Identification of Wheat Septoria tritici Resistance Genes in Wheat Germplasm Using Molecular Markers
Abstract
:1. Introduction
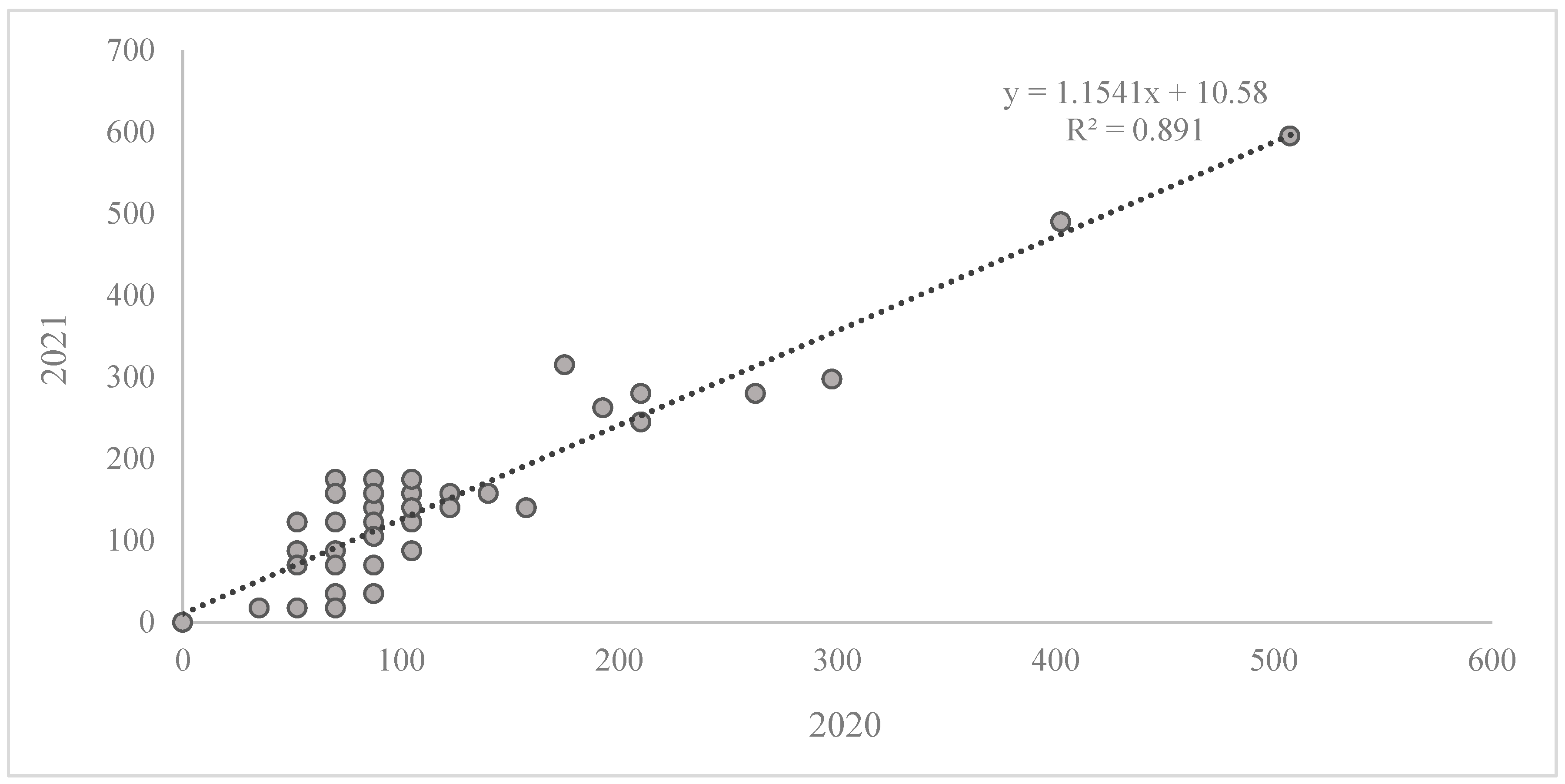
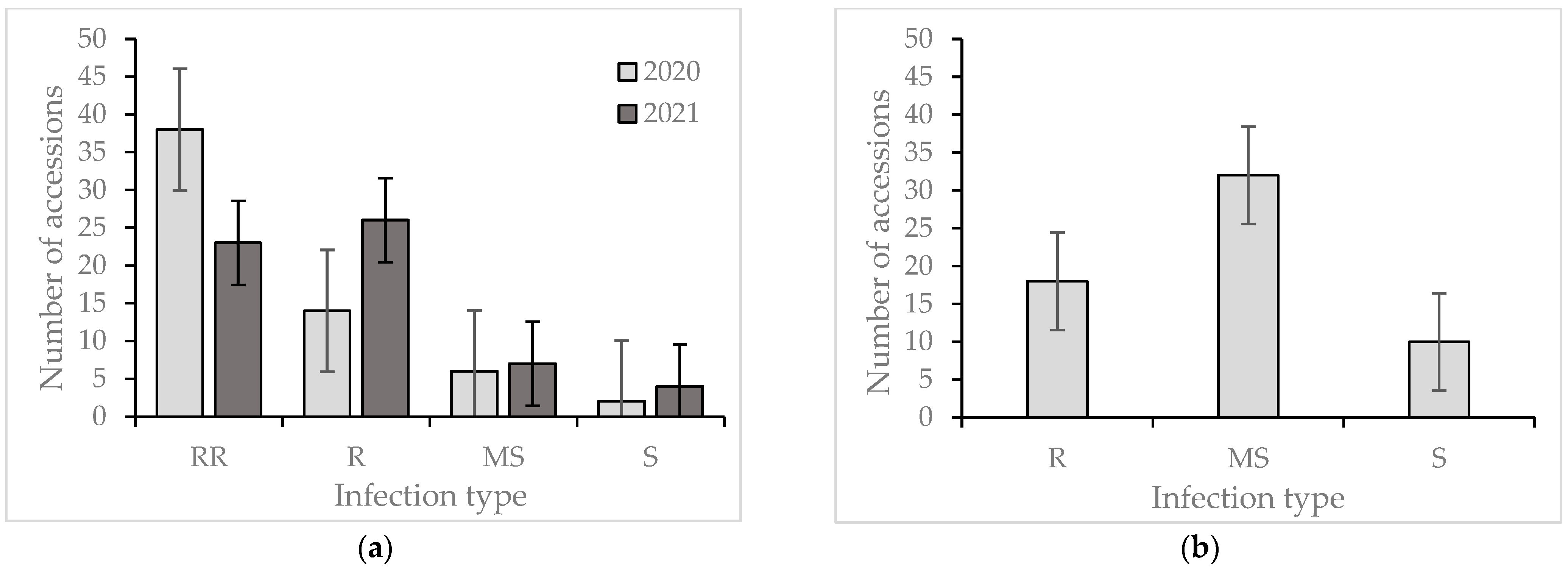
2. Results
Reaction of the Wheat Collection to Zymoseptoria tritici
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Field Evaluation of Wheat Entries against Zymoseptoria tritici
4.3. Inoculum Production and Inoculations and Seedling Resistance
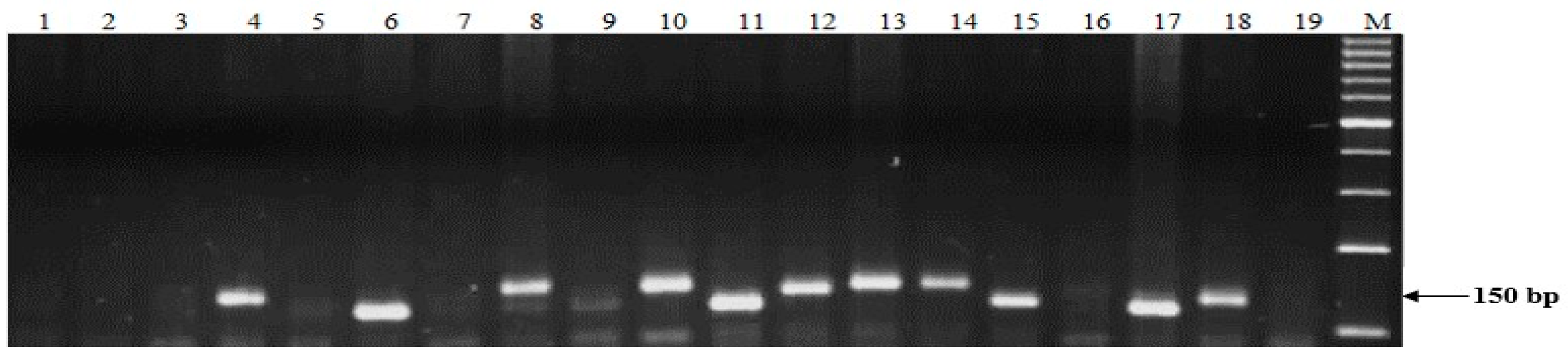
4.4. DNA Extraction and Molecular Screening of Stb Resistance Genes
4.5. Statistical Data Processing
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sharma, R.C.; Rajaram, S.; Alikulov, S.; Ziyaev, Z.; Hazratkulova, S.; Khodarahami, M.; Nazeri, S.M.; Belen, S.; Khalikulov, Z.; Mosaad, M.; et al. Improved winter wheat genotypes for Central and West Asia. Euphytica 2013, 190, 19–31. [Google Scholar] [CrossRef]
- Morgounov, A.; Akin, B.; Demir, L.; Keser, L.; Kokhmetova, A.; Martynov, S.; Orhan, S.; Özdemir, F.; Özseven, I.; Sapakhova, Z.; et al. Yield gain due to fungicide application in varieties of winter wheat (Triticum aestivum) resistant and susceptible to leaf rust. Crop Pasture Sci. 2015, 66, 649–659. [Google Scholar] [CrossRef]
- Food and Agriculture (FAO). Organization of the United Nations, Disponível em. Available online: http://www.fao.org/faostat/en/#data/CC (accessed on 24 March 2023).
