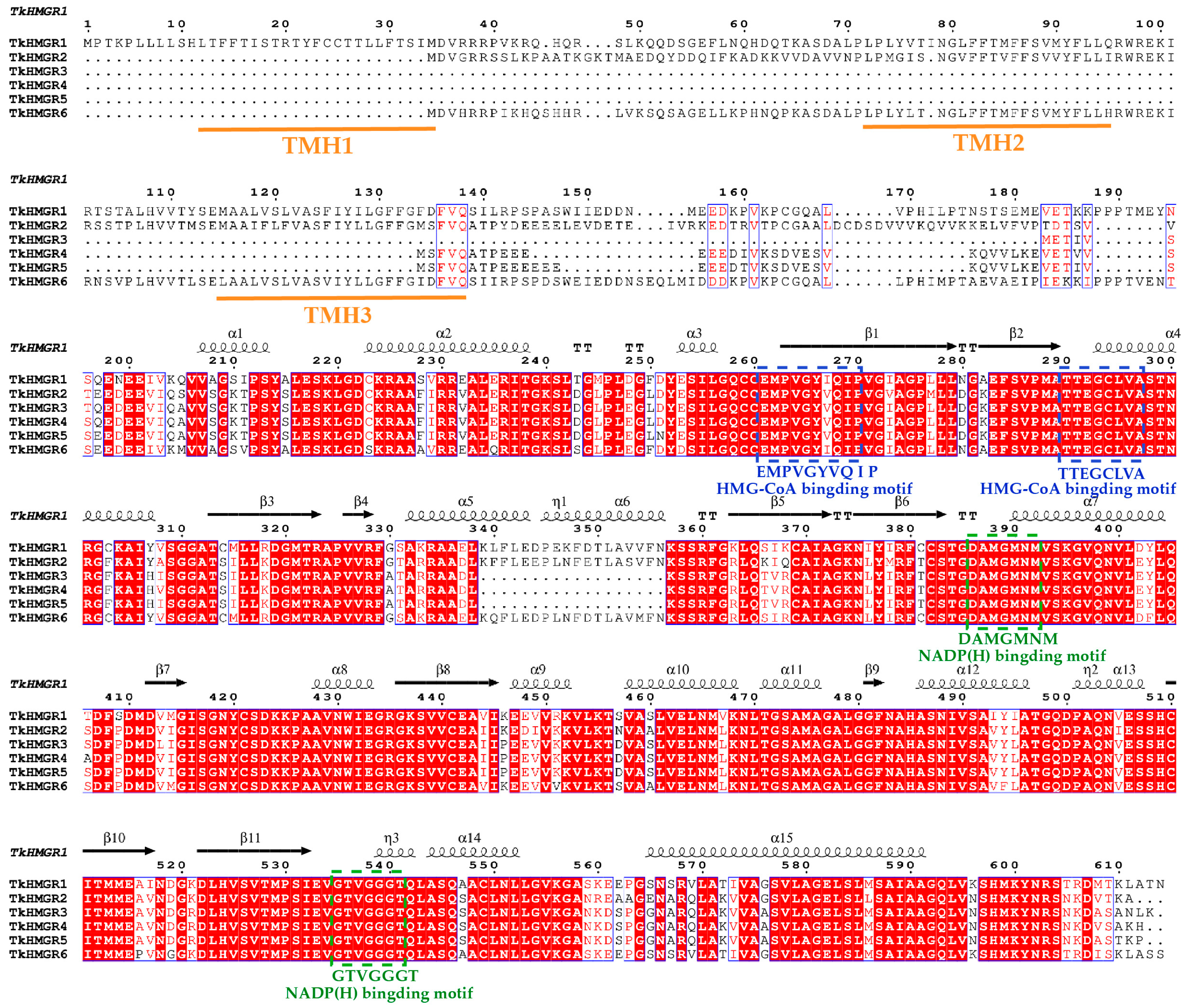
2.3. Multiple Sequence Alignments, Secondary Structure Analysis, and Multispecies Phylogenetic Analysis of HMGR Proteins in T. kok-saghyz
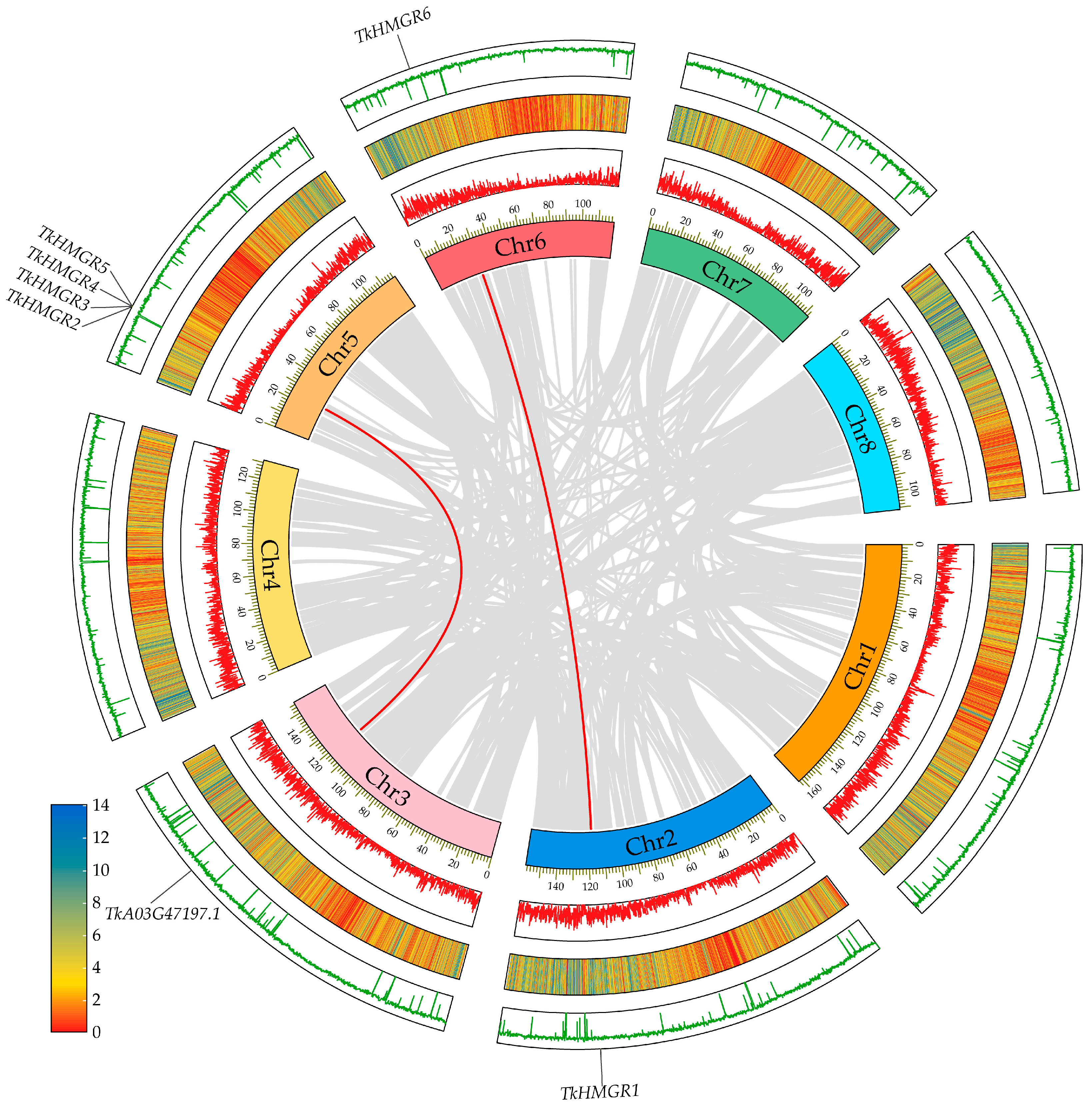
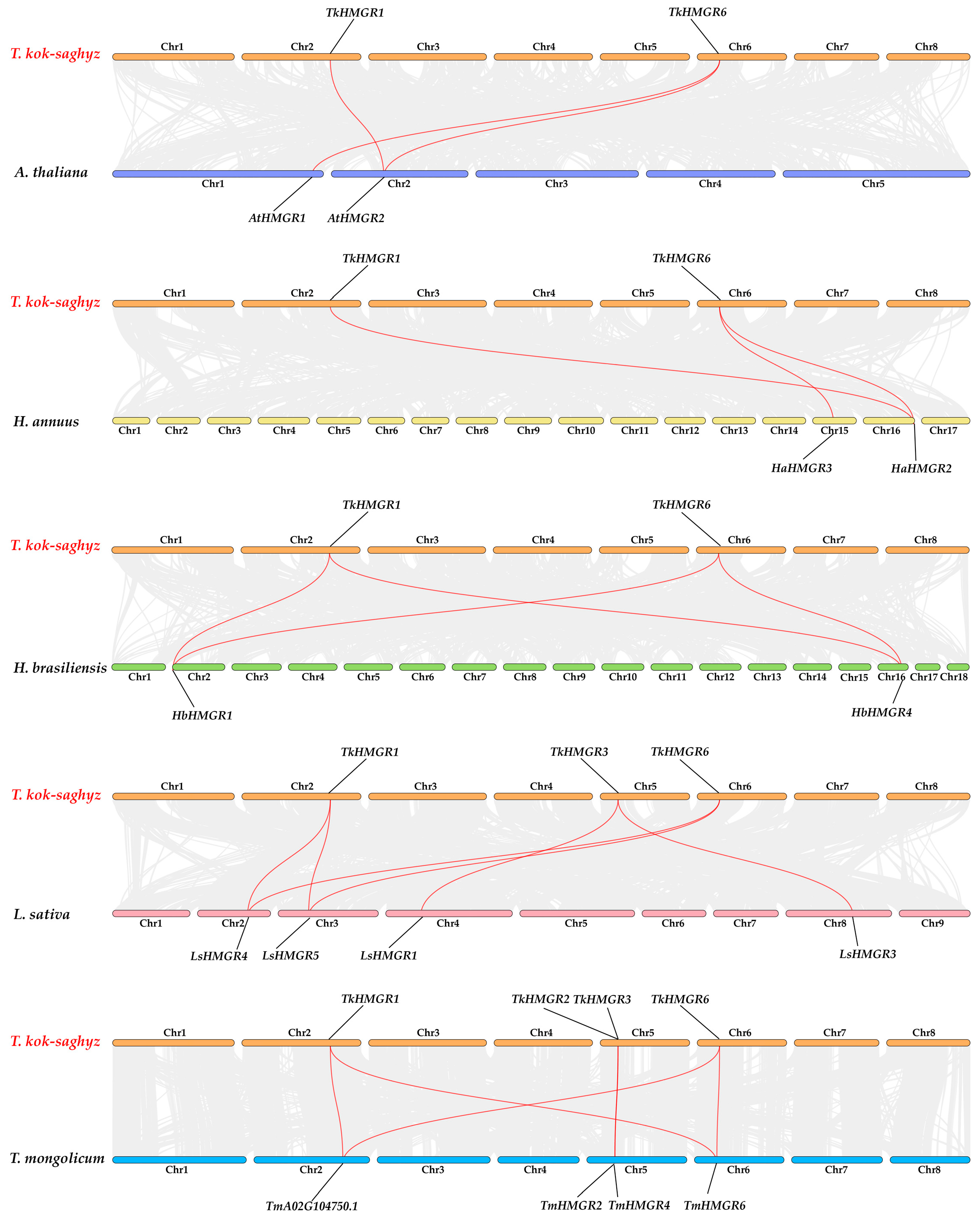
2.4. Chromosomal Localization and Collinearity Analysis in T. kok-saghyz
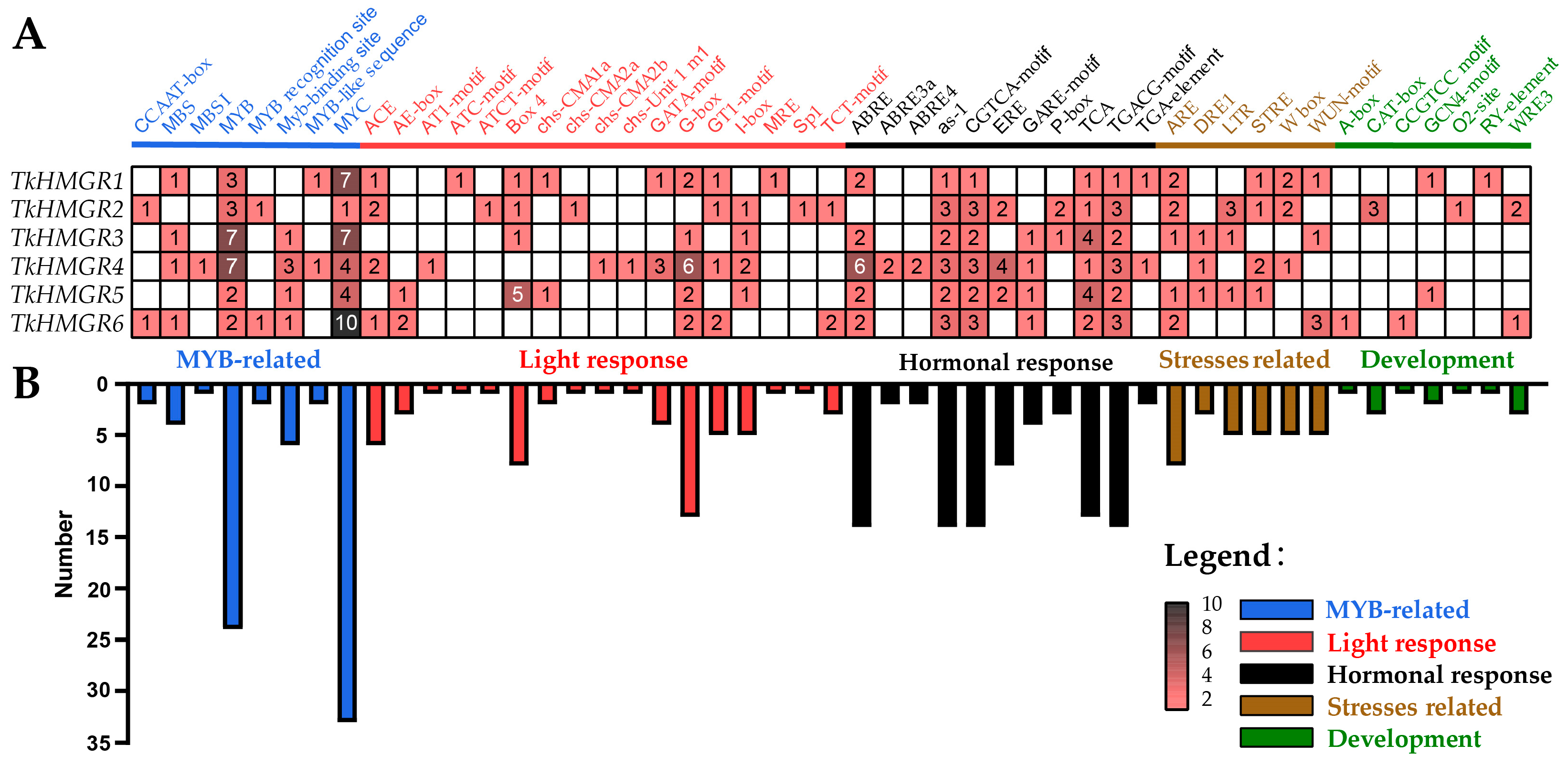
2.5. Cis-Acting Element Analysis of HMGR Gene Family in T. kok-saghyz
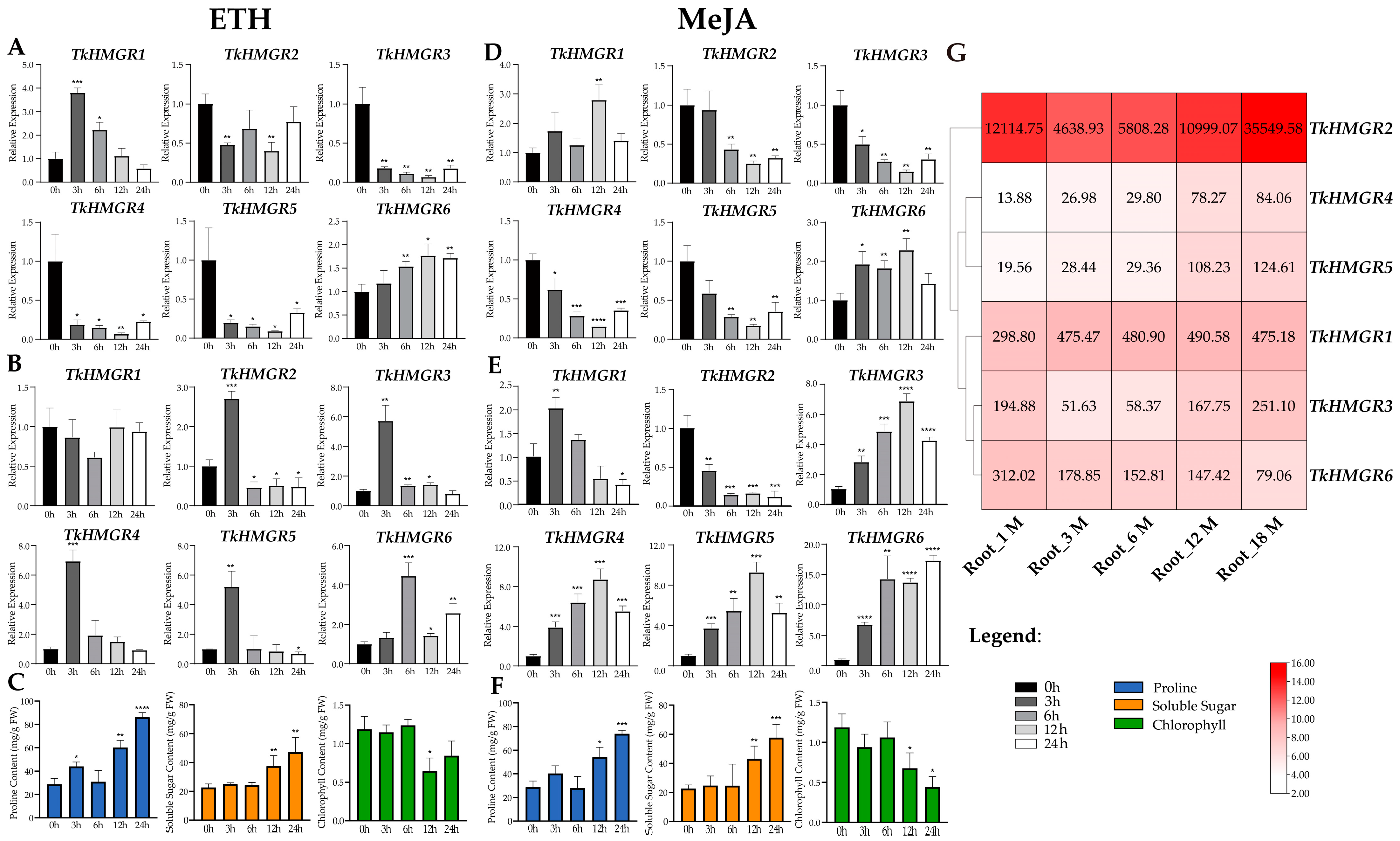
2.6. The Expression Pattern Analysis of HMGR in T. kok-saghyz and Physiological Indexes
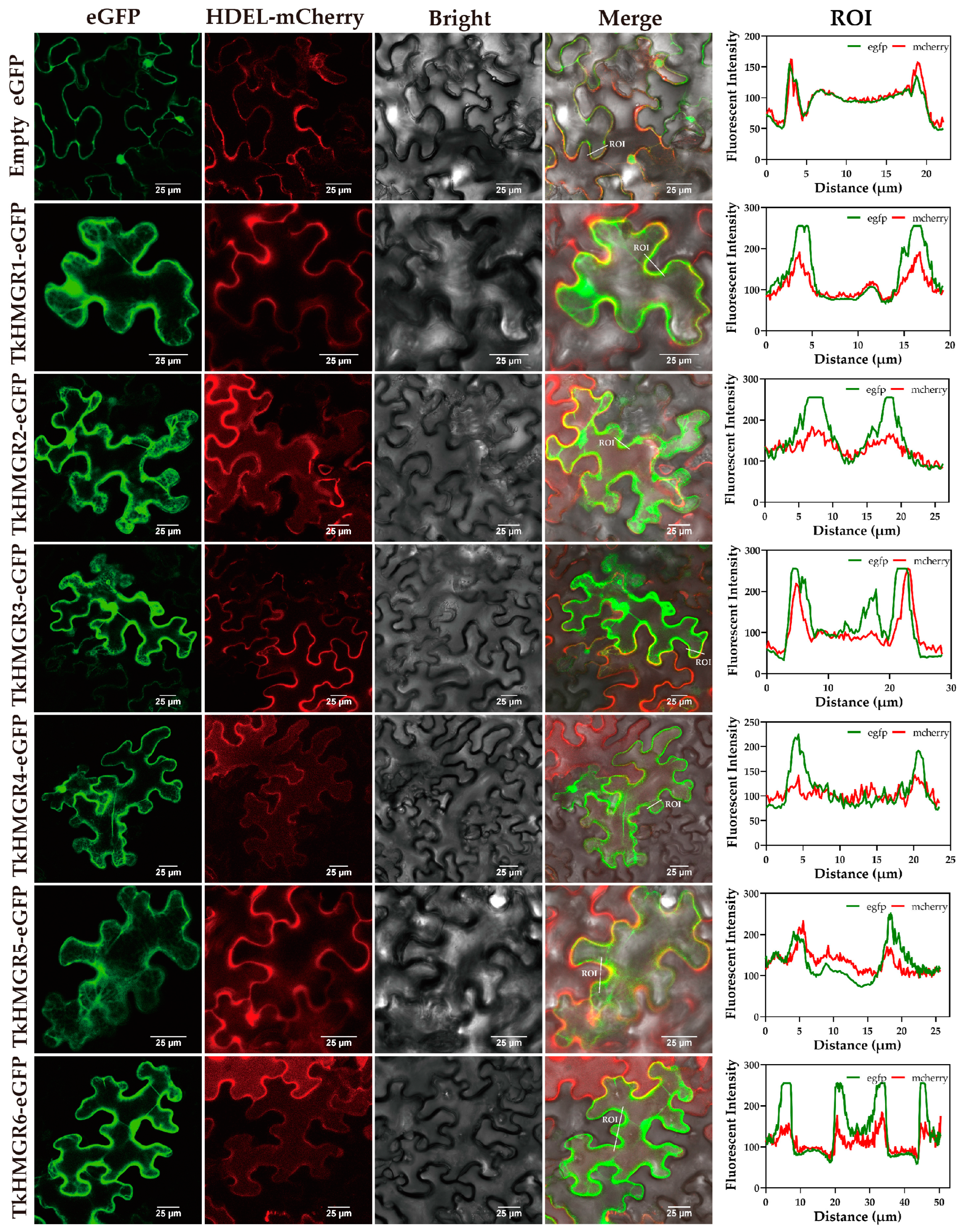
2.7. Subcellular Localization Analysis of Six TkHMGR Proteins
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of HMGR Genes in T. kok-saghyz
4.2. Phylogenetic Analysis, Gene Structure, Motif, and Conserved Domain Analysis
4.3. Multiple Sequence Alignments, Secondary Structure Prediction, and Protein 3D Structure Prediction
4.4. Collinearity Analysis and Cis-Acting Element Analysis
4.5. Plant Growth, Treatment, RNA Extraction, and qRT-PCR Analysis
4.6. RNA-Seq Analysis and Determination of Plant Physiological Indicators
4.7. Subcellular Localization of TkHMGRs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, L.; Wen, K.-S.; Ruan, X.; Zhao, Y.-X.; Wei, F.; Wang, Q. Response of Plant Secondary Metabolites to Environmental Factors. Molecules 2018, 23, 762. [Google Scholar] [CrossRef] [PubMed]
- Cherian, S.; Ryu, S.B.; Cornish, K. Natural rubber biosynthesis in plants, the rubber transferase complex, and metabolic engineering progress and prospects. Plant Biotechnol. J. 2019, 17, 2041–2061. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Ding, G.; Zhu, L.; Yu, L.; Yuan, B.; Gao, X.; Wang, D.; Sun, Y.; Liu, Y.; Li, H.; et al. Proteomic Landscape of the Mature Roots in a Rubber-Producing Grass Taraxacum kok-saghyz. Int. J. Mol. Sci. 2019, 20, 2596–2615. [Google Scholar] [CrossRef]
