Developmental Morphology, Physiology, and Molecular Basis of the Pentagram Fruit of Averrhoa carambola
Abstract
:1. Introduction
2. Results
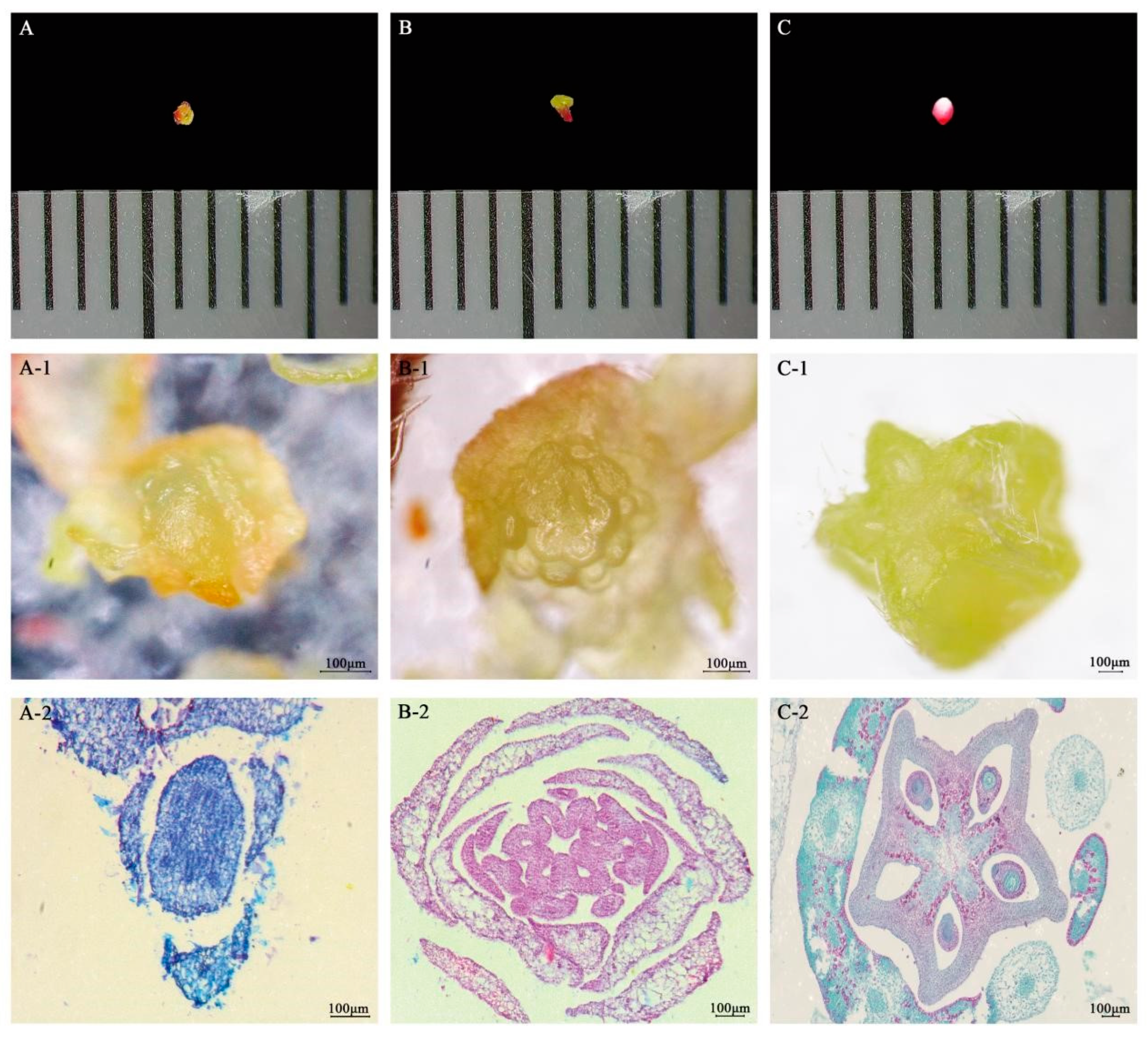
2.1. Microscopic Observation of Six Developmental Stages of Bud and Fruit of A. carambola Blossom
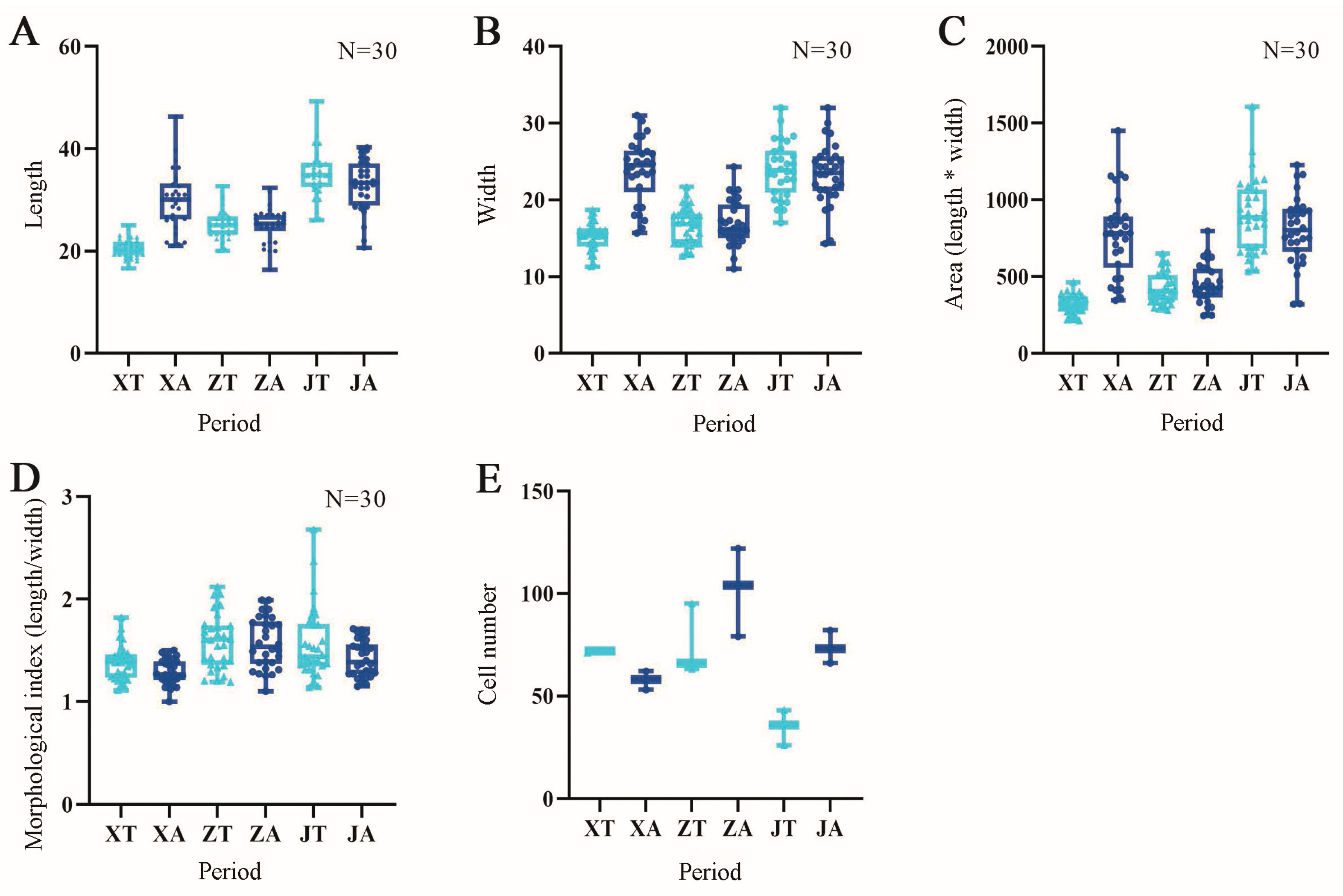
2.2. Analysis of Cell Morphology in Three Developmental Stages of the Fruit of A. carambola
2.3. Ultramicroscopic Observation of the Development Process from Bud to Blossom of the A. carambola Fruit
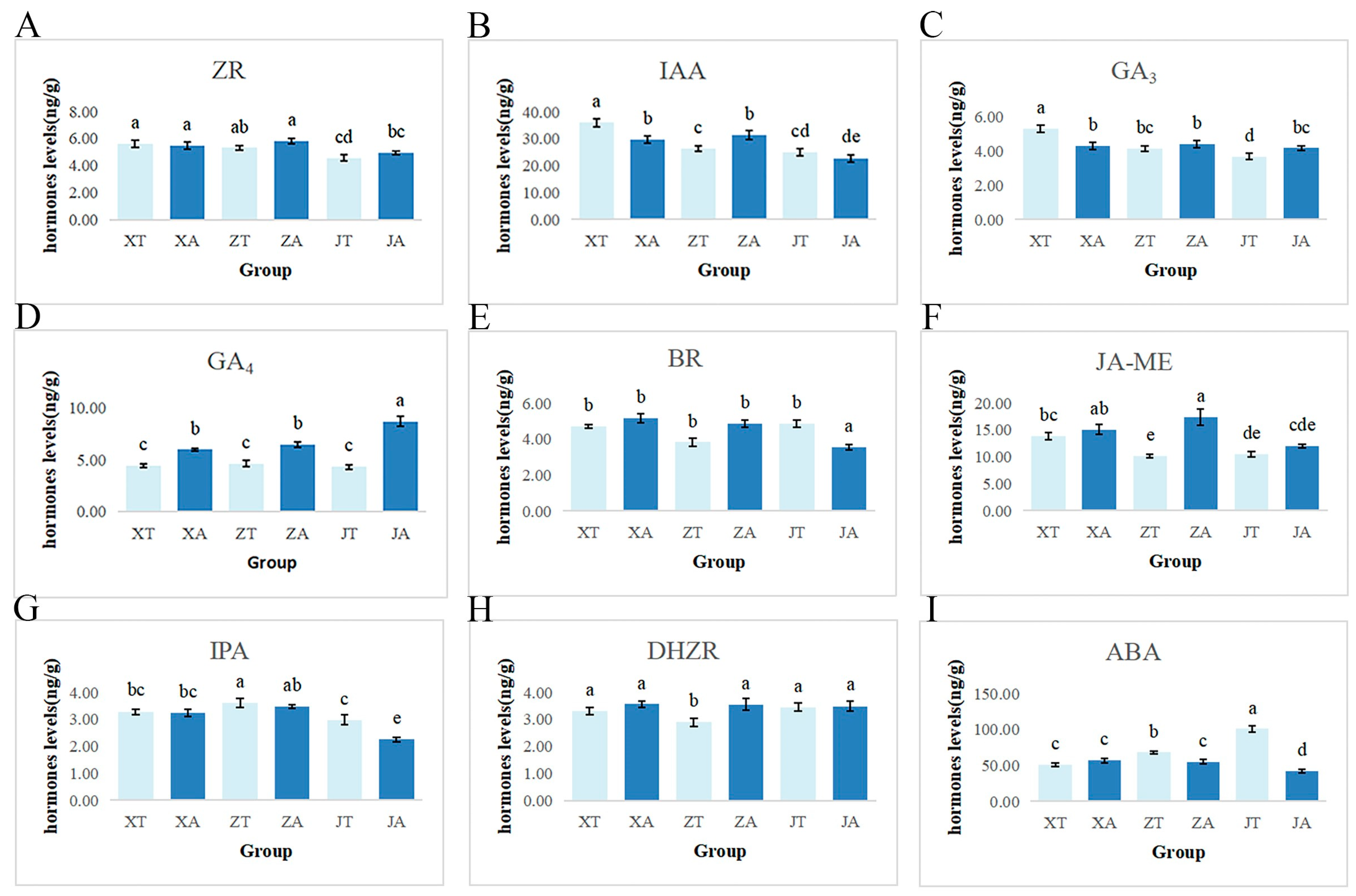
2.4. Endogenous Hormone Changes in Fruit Development of A. carambola
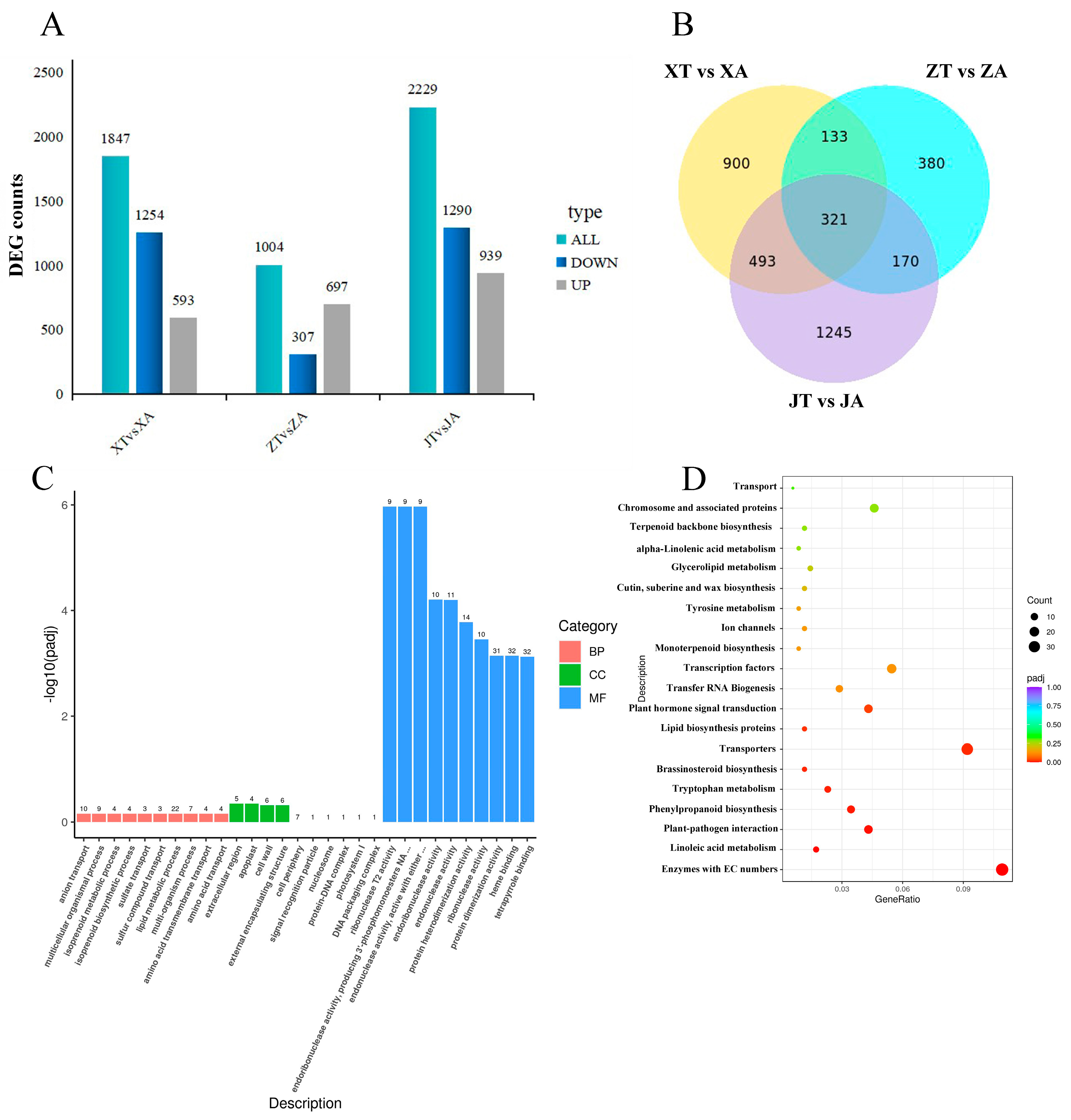
2.5. Transcriptome Sequencing Screened the Regulatory Genes Related to the Growth and Development of A. carambola Fruit Shape
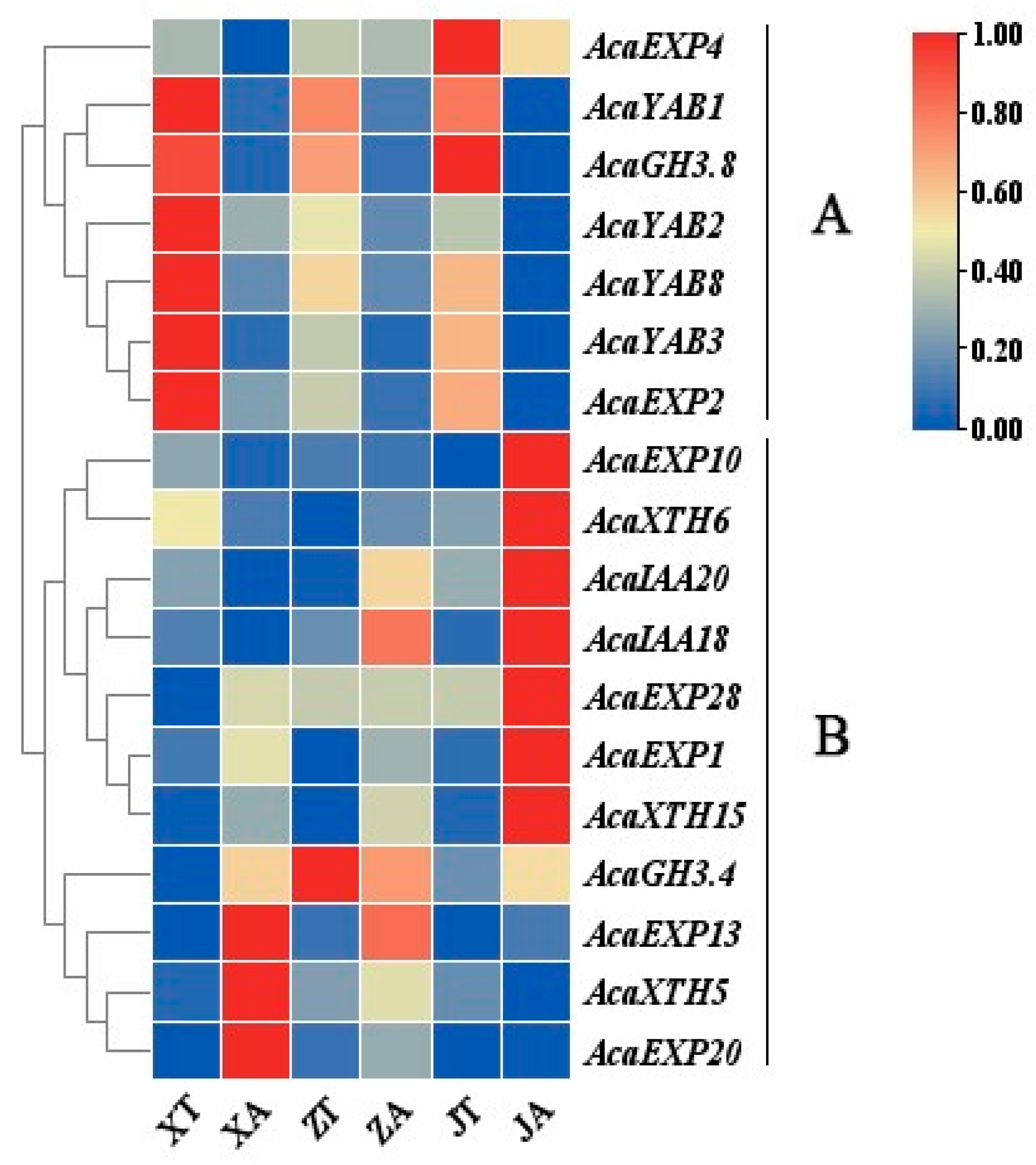
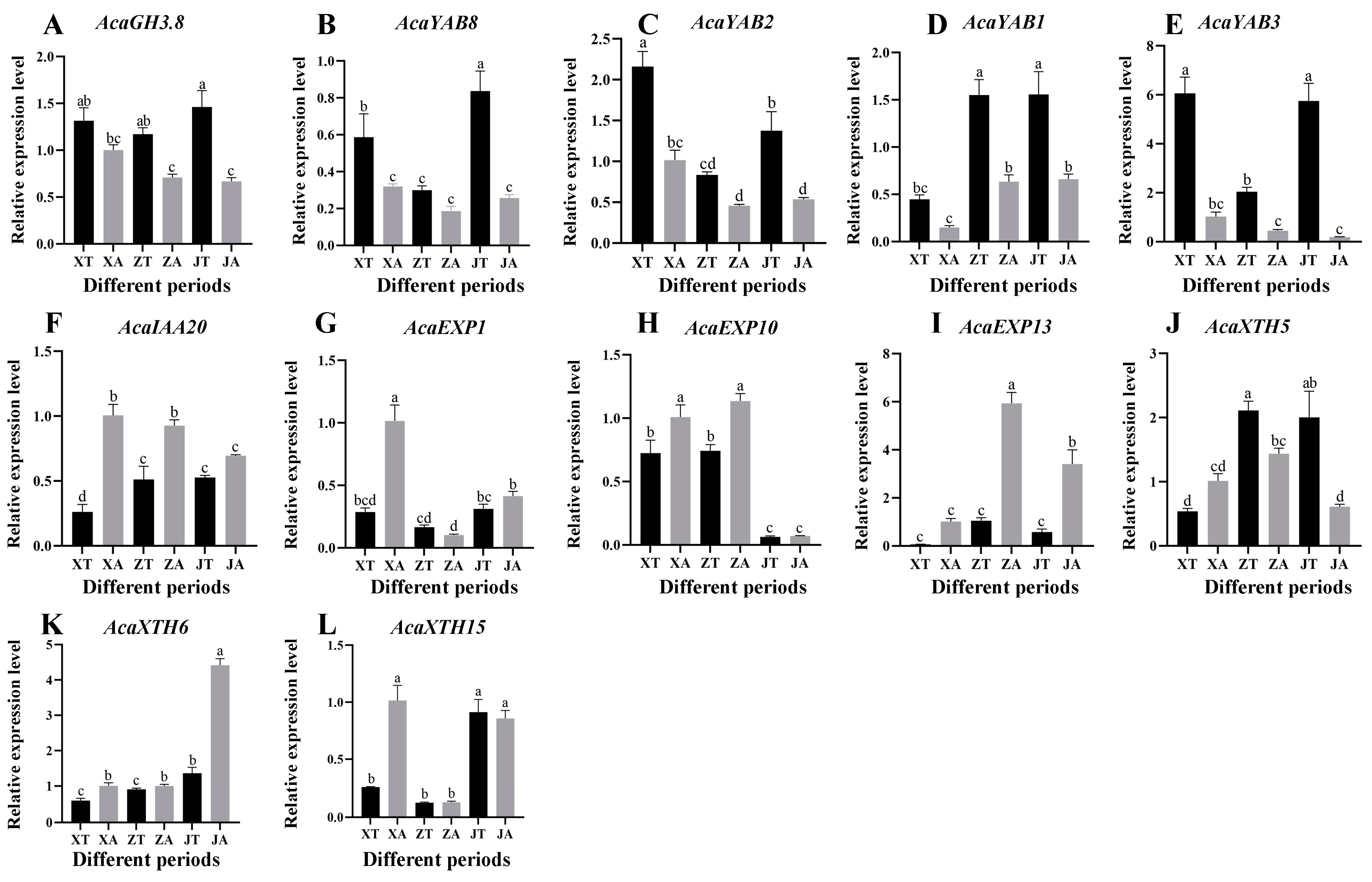
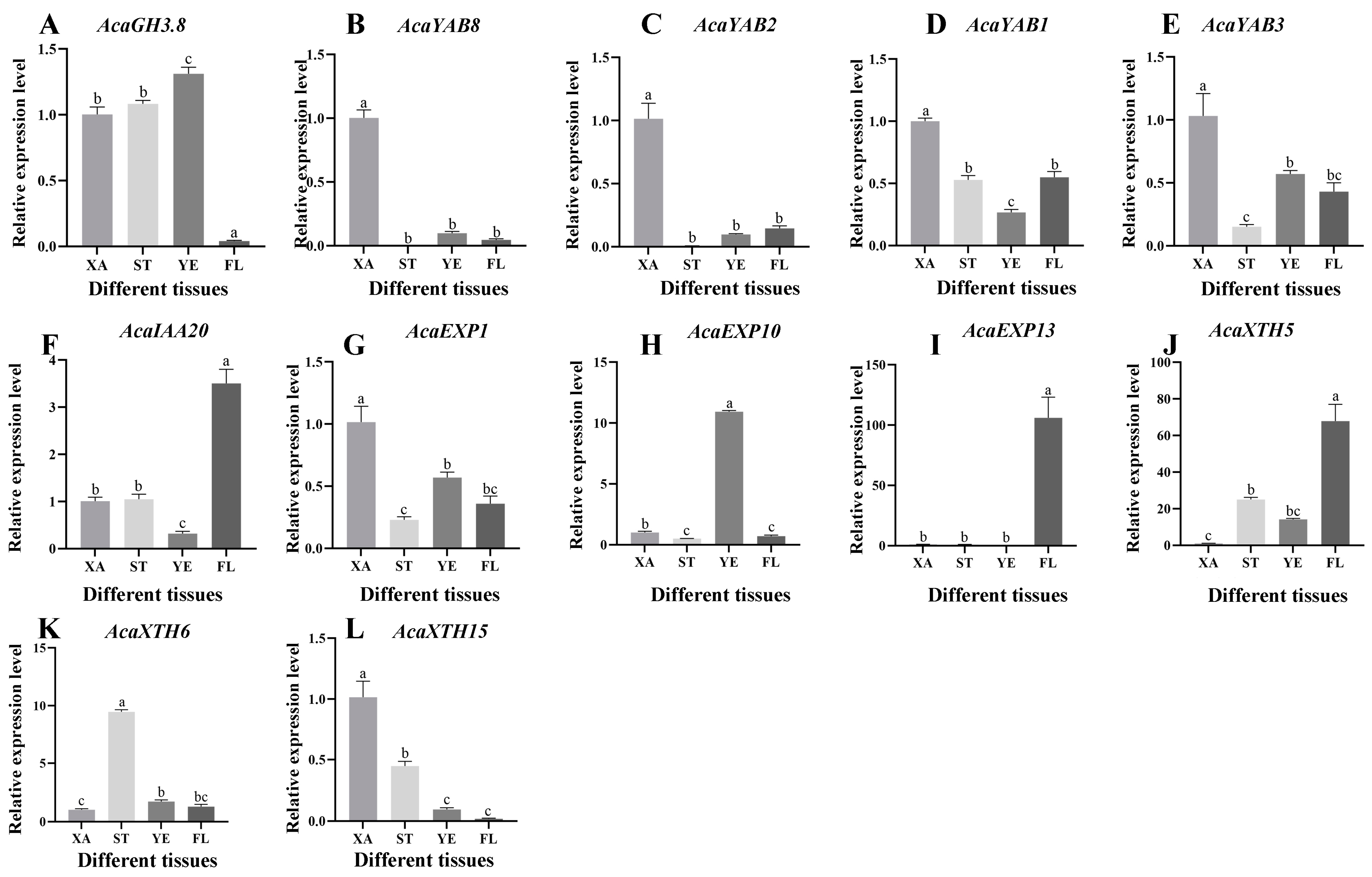
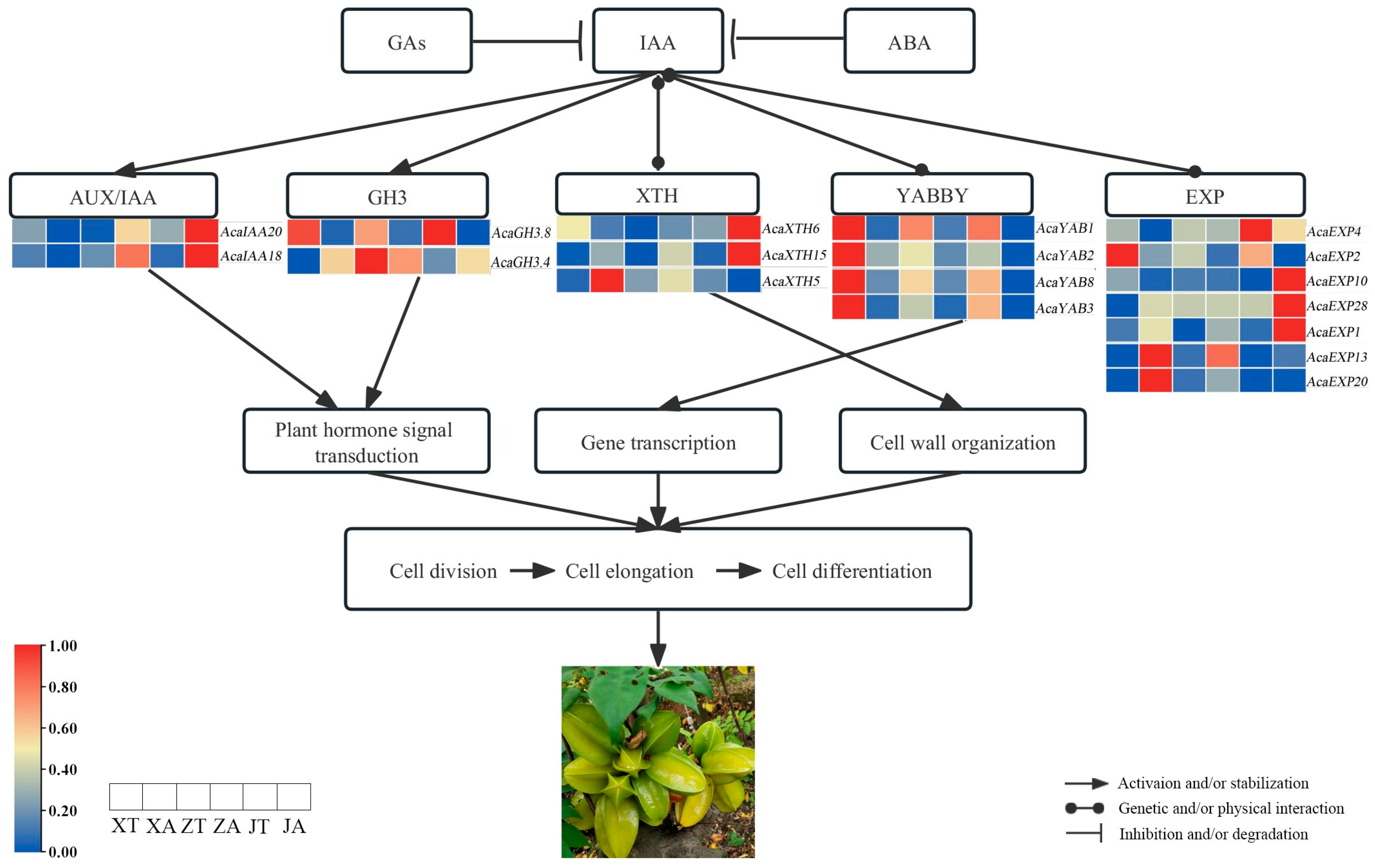
2.6. AcaAUX/IAA, AcaGH3, AcaYAB, AcaXTH and AcaEXP Gene Expression Patterns
2.7. Analysis of Expression Patterns of Related Genes
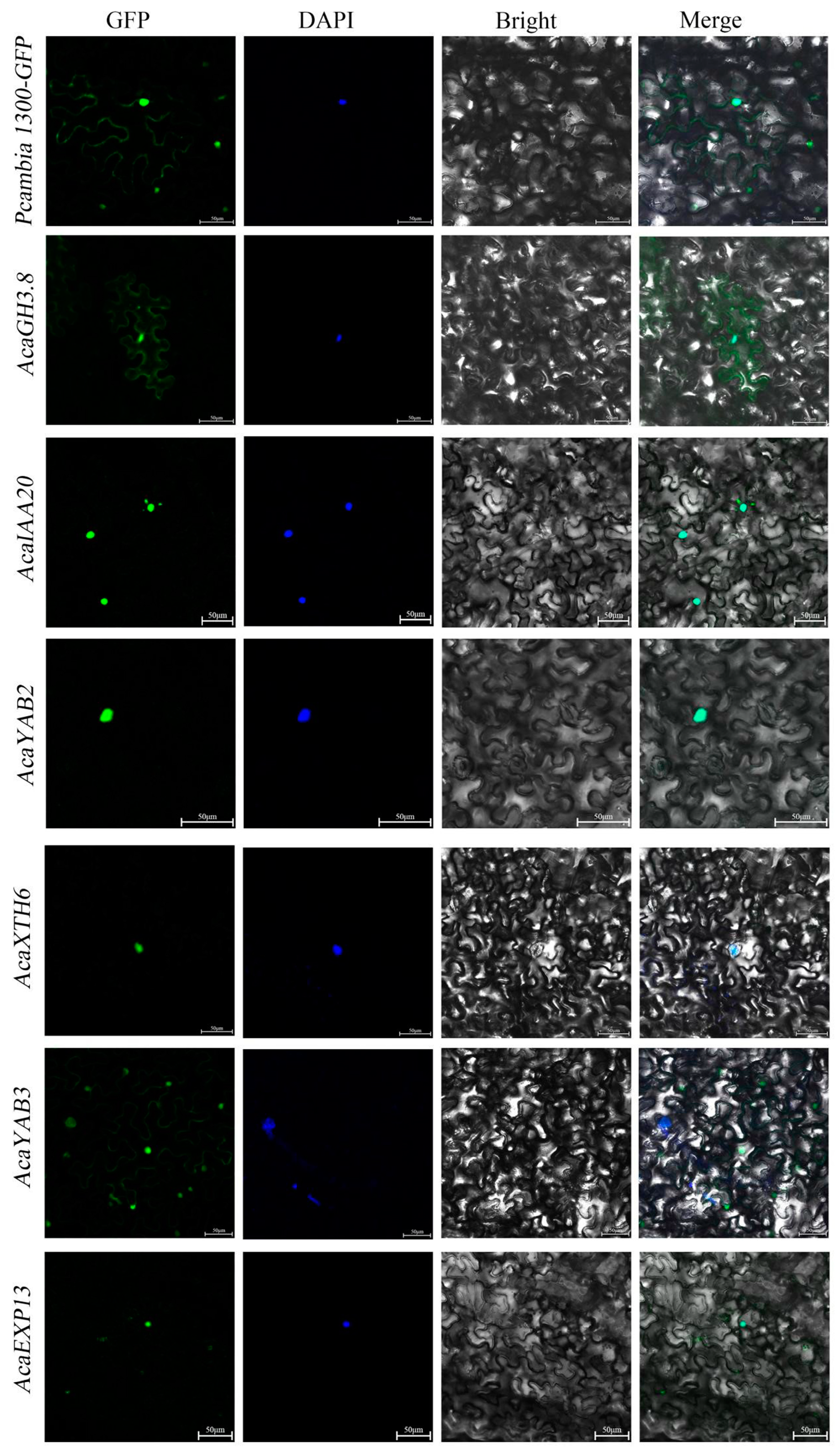
2.8. Key Gene Cloning and Subcellular Localization
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Paraffin Section Sample Preparation and Observation
4.3. SEM Sample Preparation and Observation
4.4. Image Analysis and Data Measurement
4.5. Hormone Content Determination
4.6. RNA Extraction and Transcriptomic Sequencing (RNA-seq)
4.7. qRT-PCR
4.8. Cloning and Subcellular Localization of Key Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Núñez-Elisea, R.; Crane, J.H. Selective pruning and crop removal increase early-season fruit production of carambola (Averrhoa carambola L.). Sci. Hortic. 2000, 86, 115–126. [Google Scholar] [CrossRef]
- Priadi, D.; Perdani, A.Y.; Sulistyowati, Y.; Pohan, F.N.; Mulyaningsih, E.S. Characterization of Carambola (Averrhoa Carambola L.) Plant Collection of Cibinong Plant Germplasm Garden Based on Phenotypic and Genetic Characters. Biosaintifika J. Biol. Biol. Educ. 2016, 8, 121–128. [Google Scholar] [CrossRef]