- Koyshibaev, M.K. Diseases of Wheat; FAO: Ankara, Turkey, 2018; p. 365. [Google Scholar]
- Babkenova, S.A.; Babkenov, A.T.; Pakholkova, E.V.; Kanafin, B.K. Pathogenic complexity of septoria spot disease of wheat in northern Kazakhstan. Plant Sci. Today 2020, 7, 601–606. [Google Scholar] [CrossRef]
- Eyal, Z. The Septoria tritici and Stagonospora nodorum blotch diseases of wheat. Eur. J. Plant Pathol. 1999, 105, 629–664. [Google Scholar] [CrossRef]
- Adhikari, T.B.; Mamidi, S.; Gurung, S.; Bonman, J.M. Mapping of new quantitative trait loci (QTL) for resistance to Septoria tritici blotch in spring wheat (Triticum aestivum L.). Euphytica 2015, 205, 699–706. [Google Scholar] [CrossRef]
- Kokhmetova, A.; Ali, S.; Sapakhova, Z.; Atishova, M. Identification of genotypes-carriers of resistance to tan spot Ptr ToxA and Ptr ToxB of Pyrenophora tritici-repentis in common wheat collection. Vavilov J. Genet. Breed. 2018, 22, 978–986. [Google Scholar] [CrossRef]
- Kokhmetova, A.; Atishova, M.; Madenova, A.; Kumarbayeva, M. Genotyping of wheat germplasm for resistance to toxins of tan spot Pyrenophora tritici-repentis. J. Biotechnol. 2019, 305, 53. [Google Scholar] [CrossRef]
- Kokhmetova, A.; Kovalenko, N.; Kumarbaeva, M. Pyrenophora tritici-repentis population structure in the Republic of Kazakhstan and identification of wheat germplasm resistant to tan spot. Vavilov J. Genet. Breed. 2020, 24, 722–729. [Google Scholar] [CrossRef]
- Kokhmetova, A.; Kumarbayeva, M.; Atishova, M.; Nehe, A.; Riley, I.; Morgounov, A. Identification of high-yielding wheat genotypes resistant to Pyrenophora tritici-repentis (tan spot). Euphytica 2021, 217, 97. [Google Scholar] [CrossRef]
- Lamari, L.; Strelkov, S.E.; Yahyaoui, A.; Amedov, M.; Saidov, M.; Djunusova, M.; Koichibayev, M. Virulence of Phyrenophora tritici—Repentis in the countries of the Silk Road. Can. J. Plant Pathol. 2005, 27, 383–388. [Google Scholar] [CrossRef]
- Kokhmetova, A.; Rsaliyev, S.; Atishova, M.; Kumarbayeva, M.; Malysheva, A.; Keishilov, Z.; Zhanuzak, D.; Bolatbekova, A. Evaluation of wheat germplasm for resistance to leaf rust (Puccinia triticina) and Identification of the Sources of Lr Resistance Genes Using Molecular Markers. Plants 2021, 10, 1484. [Google Scholar] [CrossRef]
- Malysheva, A.; Kokhmetova, A.; Urazaliev, R.; Kumarbayeva, M.; Keishilov, Z.; Nurzhuma, M.; Bolatbekova, A.; Kokhmetova, A. Phenotyping and Identification of Molecular Markers Associated with Leaf Rust Resistance in the Wheat Germplasm from Kazakhstan, CIMMYT and ICARDA. Plants 2023, 12, 2786. [Google Scholar] [CrossRef] [PubMed]
- Kuttibayevich, S.S.; Slyamkhanovna, T.Z.; Zharakovich, M.M.; Tontaevna, Y.A.; Mukashevna, T.O.; Amirzhanovna, I.G.; Klara, I.; Gulnara, N.; Dossym, B.; Faritovna, S.G. Classification of wheat yellow rust populations in the conditions of Kazakhstan (Puccinia striiformis West. f. sp. tritici Erikss. et Henn.). Bull. Natl. Acad. Sci. Repub. Kazakhstan 2019, 1, 263–268. [Google Scholar]
- Gultyaeva, E.; Yusov, V.; Rosova, M.; Mal’chikov, P.; Shaydayuk, E.; Kovalenko, N.; Rsaliyev, A. Evaluation of resistance of spring durum wheat germplasm from Russia and Kazakhstan to fungal foliar pathogens. Cereal Res. Commun. 2020, 48, 71–79. [Google Scholar] [CrossRef]
- Absattarova, A.; Baboev, S.; Bulatova, K.; Karabayev, M.; Koishibayev, M.; Kuklacheva, V.; Morgounov, A.; Rsaliev, S.; Sarbayev, A.; Urazaliev, R.; et al. Improvement of wheat yellow rust resistance in Kazakhstan and Uzbekistan through sub-regional cooperation. In Meeting the Challenge of Yellow Rust in Cereal Crops; Johnson, R., Yahyaoui, A., Wellings, C., Saidi, A., Ketata, H., Eds.; ICARDA: Aleppo, Syria, 2002; pp. 34–41. [Google Scholar]