- Jayashree, R.; Nazeem, P.A.; Rekha, K.; Sreelatha, S.; Thulaseedharan, A.; Krishnakumar, R.; Kala, R.G.; Vineetha, M.; Leda, P.; Jinu, U.; et al. Over-expression of 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 (hmgr1) gene under super-promoter for enhanced latex biosynthesis in rubber tree (Hevea brasiliensis Muell. Arg.). Plant Physiol. Biochem. PPB 2018, 127, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Venkatachalam, P.; Jayashree, R.; Rekha, K.; Sushmakumari, S.; Sobha, S.; Kumari Jayasree, P.; Kala, R.G.; Thulaseedharan, A. Rubber Tree (Hevea brasiliensis Muell. Arg). Methods Mol. Biol. 2006, 344, 153–164. [Google Scholar] [PubMed]
- Venkatachalam, P.; Thulaseedharan, A.; Raghothama, K. Molecular identification and characterization of a gene associated with the onset of tapping panel dryness (TPD) syndrome in rubber tree (Hevea brasiliensis Muell.) by mRNA differential display. Mol. Biotechnol. 2009, 41, 42–52. [Google Scholar] [CrossRef]
- Venkatachalam, P.; Jayasree, P.K.; Jayashree, R.; Rekha, K.; Thulaseedharan, A. Current Perspectives on Application of Biotechnology to Assist the Genetic Improvement of Rubber Tree (Hevea brasiliensis Muell. Arg.): An Overview. Funct. Plant Sci. Biotechnol. 2007, 1, 1–17. [Google Scholar]
- Krotkov, G. A review of literature onTaraxacum koksaghyz Rod. Bot. Rev. 1945, 11, 417–461. [Google Scholar] [CrossRef]
- McAssey, E.V.; Gudger, E.G.; Zuellig, M.P.; Burke, J.M. Population Genetics of the Rubber-Producing Russian Dandelion (Taraxacum kok-saghyz). PLoS ONE 2016, 11, e0146417. [Google Scholar] [CrossRef]
- Peng, C.; Chang, L.; Wang, D.; Xie, Q.; Zheng, X.; Xu, B.; Tong, Z. An efficient Agrobacterium tumefaciens-mediated transformation system of Taraxacum kok-saghyz Rodin (Russian dandelion). In Vitro Cell. Dev. Biol.—Plant 2023, 59, 413–423. [Google Scholar] [CrossRef]
- Yang, Y.; Qin, B.; Chen, Q.; Zhang, J.; Zhang, L.; Nie, Q.; Liu, S. Comparative full-length transcriptome analysis provides novel insights into the regulatory mechanism of natural rubber biosynthesis in Taraxacum kok-saghyz Rodin roots. Ind. Crops Prod. 2022, 175, 114278. [Google Scholar] [CrossRef]
- Men, X.; Wang, F.; Chen, G.-Q.; Zhang, H.-B.; Xian, M. Biosynthesis of Natural Rubber: Current State and Perspectives. Int. J. Mol. Sci. 2018, 20, 50–72. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Takahashi, S. Molecular Mechanisms of Natural Rubber Biosynthesis. Annu. Rev. Biochem. 2020, 89, 821–851. [Google Scholar] [CrossRef] [PubMed]