- Ou, J.; Zhu, Y.; Chen, H.; Zhou, J.; Chen, Y.; He, J.; Ning, L.; Pan, Z.; Gan, W. Genetic relationship analysis of 48 Averrhoa carambola L. germplasms based on SCoT marker. J. South Agric. 2019, 50, 1680–1687. [Google Scholar]
- Wu, S.; Sun, W.; Xu, Z.; Zhai, J.; Li, X.; Li, C.; Zhang, D.; Wu, X.; Shen, L.; Chen, J. The genome sequence of star fruit (Averrhoa carambola). Hortic. Res. 2020, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tian, J.; Li, S.; Liu, Y.; Feng, T.; Wang, Y.; Li, Y.; Huang, X.; Li, D. Characterization of the calmodulin/calmodulin-like protein (CAM/CML) family in Ginkgo biloba, and the influence of an ectopically expressed GbCML gene (Gb_30819) on seedling and fruit development of transgenic Arabidopsis. Plants 2022, 11, 1506. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-S.; Li, H.-L.; Peng, Z.-Z.; Liu, R.-L.; Han, Y.-C.; Wang, Y.-X.; Zhao, X.-D.; Fu, D.-Q. Composition, metabolism and postharvest function and regulation of fruit cuticle: A review. Food Chem. 2023, 411, 135449. [Google Scholar] [CrossRef]
- Ando, K.; Grumet, R. Transcriptional profiling of rapidly growing cucumber fruit by 454-pyrosequencing analysis. J. Am. Soc. Hortic. Sci. 2010, 135, 291–302. [Google Scholar] [CrossRef]
- Ando, K.; Carr, K.M.; Grumet, R. Transcriptome analyses of early cucumber fruit growth identifies distinct gene modules associated with phases of development. BMC Genom. 2012, 13, 518. [Google Scholar] [CrossRef]
- Ma, J.; Li, C.; Zong, M.; Qiu, Y.; Liu, Y.; Huang, Y.; Xie, Y.; Zhang, H.; Wang, J. CmFSI8/CmOFP13 encoding an OVATE family protein controls fruit shape in melon. J. Exp. Bot. 2022, 73, 1370–1384. [Google Scholar] [CrossRef]
- Higashi, K.; Hosoya, K.; Ezura, H. Histological analysis of fruit development between two melon (Cucumis melo L. reticulatus) genotypes setting a different size of fruit. J. Exp. Bot. 1999, 50, 1593–1597. [Google Scholar] [CrossRef]
- Olmstead, J.W.; Iezzoni, A.F.; Whiting, M.D. Genotypic differences in sweet cherry fruit size are primarily a function of cell number. J. Am. Soc. Hortic. Sci. 2007, 132, 697–703. [Google Scholar] [CrossRef]