- Kokhmetova, A.M.; Atishova, M.N. Identification of sources of resistance to wheat stem rust using molecular markers. Russ. J. Genet. Appl. Res. 2012, 2, 486–493. [Google Scholar] [CrossRef]
- Olivera, F.P.; Szabo, L.; Kokhmetova, A.; Morgunov, A.; Luster, D.G.; Jin, Y. Puccinia graminis f. sp. tritici population causing recent wheat stem rust epidemics in Kazakhstan is highly diverse and includes novel virulences. Phytopathology 2022, 112, 2403–2415. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.K.M.; Chartrain, L.; Lasserre-Zuber, P.; Saintenac, C. Genetics of resistance to Zymoseptoria tritici and applications to wheat breeding. Fungal Genet. Biol. 2015, 79, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.T.T.; Rybak, K.; Furuki, E.; Breen, S.; Solomon, P.S.; Oliver, R.P.; Tan, K.C. Differential effector gene expression underpins epistasis in a plant fungal disease. Plant J. 2016, 87, 343–354. [Google Scholar] [CrossRef]
- Friesen, T.L.; Chu, C.G.; Liu, Z.H.; Halley, S.; Faris, J.D. Host-selective toxins produced by Stagonospora nodorum confer disease susceptibility in adult wheat plants under field conditions. Theor. Appl. Genet. 2009, 118, 1489–1497. [Google Scholar] [CrossRef]
- Dreisigacker, S.; Wang, X.; Martinez Cisneros, B.A.; Jing, R.; Singh, P.K. Adult-plant resistance to Septoria tritici blotch in hexaploid spring wheat. Theor. Appl. Genet. 2015, 128, 2317–2329. [Google Scholar] [CrossRef]
- Tommasini, L.; Schnurbusch, T.; Fossati, D.; Mascher, F.; Keller, B. Association mapping of Stagonospora nodorum blotch resistance in modern European winter wheat varieties. Theor. Appl. Genet. 2007, 115, 697–708. [Google Scholar] [CrossRef]
- Adhikari, T.B.; Jackson, E.W.; Gurung, S.; Hansen, J.M.; Bonman, J.M. Association mapping of quantitative resistance to Phaeosphaeria nodorum in spring wheat landraces from the USDA National Small Grains Collection. Phytopathology 2011, 101, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.T.T.; Rybak, K.; Bertazzoni, S.; Furuki, E.; Dinglasan, E.; Hickey, L.T.; Oliver, R.P.; Tan, K.C. Novel sources of resistance to Septoria nodorum blotch in the Vavilov wheat collection identified by genome-wide association studies. Theor. Appl. Genet. 2018, 131, 1223–1238. [Google Scholar] [CrossRef] [PubMed]
- Goudemand, E.; Laurent, V.; Duchalais, L.; Tabib Ghaffary, S.M.; Kema, G.H.J.; Lonnet, P.; Margale, E.; Robert, O. Association mapping and meta-analysis: Two complementary approaches for the detection of reliable Septoria tritici blotch quantitative resistance in bread wheat (Triticum aestivum L.). Mol. Breed. 2013, 32, 563–584. [Google Scholar] [CrossRef]
- Singh, P.K.; Singh, S.; Deng, Z.; He, X.; Kehel, Z.; Singh, R.P. Characterization of QTLs for Seedling Resistance to Tan Spot and Septoria nodorum Blotch in the PBW343/Kenya Nyangumi Wheat Recombinant Inbred Lines Population. Int. J. Mol. Sci. 2019, 20, 5432. [Google Scholar] [CrossRef] [PubMed]
- Ruud, A.K.; Dieseth, J.A.; Ficke, A.; Furuki, E.; Phan, H.T.T.; Oliver, R.P.; Tan, K.C.; Lillemo, M. Genome-wide association mapping of resistance to Septoria nodorum leaf blotch in a Nordic spring wheat collection. Plant Genome 2019, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, T.B.; Cavaletto, J.R.; Dubcovsky, J.; Gieco, J.O.; Schlatter, A.R.; Goodwin, S.B. Molecular Mapping of the Stb4 gene for resistance to Septoria tritici Blotch in Wheat. Phytopathology 2004, 94, 1198–1206. [Google Scholar] [CrossRef] [PubMed]
- Olson, E.L.; Brown-Guedira, G.; Marshall, D.S.; Jin, Y.; Mergoum, M.; Lowe, I.; Dubcovsky, J. Genotyping of US wheat germplasm for presence of stem rust resistance genes Sr24, Sr36 and Sr1RSAmigo. Crop Sci. 2010, 50, 668–675. [Google Scholar] [CrossRef]