- Wititsuwannakul, R.; Pasitkul, P.; Jewtragoon, P.; Wititsuwannakul, D. Hevea latex lectin binding protein in C-serum as an anti-latex coagulating factor and its role in a proposed new model for latex coagulation. Phytochemistry 2008, 69, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Kirby, J.; Keasling, J.D. Biosynthesis of plant isoprenoids: Perspectives for microbial engineering. Annu. Rev. Plant Biol. 2009, 60, 335–355. [Google Scholar] [CrossRef]
- Stermer, B.A.; Bianchini, G.M.; Korth, K.L. Regulation of HMG-CoA reductase activity in plants. J. Lipid Res. 1994, 35, 1133–1140. [Google Scholar] [CrossRef]
- Bach, T.J. Hydroxymethylglutaryl-CoA reductase, a key enzyme in phytosterol synthesis? Lipids 1986, 21, 82–88. [Google Scholar] [CrossRef]
- Leivar, P.; González, V.M.; Castel, S.; Trelease, R.N.; López-Iglesias, C.; Arró, M.; Boronat, A.; Campos, N.; Ferrer, A.; Fernàndez-Busquets, X. Subcellular localization of Arabidopsis 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Plant Physiol. 2005, 137, 57–69. [Google Scholar] [CrossRef]
- Campos, N.; Boronat, A. Targeting and topology in the membrane of plant 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Cell 1995, 7, 2163–2174. [Google Scholar]
- Li, W.; Liu, W.; Wei, H.; He, Q.; Chen, J.; Zhang, B.; Zhu, S. Species-specific expansion and molecular evolution of the 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) gene family in plants. PLoS ONE 2014, 9, e94172. [Google Scholar] [CrossRef]
- Burg, J.S.; Espenshade, P.J. Regulation of HMG-CoA reductase in mammals and yeast. Prog. Lipid Res. 2011, 50, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Gardner, R.G.; Hampton, R.Y. A highly conserved signal controls degradation of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase in eukaryotes. J. Biol. Chem. 1999, 274, 31671–31678. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Caelles, C.; Ferrer, A.; Balcells, L.; Hegardt, F.G.; Boronat, A. Isolation and structural characterization of a cDNA encoding Arabidopsis thaliana 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Mol. Biol. 1989, 13, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.H.; Lee, S.W.; Kim, Y.M.; Hwang, Y.S. Molecular characterization of Hmg2 gene encoding a 3-hydroxy-methylglutaryl-CoA reductase in rice. Mol. Cells 2001, 11, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Loguercio, L.L.; Scott, H.C.; Trolinder, N.L.; Wilkins, T.A. Hmg-coA reductase gene family in cotton (Gossypium hirsutum L.): Unique structural features and differential expression of hmg2 potentially associated with synthesis of specific isoprenoids in developing embryos. Plant Cell Physiol. 1999, 40, 750–761. [Google Scholar] [CrossRef]
- Aoyagi, K.; Beyou, A.; Moon, K.; Fang, L.; Ulrich, T. Isolation and characterization of cDNAs encoding wheat 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Physiol. 1993, 102, 623–628. [Google Scholar] [CrossRef]
- Venkatachalam, P.; Priya, P.; Jayashree, R.; Rekha, K.; Thulaseedharan, A. Molecular cloning and characterization of a 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 (hmgr1) gene from rubber tree (Hevea brasiliensis Muell. Arg.): A key gene involved in isoprenoid biosynthesis. Physiol. Mol. Biol. Plants 2009, 15, 133–143. [Google Scholar] [CrossRef]
- Schaller, H.; Grausem, B.; Benveniste, P.; Chye, M.L.; Tan, C.T.; Song, Y.H.; Chua, N.H. Expression of the Hevea brasiliensis (H.B.K.) Mull. Arg. 3-Hydroxy-3-Methylglutaryl-Coenzyme A Reductase 1 in Tobacco Results in Sterol Overproduction. Plant Physiol. 1995, 109, 761–770. [Google Scholar] [CrossRef]
- Pütter, K.M.; van Deenen, N.; Unland, K.; Prüfer, D.; Schulze Gronover, C. Isoprenoid biosynthesis in dandelion latex is enhanced by the overexpression of three key enzymes involved in the mevalonate pathway. BMC Plant Biol. 2017, 17, 88. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Lee, O.R.; Oh, J.Y.; Jang, M.-G.; Yang, D.-C. Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme a reductase encoding genes in triterpene saponin-producing ginseng. Plant Physiol. 2014, 165, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Zhuge, Q.; Li, D.; Wang, P.; Li, L.; Sun, W.; Xu, C.; Movahedi, A.; Wei, H. Overexpression of PtHMGR enhances drought and salt tolerance of poplar. Ann. Bot. 2020, 125, 785–803. [Google Scholar]