- Scorzal, R.; May, L.G.; Purnell, B.; Upchurch, B. Differences in number and area of mesocarp cells between small-and large-fruited peach cultivars. J. Am. Soc. Hortic. Sci. 1991, 116, 861–864. [Google Scholar] [CrossRef]
- Zhang, C.; Tanabe, K.; Tani, H.; Nakajima, H.; Mori, M.; Sakuno, E. Biologically active gibberellins and abscisic acid in fruit of two late-maturing Japanese pear cultivars with contrasting fruit size. J. Am. Soc. Hortic. Sci. 2007, 132, 452–458. [Google Scholar] [CrossRef]
- Tian, Z.; Wu, B.; Liu, J.; Zhang, L.; Wu, T.; Wang, Y.; Han, Z.; Zhang, X. Genetic variations in MdSAUR36 participate in the negative regulation of mesocarp cell division and fruit size in Malus species. Mol. Breed. 2024, 44, 1. [Google Scholar] [CrossRef]
- Liu, X.; Pan, Y.; Liu, C.; Ding, Y.; Wang, X.; Cheng, Z.; Meng, H. Cucumber fruit size and shape variations explored from the aspects of morphology, histology, and endogenous hormones. Plants 2020, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Fan, X.; Liu, C.; Fang, J. Anatomical berry characteristics during the development of grape berries with different shapes. Hortic. Plant J. 2021, 7, 295–306. [Google Scholar] [CrossRef]
- Li, Y.-H.; Zhang, Z.; Sun, G.-M. Changes in cell number and cell size during pineapple (Ananas comosus L.) fruit development and their relationship with fruit size. Aust. J. Bot. 2010, 58, 673–678. [Google Scholar] [CrossRef]
- Sachs, T. Cell polarity and tissue patterning in plants. Development 1991, 113, 83–93. [Google Scholar] [CrossRef]
- Zumajo-Cardona, C.; Vasco, A.; Ambrose, B.A. The evolution of the KANADI gene family and leaf development in lycophytes and ferns. Plants 2019, 8, 313. [Google Scholar] [CrossRef]
- Bürstenbinder, K.; Savchenko, T.; Müller, J.; Adamson, A.W.; Stamm, G.; Kwong, R.; Zipp, B.J.; Dinesh, D.C.; Abel, S. Arabidopsis calmodulin-binding protein IQ67-domain 1 localizes to microtubules and interacts with kinesin light chain-related protein-1. J. Biol. Chem. 2013, 288, 1871–1882. [Google Scholar] [CrossRef]