- Babkenova, S.A.; Babkenov, A.T.; Kolomiets, T.M.; Skolotneva, E.S.; Divashuk, M.G. Molecular genetic tagging of wheat varieties genes resistant to Septoria tritici in northern Kazakhstan. Int. J. Green Pharm. 2017, 11, 430–437. [Google Scholar]
- Pakholkova, E.V.; Sal’nikova, N.N.; Kurkova, N.A. Genetic structure of regional population of Mycosphaerella graminicola (Septoria tritici). The pathogen of Septoria disease in wheat (Triticum aestivum L.). Agric. Biol. 2016, 51, 722–730. [Google Scholar]
- Brown, J.K.M.; Kema, G.H.J.; Forrer, H.R.; Verstappen, E.C.P.; Arraiano, L.S.; Brading, P.A.; Foster, E.M.; Fried, P.M.; Jenny, E. Resistance of wheat cultivars and breeding lines to Septoria tritici blotch caused by isolates of Mycosphaerella graminicola in field trials. Plant Pathol. 2001, 50, 325–338. [Google Scholar] [CrossRef]
- Chartrain, L.; Brading, P.A.; Makepeace, J.C.; Brown, J.K.M. Sources of resistance to Septoria tritici blotch and implications for wheat breeding. Plant Pathol. 2004, 53, 454–460. [Google Scholar] [CrossRef]
- Parker, S.R.; Welham, S.; Paveley, N.D.; Foulkes, J.; Scott, R.K. Tolerance of Septoria leaf blotch in winter wheat. Plant Pathol. 2004, 53, 1–10. [Google Scholar] [CrossRef]
- Arraiano, L.S.; Brown, J.K.M. Identification of isolate-specific and partial resistance to Septoria tritici blotch in 238 European wheat cultivars and breeding lines. Plant Pathol. 2006, 55, 726–738. [Google Scholar] [CrossRef]
- Arraiano, L.S.; Brown, J.K.M. Sources of resistance and susceptibility to Septoria tritici blotch of wheat. Mol. Plant Pathol. 2017, 18, 276–292. [Google Scholar] [CrossRef] [PubMed]
- Ajaz, S.; Benbow, H.R.; Christodoulou, T.; Uauy, C.; Doohan, F.M. Evaluation of the susceptibility of modern, wild, ancestral, and mutational wheat lines to Septoria tritici blotch disease. Plant Pathol. 2021, 70, 1123–1137. [Google Scholar] [CrossRef]
- Kema, G.H.J.; van Ginkel, M. Global Insights into the Septoria and Stagonospora Diseases in Cereals. In Proceedings of the 6th International Symposium on Septoria and Stagonospora Diseases in Cereals, Tunis, Tunisia, 12 December 2003; pp. 131–140. [Google Scholar]
- Zeleneva, Y.V.; Konkova, E.A. Soft wheat cultivars grown in the Saratov region and their resistance to Septoria blotch. Vavilov J. Genet. Breed. 2023, 27, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Tyryshkin, L.G.; Ershova, M.A. Inheritance of resistance to Septoria blotch in common wheat sample MN81330. Russ. J. Genet. 2004, 40, 454–457. [Google Scholar] [CrossRef]
- Toropova, E.Y.; Kazakova, O.A.; Piskarev, V.V. Septoria blotch epidemic process on spring wheat varieties. Vavilov J. Genet. Breed. 2020, 24, 139–148. [Google Scholar] [CrossRef]
- Kolomiets, T.M.; Pankratova, L.F.; Pakholkova, E.V. Wheat (Triticum L.) cultivars from GRIN collection (USA) selected for durable resistance to Septoria tritici and Stagonospora nodorum Blotch. Agric. Biol. 2017, 52, 561–569. [Google Scholar] [CrossRef]
- Kolomiets, T.M.; Shamanin, V.P.; Pakholkova, E.V.; Pankratova, L.F.; Salnikova, N.N.; Shepelev, S.S.; Pototskaya, I.V.; Abugalieva, A.; Morgunov, A.I. Screening of hexaploid synthetic wheat lines and spring bread wheat variety for resistance to Septoria blotch. Bull. Omsk. State Agrar. Univ. 2018, 3, 13–26. (In Russian) [Google Scholar]
- Postnikova, E.N.; Khasanov, B.A. Tan Spot in Central Asia—Helminthosporium Blights of Wheat: Spot Bloch and Tan Spot; CIMMYT-UCL: Veracruz, Mexico, 1997; pp. 107–114. [Google Scholar]
- Zeleneva, Y.V.; Sudnikova, V.P.; Kovalenko, N.M.; Gusev, I.V. Resistance of spring bread wheat cultivars and lines to Septoria leaf blotch, tan spot, and spot blotch pathogens. Proc. Appl. Bot. Genet. Breed. 2023, 184, 196–206. (In Russian) [Google Scholar] [CrossRef]