- Choi, D.; Ward, B.L.; Bostock, R.M. Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes in response to Phytophthora infestans and to its elicitor arachidonic acid. Plant Cell 1992, 4, 1333–1344. [Google Scholar] [PubMed]
- Zheng, T.; Guan, L.; Yu, K.; Haider, M.S.; Nasim, M.; Liu, Z.; Li, T.; Zhang, K.; Jiu, S.; Jia, H.; et al. Expressional diversity of grapevine 3-Hydroxy-3-methylglutaryl-CoA reductase (VvHMGR) in different grapes genotypes. BMC Plant Biol. 2021, 21, 279. [Google Scholar] [CrossRef]
- Lv, D.; Zhang, Y. Isolation and functional analysis of apple MdHMGR1 and MdHMGR4 gene promoters in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 2016, 129, 133–143. [Google Scholar] [CrossRef]
- Lv, D.M.; Zhang, T.T.; Deng, S.; Zhang, Y.H. Functional analysis of the Malus domestica MdHMGR2 gene promoter in transgenic Arabidopsis thaliana. Biol. Plant. 2016, 60, 667–676. [Google Scholar] [CrossRef]
- Lin, T.; Xu, X.; Du, H.; Fan, X.; Chen, Q.; Hai, C.; Zhou, Z.; Su, X.; Kou, L.; Gao, Q.; et al. Extensive sequence divergence between the reference genomes of Taraxacum kok-saghyz and Taraxacum mongolicum. Sci. China. Life Sci. 2022, 65, 515–528. [Google Scholar] [CrossRef]
- Istvan, E.S.; Deisenhofer, J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science 2001, 292, 1160–1164. [Google Scholar] [CrossRef]
- Friesen, J.A.; Rodwell, V.W. The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases. Genome Biol. 2004, 5, 248. [Google Scholar] [CrossRef]
- Li, W.; Yang, W.; Wang, X.-J. Pseudogenes: Pseudo or real functional elements? J. Genet. Genom. 2013, 40, 171–177. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, Z.; Li, W.; Zhu, W.; Ren, Z.; Wang, Z.; Li, L.; Jia, L.; Zhu, S.; Ma, Z. Genome-Wide Identification and Comparative Analysis of the 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase (HMGR) Gene Family in Gossypium. Molecules 2018, 23, 193. [Google Scholar] [CrossRef]