- Levy, M.; Wang, Q.; Kaspi, R.; Parrella, M.P.; Abel, S. Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 2005, 43, 79–96. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Xiao, H.; Cabrera, A.; Meulia, T.; van der Knaap, E. SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiol. 2011, 157, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Handa, A.K. Hormonal regulation of tomato fruit development: A molecular perspective. J. Plant Growth Regul. 2005, 24, 67–82. [Google Scholar] [CrossRef]
- Wang, Y.; Clevenger, J.P.; Illa-Berenguer, E.; Meulia, T.; van der Knaap, E.; Sun, L. A comparison of sun, ovate, fs8. 1 and auxin application on tomato fruit shape and gene expression. Plant Cell Physiol. 2019, 60, 1067–1081. [Google Scholar] [CrossRef]
- Chen, S.; Wang, X.-J.; Tan, G.-F.; Zhou, W.-Q.; Wang, G.-L. Gibberellin and the plant growth retardant Paclobutrazol altered fruit shape and ripening in tomato. Protoplasma 2020, 257, 853–861. [Google Scholar] [CrossRef]
- Wang, H.; Jones, B.; Li, Z.; Frasse, P.; Delalande, C.; Regad, F.; Chaabouni, S.; Latche, A.; Pech, J.-C.; Bouzayen, M. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 2005, 17, 2676–2692. [Google Scholar] [CrossRef]
- Goetz, M.; Hooper, L.C.; Johnson, S.D.; Rodrigues, J.C.M.; Vivian-Smith, A.; Koltunow, A.M. Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol. 2007, 145, 351–366. [Google Scholar] [CrossRef]
- Shi, J.; Dong, J.; Xue, J.; Wang, H.; Yang, Z.; Jiao, Y.; Xu, L.; Huang, H. Model for the role of auxin polar transport in patterning of the leaf adaxial–abaxial axis. Plant J. 2017, 92, 469–480. [Google Scholar] [CrossRef]
- Smulders, M.J.; Croes, A.F.; Wullems, G.J. Polar transport of 1-naphthaleneacetic acid determines the distribution of flower buds on explants of tobacco. Plant Physiol. 1988, 88, 752–756. [Google Scholar] [CrossRef]
- Audran-Delalande, C.; Bassa, C.; Mila, I.; Regad, F.; Zouine, M.; Bouzayen, M. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol. 2012, 53, 659–672. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, L.; Gong, L.; Nie, F.-J.; Yang, J.-W.; Liu, X.; Yang, W.-J.; Zhang, G.-H.; Xie, R.-X.; Zhang, L. Genome-wide Identification of Potato WOX Gene Family and Its Expression Analysis in in vitro Regeneration and Abiotic Stress. Biotechnol. Bull. 2024, 40, 170. [Google Scholar]