- Haque, A.; Shaheen, T.; Gulzar, T.; Rahman, M.U.; Jalal, F.; Sattar, S.; Ehsan, B.; Iqbal, Z.; Younas, M. Study of rust resistance genes in wheat germplasm with DNA markers. Bioinformation 2014, 10, 371–377. [Google Scholar] [CrossRef]
- Dospekhov, B.A. Methods of Field Experience (With the Basics of Statistical Processing of Research Results), 5th ed.; Kolos: Kovalivka, Ukraine, 1985. [Google Scholar]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Saari, E.E.; Prescott, J.M. A scale for appraising the foliar intensity of wheat diseases. Plant Dis. Rep. 1975, 59, 377–380. [Google Scholar]
- Pyzhikova, G.V.; Karaseva, E.V. Methods of studying Septoria pathogens on isolated wheat leaves. Selskokhozyaystvennaya biologiya. 1985, 12, 112–114. [Google Scholar]
- Scala, V.; Pietricola, C.; Farina, V.; Beccaccioli, M.; Zjalic, S.; Quaranta, F.; Fornara, M.; Zaccaria, M.; Momeni, B.; Reverberi, M.; et al. Tramesan Elicits Durum Wheat Defense against the Septoria Disease Complex. Biomolecules 2020, 10, 608. [Google Scholar] [CrossRef]
- Fagundes, W.C.; Haueisen, J.; Stukenbrock, E.H. Dissecting the Biology of the Fungal Wheat Pathogen Zymoseptoria tritici: A Laboratory Workflow. Curr. Protoc. Microbiol. 2020, 59, e128. [Google Scholar] [CrossRef]
- Kolomiets, T.M.; Pakholkova, E.V.; Dubovaya, L.P. Selection of the Initial Material for the Creation of Wheat Cultivars with Long-Term Resistance to Septoria Blotch; Pechatnyy Gorod: Moscow, Russia, 2017; p. 56. [Google Scholar]
- Riede, C.; Anderson, J. Linkage of RFLP markers to an aluminum tolerance gene in wheat. Crop Sci. 1996, 36, 905–909. [Google Scholar] [CrossRef]
- Chen, X.; Line, R.; Leung, H. Genome scanning for resistance-gene analogs in rice, barley, and wheat by high-resolution electrophoresis. Theor. Appl. Genet. 1998, 97, 345–355. [Google Scholar] [CrossRef]
- Adhikari, T.B.; Wallwork, H.; Goodwin, S.B. Microsatellite markers linked to the Stb2 and Stb3 Genes for Resistance to Septoria Tritici Blotch in Wheat. Crop Sci. 2004, 44, 1403–1411. [Google Scholar] [CrossRef]
- Röder, M.S.; Plaschke, J.; König, S.U.; Börner, A.; Sorrells, M.E.; Tanksley, S.D.; Ganal, M.W. Abundance, variability and chromosomal location of microsatellites in wheat. Mol. Genet. Genom. 1995, 246, 327–333. [Google Scholar] [CrossRef]
- McCartney, C.A.; Brûlé-Babel, A.L.; Lamari, L.; Somers, D.J. Chromosomal location of a race-specific resistance gene to Mycosphaerella graminicola in the spring wheat ST6. Theor. Appl. Genet. 2003, 107, 1181–1186. [Google Scholar] [CrossRef]
- Wilcoxson, R.; Skovmand, B.; Atif, A. Evaluation of wheat cultivars ability to retard development of stem rust. Ann. Appl. Biol. 1974, 80, 275–281. [Google Scholar] [CrossRef]
- Aphalo, P.J. OpenIntro Statistics, by David M. Diez, Christopher D. Barr and Mine Cetinkaya-Rundel. UV4Plants Bull. 2016, 51–53. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; The R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: http://www.R-project.org/ (accessed on 21 April 2023).
| Trait | Factor | SS | df | MS | F-Value | %SS | hb2, % |
|---|---|---|---|---|---|---|---|
| AUDPC, field | Genotype | 1,197,935 | 59 | 20,304 | 25.72 *** | 94.44 | 0.94 |
| Year | 24,013 | 1 | 24,013 | 30.42 *** | 1.89 | ||
| Residuals | 46,578 | 59 | 46,578 | 3.67 | |||
| Z. tritici, seedling | Genotype | 134.87 | 59 | 2.2859 | 11.82 *** | 74.24 | 0.74 |
| Replication | 1.17 | 4 | 0.2917 | 1.50 | 0.64 | ||
| Residuals | 45.63 | 236 | 0.1934 | 25.12 |
| Trait | Min | Max | Mean | SE | Variance | SD | Median | Mode | CV |
|---|---|---|---|---|---|---|---|---|---|
| AUDPC 2020 | 0 | 507.5 | 114.92 | 11.87 | 8454.23 | 91.95 | 87.5 | 70 | 80.01 |