- Wang, X.; Wang, C.; Yang, M.; Jie, W.; Fazal, A.; Fu, J.; Yin, T.; Cai, J.; Liu, B.; Lu, G.; et al. Genome-Wide Comparison and Functional Characterization of HMGR Gene Family Associated with Shikonin Biosynthesis in Lithospermum erythrorhizon. Int. J. Mol. Sci. 2023, 24, 12532. [Google Scholar] [CrossRef]
- Whitley, P.; Grau, B.; Gumbart, J.C.; Martínez-Gil, L.; Mingarro, I. Folding and Insertion of Transmembrane Helices at the ER. Int. J. Mol. Sci. 2021, 22, 12778. [Google Scholar] [CrossRef]
- Liao, Z.; Tan, Q.; Chai, Y.; Zuo, K.; Chen, M.; Gong, Y.; Wang, P.; Pi, Y.; Tan, F.; Sun, X.; et al. Cloning and characterisation of the gene encoding HMG-CoA reductase from Taxus media and its functional identification in yeast. Funct. Plant Biol. 2004, 31, 73–81. [Google Scholar] [CrossRef]
- Leichner, G.S.; Avner, R.; Harats, D.; Roitelman, J. Metabolically regulated endoplasmic reticulum-associated degradation of 3-hydroxy-3-methylglutaryl-CoA reductase: Evidence for requirement of a geranylgeranylated protein. J. Biol. Chem. 2011, 286, 32150–32161. [Google Scholar] [CrossRef]
- Lallemand, T.; Leduc, M.; Landès, C.; Rizzon, C.; Lerat, E. An Overview of Duplicated Gene Detection Methods: Why the Duplication Mechanism Has to Be Accounted for in Their Choice. Genes 2020, 11, 1046. [Google Scholar] [CrossRef]
- Jiang, C.; Dai, J.; Han, H.; Wang, C.; Zhu, L.; Lu, C.; Chen, H. Determination of thirteen acidic phytohormones and their analogues in tea (Camellia sinensis) leaves using ultra high performance liquid chromatography tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2020, 1149, 122144. [Google Scholar] [CrossRef]
- Yu, L.; Yuan, B.; Wang, L.; Sun, Y.; Ding, G.; Souleymane, O.A.; Zhang, X.; Xie, Q.; Wang, X. Identification and Characterization of Glycoproteins and Their Responsive Patterns upon Ethylene Stimulation in the Rubber Latex. Int. J. Mol. Sci. 2020, 21, 5282. [Google Scholar] [CrossRef]
- He, H.; Wang, J.; Meng, Z.; Dijkwel, P.P.; Du, P.; Shi, S.; Dong, Y.; Li, H.; Xie, Q. Genome-Wide Analysis of the SRPP/REF Gene Family in Taraxacum kok-saghyz Provides Insights into Its Expression Patterns in Response to Ethylene and Methyl Jasmonate Treatments. Int. J. Mol. Sci. 2024, 25, 6864. [Google Scholar] [CrossRef]