- Terol, J.; Domingo, C.; Talón, M. The GH3 family in plants: Genome wide analysis in rice and evolutionary history based on EST analysis. Gene 2006, 371, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L. The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 2000, 3, 17–22. [Google Scholar] [CrossRef]
- Singh, V.K.; Jain, M.; Garg, R. Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes. Front. Plant Sci. 2015, 5, 789. [Google Scholar] [CrossRef] [PubMed]
- Cano, A.; Sánchez-García, A.B.; Albacete, A.; González-Bayón, R.; Justamante, M.S.; Ibáñez, S.; Acosta, M.; Pérez-Pérez, J.M. Enhanced conjugation of auxin by GH3 enzymes leads to poor adventitious rooting in carnation stem cuttings. Front. Plant Sci. 2018, 9, 566. [Google Scholar] [CrossRef]
- Domingo, C.; Andrés, F.; Tharreau, D.; Iglesias, D.J.; Talón, M. Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant-Microbe Interact. 2009, 22, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-W.; Li, C.-H.; Cao, J.; Zhang, Y.-C.; Zhang, S.-Q.; Xia, Y.-F.; Sun, D.-Y.; Sun, Y. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 2009, 151, 1889–1901. [Google Scholar] [CrossRef]
- Ding, X.; Cao, Y.; Huang, L.; Zhao, J.; Xu, C.; Li, X.; Wang, S. Activation of the indole-3-acetic acid–amido synthetase GH3-8 suppresses expansin expression and promotes salicylate-and jasmonate-independent basal immunity in rice. Plant Cell 2008, 20, 228–240. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 1999, 126, 2387–2396. [Google Scholar] [CrossRef]
- Ye, Q. Anatomical Analysis during Fruit Development and Isolation of Fruit Shaping-Related Gene in Vaccinium corymbosum. Master’s Thesis, Zhejiang Normal University, Jinhua, China, 2017. [Google Scholar]
- Biswas, M.K.; Chai, L.; Qiang, X.; Deng, X. Generation, functional analysis and utility of Citrus grandis EST from a flower-derived cDNA library. Mol. Biol. Rep. 2012, 39, 7221–7235. [Google Scholar] [CrossRef]