| AUDPC 2021 | 0 | 595 | 143.21 | 14.51 | 12,639.21 | 112.42 | 122.5 | 70 | 78.50 |
| Z. tritici, seedling | 1 | 3.6 | 2.23 | 0.09 | 0.457175 | 0.68 | 2.4 | 2.6 | 30.28 |
| Name of Variety | Origin | Type | Final Score 2020 (Field) | AUDPC 2020 (Field) | Final Score 2021 (Field) | AUDPC 2021 (Field) | Z. tritici (Seedling) | Stb Genes |
|---|---|---|---|---|---|---|---|---|
| Saratovskaya 70 | RU | Spring | 15 | 175 | 40 | 315.0 | 2.6 | none |
| Severyanka | RU | Spring | 5 | 52.5 | 10 | 70.0 | 2 | none |
| Saratovskaya 29 | RU | Spring | 15 | 105 | 20 | 157.5 | 2 | none |
| Saratovskaya 42 | RU | Spring | 15 | 87.5 | 25 | 175.0 | 1.2 | none |
| Saratovskaya 55 | RU | Spring | 10 | 70 | 20 | 175.0 | 1.6 | Stb8 |
| Albidum 31 | RU | Spring | 5 | 70 | 10 | 70.0 | 3.6 | Stb4 |
| Rosinka 3 | RU | Spring | 10 | 87.5 | 10 | 70.0 | 2.6 | Stb8 |
| Omskaya 18 | RU | Spring | 0 | 0 | 0 | 0.0 | 2.4 | Stb7, Stb2 |
| Omskaya 19 | RU | Spring | 10 | 87.5 | 20 | 157.5 | 2.2 | none |
| Omskaya 28 | RU | Spring | 5 | 35 | 5 | 17.5 | 2.6 | none |
| Omskaya 29 | RU | Spring | 5 | 70 | 10 | 70.0 | 1.4 | Stb4 |
| Omskaya 35 | RU | Spring | 5 | 52.5 | 15 | 87.5 | 1.4 | Stb2 |
| Omskaya 36 | RU | Spring | 5 | 70 | 10 | 70.0 | 2.6 | Stb7 |
| Mironovskaya 808 | RU | Winter | 10 | 105 | 10 | 122.5 | 3 | Stb8 |
| Pamyati Azieva | RU | Spring | 5 | 52.5 | 10 | 70.0 | 2.6 | Stb2 |
| KR11-13 | CIMMYT-ICARDA-IWWIP | Winter | 10 | 122.5 | 15 | 140.0 | 2.4 | Stb4 |
| KR12-9011 | CIMMYT-ICARDA-IWWIP | Winter | 15 | 105 | 20 | 157.5 | 3 | Stb8 |
| KR11-40 | CIMMYT-ICARDA-IWWIP | Winter | 10 | 87.5 | 10 | 140.0 | 2 | none |
| KR12-9012 | CIMMYT-ICARDA-IWWIP | Winter | 15 | 87.5 | 20 | 157.5 | 3 | Stb7 |
| KR11-9025 | CIMMYT-ICARDA-IWWIP | Winter | 15 | 105 | 10 | 122.5 | 1.4 | none |
| KR12-5001 | CIMMYT-ICARDA-IWWIP | Winter | 5 | 52.5 | 15 | 122.5 | 2.4 | none |
| KR12-07 | CIMMYT-ICARDA-IWWIP | Winter | 20 | 157.5 | 15 | 140.0 | 1.4 | none |
| KR12-5035 | CIMMYT-ICARDA-IWWIP | Winter | 10 | 70 | 15 | 122.5 | 2.8 | none |
| KR12-09 | CIMMYT-ICARDA-IWWIP | Winter | 5 | 70 | 10 | 70.0 | 2.2 | none |
| KR12-5075 | CIMMYT-ICARDA-IWWIP | Winter | 30 | 210 | 30 | 245.0 | 3 | Stb8 |
| KR11-03 | CIMMYT-ICARDA-IWWIP | Winter | 10 | 87.5 | 10 | 122.5 | 1 | Stb2 |
| KR12-10 | CIMMYT-ICARDA-IWWIP | Winter | 10 | 122.5 | 15 | 140.0 | 2.4 | Stb2 |
| KR11-9014 | CIMMYT-ICARDA-IWWIP | Winter | 30 | 297.5 | 25 | 297.5 | 1 | Stb2 |
| KR12-18 | CIMMYT-ICARDA-IWWIP | Winter | 5 | 105 | 15 | 140.0 | 2 | Stb4 |
| KR11-26 | CIMMYT-ICARDA-IWWIP | Winter | 20 | 192.5 | 30 | 262.5 | 2.6 | Stb4 |
| SOMO/SORA/ACTS5 | CIMMYT—SEPTMON | Spring | 10 | 70 | 20 | 122.5 | 3 | Stb2 |
| P83-5112/V82274 | CIMMYT—SEPTMON | Spring | 15 | 140 | 20 | 157.5 | 1.6 | none |
| DOMOJNJA | CIMMYT—SEPTMON | Spring | 10 | 70 | 15 | 87,5 | 2 | none |
| NANJTNG 82149 KAUZ | CIMMYT—SEPTMON | Spring | 10 | 87.5 | 20 | 122.5 | 1.2 | none |
| CROC_1/AE.SQUARROSA-205//BORL95/3/2/*Milan | CIMMYT—SEPTMON | Spring | 5 | 52.5 | 10 | 70.0 | 1.6 | Stb8 |
| TALHUENJNJA | CIMMYT—SEPTMON | Spring | 15 | 122.5 | 20 | 157.5 | 2.4 | none |
| JAC161/TEMU51.80 | CIMMYT—SEPTMON | Spring | 5 | 70 | 20 | 157.5 | 1 | none |
| GAN/AE.437 SQUARROSA | CIMMYT—SEPTMON | Spring | 5 | 70 | 10 | 70.0 | 2.4 | none |
| EFED/22150 | CIMMYT—SEPTMON | Spring | 15 | 105 | 15 | 87.5 | 2.6 | Stb2, Stb8 |
| BR14/CEP847-2 | CIMMYT—SEPTMON | Spring | 15 | 87.5 | 20 | 105.0 | 1.6 | none |
| ECHA/LI115 | CIMMYT—SEPTMON | Spring | 10 | 105 | 15 | 140.0 | 1.8 | none |
| TOO11/TOOOO7 | CIMMYT—SEPTMON | Spring | 10 | 52.5 | 15 | 87.5 | 2.8 | none |
| F133/SHA5//OPATA | CIMMYT—SEPTMON | Spring | 10 | 70 | 15 | 87.5 | 2.6 | none |