- Abrahám, E.; Hourton-Cabassa, C.; Erdei, L.; Szabados, L. Methods for determination of proline in plants. Methods Mol. Biol. 2010, 639, 317–331. [Google Scholar]
- Verbruggen, N.; Hermans, C. Proline accumulation in plants: A review. Amino Acids 2008, 35, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Rosa, M.; Prado, C.; Podazza, G.; Interdonato, R.; González, J.A.; Hilal, M.; Prado, F.E. Soluble sugars--metabolism, sensing and abiotic stress: A complex network in the life of plants. Plant Signal. Behav. 2009, 4, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sun, Z.; Lu, S.; Omasa, K. Estimation of the leaf chlorophyll content using multiangular spectral reflectance factor. Plant Cell Environ. 2019, 42, 3152–3165. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Ma, J.; Ding, G.; Yuan, B.; Wang, Y.; He, L.; Han, Y.; Cao, A.; Li, R.; Zhang, W. Transcriptomics and proteomics profiles of Taraxacum kok-saghyz roots revealed different gene and protein members play different roles for natural rubber biosynthesis. Ind. Crops Prod. 2022, 181, 114776–114789. [Google Scholar] [CrossRef]
- Leivar, P.; Antolín-Llovera, M.; Ferrero, S.; Closa, M.; Arró, M.; Ferrer, A.; Boronat, A.; Campos, N. Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A. Plant Cell 2011, 23, 1494–1511. [Google Scholar] [CrossRef]
- Robertlee, J.; Kobayashi, K.; Tang, J.; Suzuki, M.; Muranaka, T. Evidence that the Arabidopsis thaliana 3-hydroxy-3-methylglutaryl-CoA reductase 1 is phosphorylated at Ser577 in planta. Plant Biotechnol. 2018, 35, 1–7. [Google Scholar] [CrossRef]
- Nieto, B.; Forés, O.; Arró, M.; Ferrer, A. Arabidopsis 3-hydroxy-3-methylglutaryl-CoA reductase is regulated at the post-translational level in response to alterations of the sphingolipid and the sterol biosynthetic pathways. Phytochemistry 2009, 70, 53–59. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene ID 1 | CDS Length (bp) | Protein Length (aa) | MW (kDa) 2 | pI 3 | Predicted Location(s) | Number of Predicted TMHs 4 |
|---|---|---|---|---|---|---|---|
| TkHMGR1 | TkA02G464040 | 1845 | 614 | 66.688 | 8.05 | Endoplasmic reticulum | 3 |
| TkHMGR2 | TkA05G087240 | 1749 | 582 | 62.642 | 5.58 | Endoplasmic reticulum | 2 |