- Xiang, J.; Liu, R.; Li, T.; Han, L.; Zou, Y.; Xu, T.; Wei, J.; Wang, Y.; Xu, Y. Isolation and characterization of two VpYABBY genes from wild Chinese Vitis pseudoreticulata. Protoplasma 2013, 250, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Da, X.; Yu, K.; Shen, S.; Zhang, Y.; Wu, J.; Yi, H. Identification of differentially expressed genes in a spontaneous altered leaf shape mutant of the navel orange [Citrus sinensis (L.) Osbeck]. Plant Physiol. Biochem. 2012, 56, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Miedes, E.; Zarra, I.; Hoson, T.; Herbers, K.; Sonnewald, U.; Lorences, E. Xyloglucan endotransglucosylase and cell wall extensibility. J. Plant Physiol. 2011, 168, 196–203. [Google Scholar] [CrossRef]
- Fry, S.; Smith, R.; Renwick, K.; Martin, D.; Hodge, S.; Matthews, K. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 1992, 282, 821–828. [Google Scholar] [CrossRef]
- Miedes, E.; Herbers, K.; Sonnewald, U.; Lorences, E.P. Overexpression of a cell wall enzyme reduces xyloglucan depolymerization and softening of transgenic tomato fruits. J. Agric. Food Chem. 2010, 58, 5708–5713. [Google Scholar] [CrossRef]
- Witasari, L.D.; Huang, F.C.; Hoffmann, T.; Rozhon, W.; Fry, S.C.; Schwab, W. Higher expression of the strawberry xyloglucan endotransglucosylase/hydrolase genes Fv XTH 9 and Fv XTH 6 accelerates fruit ripening. Plant J. 2019, 100, 1237–1253. [Google Scholar] [CrossRef]
- Yokoyama, R.; Nishitani, K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 2001, 42, 1025–1033. [Google Scholar] [CrossRef]
- Xu, W.; Campbell, P.; Vargheese, A.K.; Braam, J. The Arabidopsis XET-related gene family: Environmental and hormonal regulation of expression. Plant J. 1996, 9, 879–889. [Google Scholar] [CrossRef]
- Huang, J.; Chen, G.-Z.; Ahmad, S.; Wang, Q.; Tu, S.; Shi, X.-L.; Hao, Y.; Zhou, Y.-Z.; Lan, S.-R.; Liu, Z.-J. Identification, Molecular Characteristics, and Evolution of YABBY Gene Family in Melastoma dodecandrum. Int. J. Mol. Sci. 2023, 24, 4174. [Google Scholar] [CrossRef]
- Bohner, J.; Bangerth, F. Cell number, cell size and hormone levels in semi-isogenic mutants of Lycopersicon pimpinellifolium differing in fruit size. Physiol. Plant. 1988, 72, 316–320. [Google Scholar] [CrossRef]