| TRAP#1/BOW | CIMMYT—SEPTMON | Spring | 5 | 52.5 | 10 | 70.0 | 2.4 | Stb4 |
| EFED/F5.83 7792(BAJAS) | CIMMYT—SEPTMON | Spring | 5 | 52.5 | 10 | 70.0 | 2.4 | Stb7, Stb4 |
| Batyr | KZ | Winter | 10 | 70 | 10 | 35.0 | 2.8 | none |
| Bayandy | KZ | Winter | 0 | 52.5 | 5 | 17.5 | 2.6 | none |
| Botagoz | KZ | Winter | 10 | 87.5 | 10 | 35.0 | 3 | none |
| Celinnaya 50 | KZ | Spring | 15 | 105 | 25 | 175.0 | 1.6 | none |
| Derbes | KZ | Winter | 10 | 157.5 | 15 | 140.0 | 2.8 | none |
| Egemen | KZ | Winter | 45 | 507.5 | 50 | 595.0 | 2.6 | Stb2, Stb8 |
| Kazakhstanskaya 4 | KZ | Spring | 25 | 210 | 30 | 280.0 | 3.6 | none |
| Kokbiday | KZ | Winter | 5 | 70 | 5 | 17.5 | 3 | Stb8 |
| Kupava/Avocet (S)-1 | KZ | Winter | 15 | 87.5 | 20 | 157.5 | 2.6 | Stb8 |
| Kyzylbiday | KZ | Winter | 30 | 402.5 | 40 | 490.0 | 2.4 | none |
| Manshyk | KZ | Winter | 40 | 402.5 | 40 | 490.0 | 1 | none |
| Samgay | KZ | Spring | 5 | 70 | 10 | 70.0 | 2.6 | none |
| Sapaly | KZ | Winter | 10 | 87.5 | 10 | 70.0 | 1.6 | none |
| Steklovidnaya 24 | KZ | Winter | 35 | 262.5 | 30 | 280.0 | 1 | none |
| Sultan 2 | KZ | Winter | 10 | 122.5 | 15 | 140.0 | 3 | Stb2 |
| Controls | ||||||||
| Morocco | MA | Winter | 50 | 595 | 60 | 665 | 3.8 | none |
| Veranopolis (Stb2) | Brazil | Spring | 5 | 52.5 | 10 | 70 | 1 | Stb2 |
| Tadinia (Stb4) | USA | Spring | 5 | 70 | 10 | 70.0 | 1.6 | Stb4 |
| CS (Synthetic 7D) (Stb5) | USA | Spring | 5 | 52.5 | 10 | 70.0 | 1.4 | Stb5 |
| Estanzuela Federal (Stb7) | Uruguay | Spring | 5 | 52.5 | 10 | 70.0 | 1 | Stb7 |
| Synthetic W-7984 (Stb8) | USA | Winter | 5 | 52.5 | 10 | 70.0 | 1.2 | Stb8 |
| Gen | Chr | Type of Marker | Primer Name | Sequence of Primers 5′-3′ | Annealing Temperature, °C | Fragment Size, b.p | Reference |
|---|---|---|---|---|---|---|---|
| Stb2 | 3BS | SSR | WMS389-L | 5′-ATC ATG TCG ATC TCC TTG ACG-3′ | 60 | 150 | [58] |
| WMS389-R | 5′-CAT GCA CAT TTA GCA GAT-3′ | ||||||
| Stb4 | 6D | SSR | WMS111-L | 5′-ACC TGA TCA GAT CCC ACT CG-3′ | 55 | 210/220 | [30] |
| WMS111-R | 5′-TTC GTA GGC TCT CTC CGA CTG-3′ | ||||||
| Stb5 | 7DS | SSR | WMS44-L | 5′-GTT GAG CTT TTC AGT TCG GC-3′ | 60 | 182 | [59] |
| WMS44-R | 5′-ACT GGC ATC CAC TGA AGC TG-3′ | ||||||
| Stb7 | 4AL | SSR | WMC313-L | 5′-GCA GTC TAA TTA TCT GCT GG CG-3′ 5′-GGG TCC TTG TCT ACT CAT GT CT-3′ | 51 | 197 | [60] |
| WMC313-R | |||||||
| Stb8 | 7BL | SSR | WMS577-L | 5’-ATG GCA TAA TTT GGT GAA AT TG-3 5′-TGT TTC AAG CCC AAC TTC TA TT-3′ | 55 | 180 | [20] |
| WMS577-R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokhmetova, A.; Bolatbekova, A.; Zeleneva, Y.; Malysheva, A.; Bastaubayeva, S.; Bakhytuly, K.; Dutbayev, Y.; Tsygankov, V. Identification of Wheat Septoria tritici Resistance Genes in Wheat Germplasm Using Molecular Markers. Plants 2024, 13, 1113. https://doi.org/10.3390/plants13081113
Kokhmetova A, Bolatbekova A, Zeleneva Y, Malysheva A, Bastaubayeva S, Bakhytuly K, Dutbayev Y, Tsygankov V. Identification of Wheat Septoria tritici Resistance Genes in Wheat Germplasm Using Molecular Markers. Plants. 2024; 13(8):1113. https://doi.org/10.3390/plants13081113
Chicago/Turabian StyleKokhmetova, Alma, Ardak Bolatbekova, Yuliya Zeleneva, Angelina Malysheva, Sholpan Bastaubayeva, Kanat Bakhytuly, Yerlan Dutbayev, and Vladimir Tsygankov. 2024. "Identification of Wheat Septoria tritici Resistance Genes in Wheat Germplasm Using Molecular Markers" Plants 13, no. 8: 1113. https://doi.org/10.3390/plants13081113
APA StyleKokhmetova, A., Bolatbekova, A., Zeleneva, Y., Malysheva, A., Bastaubayeva, S., Bakhytuly, K., Dutbayev, Y., & Tsygankov, V. (2024). Identification of Wheat Septoria tritici Resistance Genes in Wheat Germplasm Using Molecular Markers. Plants, 13(8), 1113. https://doi.org/10.3390/plants13081113