| TkHMGR3 | TkA05G087830 | 1221 | 406 | 42.694 | 7.50 | Endoplasmic reticulum | 0 |
| TkHMGR4 | TkA05G087870 | 1311 | 436 | 46.061 | 5.68 | Endoplasmic reticulum | 0 |
| TkHMGR5 | TkA05G087940 | 1320 | 439 | 46.443 | 5.32 | Endoplasmic reticulum | 0 |
| TkHMGR6 | TkA06G113340 | 1761 | 586 | 62.902 | 6.65 | Endoplasmic reticulum | 2 |
| Protein Name | α-Helix | β-Turn | Extended Strand | Random Coil |
|---|---|---|---|---|
| TkHMGR1 | 243 (39.58%) | 36 (5.86%) | 96 (15.64%) | 239 (38.93%) |
| TkHMGR2 | 257 (44.16%) | 38 (6.53%) | 94 (16.15%) | 193 (33.16%) |
| TkHMGR3 | 197 (48.52%) | 27 (6.65%) | 70 (17.24%) | 112 (27.59%) |
| TkHMGR4 | 213 (48.85%) | 23 (5.28%) | 69 (15.83%) | 131 (30.05%) |
| TkHMGR5 | 215 (48.97%) | 25 (5.69%) | 69 (15.72%) | 130 (29.61%) |
| TkHMGR6 | 252 (43%) | 30 (5.12%) | 81 (13.82%) | 223 (38.05%) |
| Gene ID | Duplication Type(s) |
|---|---|
| TkHMGR1 | WGD or Segmental |
| TkHMGR2 | WGD or Segmental |
| TkHMGR3 | Tandem |
| TkHMGR4 | Tandem |
| TkHMGR5 | Tandem |
| TkHMGR6 | WGD or Segmental |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, P.; He, H.; Wang, J.; Wang, L.; Meng, Z.; Jin, X.; Zhang, L.; Wang, F.; Li, H.; Xie, Q. Genome-Wide Identification and Characterization of the HMGR Gene Family in Taraxacum kok-saghyz Provide Insights into Its Regulation in Response to Ethylene and Methyl Jsamonate Treatments. Plants 2024, 13, 2646. https://doi.org/10.3390/plants13182646
Du P, He H, Wang J, Wang L, Meng Z, Jin X, Zhang L, Wang F, Li H, Xie Q. Genome-Wide Identification and Characterization of the HMGR Gene Family in Taraxacum kok-saghyz Provide Insights into Its Regulation in Response to Ethylene and Methyl Jsamonate Treatments. Plants. 2024; 13(18):2646. https://doi.org/10.3390/plants13182646
Chicago/Turabian StyleDu, Pingping, Huan He, Jiayin Wang, Lili Wang, Zhuang Meng, Xiang Jin, Liyu Zhang, Fei Wang, Hongbin Li, and Quanliang Xie. 2024. "Genome-Wide Identification and Characterization of the HMGR Gene Family in Taraxacum kok-saghyz Provide Insights into Its Regulation in Response to Ethylene and Methyl Jsamonate Treatments" Plants 13, no. 18: 2646. https://doi.org/10.3390/plants13182646