- Niu, H.; Liu, X.; Tong, C.; Wang, H.; Li, S.; Lu, L.; Pan, Y.; Zhang, X.; Weng, Y.; Li, Z. The WUSCHEL-related homeobox1 gene of cucumber regulates reproductive organ development. J. Exp. Bot. 2018, 69, 5373–5387. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K.; Ostin, A.; Lioussanne, L.; Sandberg, G. Developmental regulation of indole-3-acetic acid turnover in Scots pine seedlings. Plant Physiol. 2001, 125, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.; Lee, S.H.; Cho, H.-T. P-Glycoprotein4 displays auxin efflux transporter–like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 2007, 19, 3930–3943. [Google Scholar] [CrossRef]
- Ilegems, M.; Douet, V.; Meylan-Bettex, M.; Uyttewaal, M.; Brand, L.; Bowman, J.L.; Stieger, P.A. Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development 2010, 137, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hao, N.; Li, H.; Ge, D.; Du, Y.; Liu, R.; Wen, C.; Li, Y.; Zhang, X.; Wu, T. PINOID is required for lateral organ morphogenesis and ovule development in cucumber. J. Exp. Bot. 2019, 70, 5715–5730. [Google Scholar] [CrossRef]
- Sato, A.; Yamamoto, K.T. Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol. Plant. 2008, 133, 397–405. [Google Scholar] [CrossRef]
- Takase, T.; Nakazawa, M.; Ishikawa, A.; Kawashima, M.; Ichikawa, T.; Takahashi, N.; Shimada, H.; Manabe, K.; Matsui, M. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004, 37, 471–483. [Google Scholar] [CrossRef]
- Böttcher, C.; Keyzers, R.A.; Boss, P.K.; Davies, C. Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J. Exp. Bot. 2010, 61, 3615–3625. [Google Scholar] [CrossRef]
- Liu, C.; Xu, Z.; Chua, N.-h. Auxin polar transport is essential for the establishment of bilateral symmetry during early plant embryogenesis. Plant Cell 1993, 5, 621–630. [Google Scholar] [CrossRef]
- Rose, J.K.; Braam, J.; Fry, S.C.; Nishitani, K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002, 43, 1421–1435. [Google Scholar] [CrossRef]
- Millar, D.J.; Whitelegge, J.P.; Bindschedler, L.V.; Rayon, C.; Boudet, A.M.; Rossignol, M.; Borderies, G.; Bolwell, G.P. The cell wall and secretory proteome of a tobacco cell line synthesising secondary wall. Proteomics 2009, 9, 2355–2372. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.-T.; Cosgrove, D.J. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 9783–9788. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Pang, S.; Ma, Y.; Deng, L.; He, S.; Yi, S.; Lv, Q.; Zheng, Y. The ARF, AUX/IAA and GH3 gene families in citrus: Genome-wide identification and expression analysis during fruitlet drop from abscission zone A. Mol. Genet. Genom. 2015, 290, 2089–2105. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, S.; Guan, X.; Chen, L.; He, Y.; Wang, J.; Lu, G. Genome-wide identification and transcriptional profiling analysis of auxin response-related gene families in cucumber. BMC Res. Notes 2014, 7, 218. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.H.; Jacobson, K.A.; Rose, J.; Zeller, R. Paraffin embedding tissue samples for sectioning. CSH Protoc. 2008, 2008, pdb.prot4989. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, J.; Wang, Z.; Zhu, Q.; Wang, W. Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol. 2001, 127, 315–323. [Google Scholar] [CrossRef]
- Love, M.; Anders, S.; Huber, W. Differential analysis of count data—The DESeq2 package. Genome Biol. 2014, 15, 10–1186. [Google Scholar]
- Li, X.; Zhu, Y.; Zhao, Y.; Chen, L.; Ren, H.; Wu, S.; Zhai, J. Selection and Validation of Reference Genes of Averrhoa carambola by Quantitative Real-time PCR. Mol. Plant Breed. 2021, 22, 789–799. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tuo, W.; Wu, C.; Wang, X.; Yang, Z.; Xu, L.; Shen, S.; Zhai, J.; Wu, S. Developmental Morphology, Physiology, and Molecular Basis of the Pentagram Fruit of Averrhoa carambola. Plants 2024, 13, 2696. https://doi.org/10.3390/plants13192696
Tuo W, Wu C, Wang X, Yang Z, Xu L, Shen S, Zhai J, Wu S. Developmental Morphology, Physiology, and Molecular Basis of the Pentagram Fruit of Averrhoa carambola. Plants. 2024; 13(19):2696. https://doi.org/10.3390/plants13192696
Chicago/Turabian StyleTuo, Wanli, Chunmei Wu, Xuexuan Wang, Zirui Yang, Lianhuan Xu, Siyuan Shen, Junwen Zhai, and Shasha Wu. 2024. "Developmental Morphology, Physiology, and Molecular Basis of the Pentagram Fruit of Averrhoa carambola" Plants 13, no. 19: 2696. https://doi.org/10.3390/plants13192696





