Identification and Characterization of Mitogen-Activated Protein Kinase (MAPK) Genes in Sunflower (Helianthus annuus L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Retrieval and Identification of Putative MAP Kinase Cascade Genes
2.2. Phylogenetic Tree Construction and Homology Assessment
2.3. Chromosomal Locations and Gene Structure
2.4. Nomenclature of MPKs and MKKs
2.5. Expression Analysis and miRNA Prediction of Sunflower MPKs and MKKs
2.6. Tajima’s Relative Rate and Neutrality Test
3. Results
3.1. The Diversity of MPK and MKK Genes in Sunflower Relative to Other Species
3.2. Gene Location, Subcellular Localization and Structural Variation of MPKs and MKKs in H. annuus
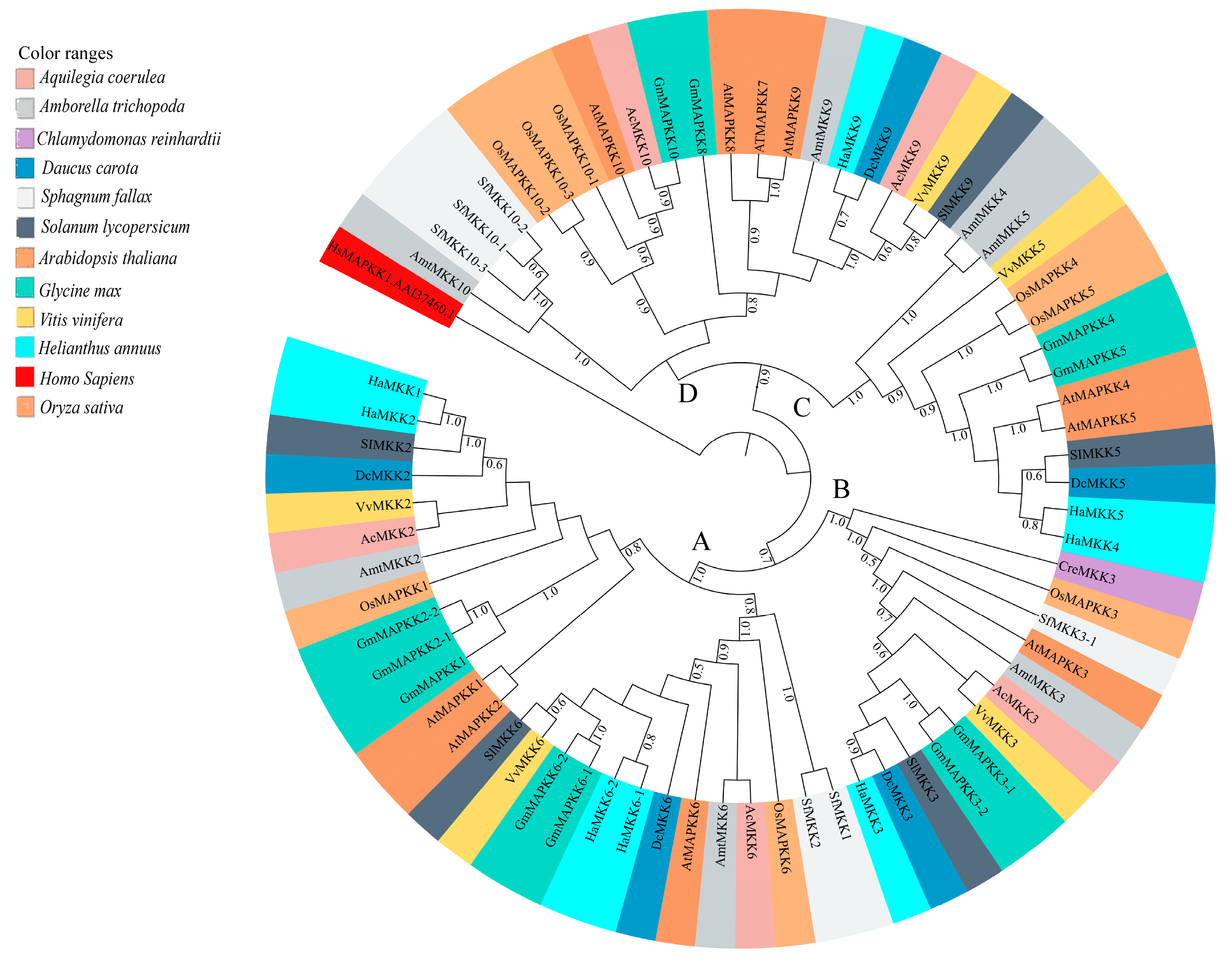
3.3. Phylogenetic Analyses
3.3.1. MPKs
3.3.2. MKKs
3.4. Expression Analysis and miRNA Prediction of Sunflower MPKs and MKKs
3.5. Tajima’s Relative Rate and Neutrality Tests on MPKs and MKKs
4. Discussion
4.1. Nomenclature of MPKs and MKKs
4.2. Diversity and the Phylogenetic Relationship of MPKs
4.3. Diversity and Phylogenetic Relationship of MKKs
4.4. Expression Analysis and miRNA Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mohanta, T.K.; Arora, P.K.; Mohanta, N.; Parida, P.; Bae, H. Identification of new members of the MAPK gene family in plants shows diverse conserved domains and novel activation loop variants. BMC Genom. 2015, 16, 58. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cole, P.A. Catalytic mechanisms and regulation of protein kinases. In Methods Enzymolgy; Elsevier: Amsterdam, The Netherlands, 2014; Volume 548, pp. 1–21. [Google Scholar]
- Ichimura, K.; Shinozaki, K.; Tena, G.; Sheen, J.; Henry, Y.; Champion, A.; Kreis, M.; Zhang, S.; Hirt, H.; Wilson, C. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar]
- Hamel, L.-P.; Nicole, M.-C.; Sritubtim, S.; Morency, M.-J.; Ellis, M.; Ehlting, J.; Beaudoin, N.; Barbazuk, B.; Klessig, D.; Lee, J. Ancient signals: Comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006, 11, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Sturgill, T.W.; Ray, L.B. Muscle proteins related to microtubule associated protein-2 are substrates for an insulin-stimulatable kinase. Biochem. Biophys. Res. Commun. 1986, 134, 565–571. [Google Scholar] [CrossRef]
- Duerr, B.; Gawienowski, M.; Ropp, T.; Jacobs, T. MsERK1: A mitogen-activated protein kinase from a flowering plant. Plant Cell 1993, 5, 87–96. [Google Scholar] [CrossRef]
- Stafstrom, J.P.; Altschuler, M.; Anderson, D.H. Molecular cloning and expression of a MAP kinase homologue from pea. Plant Mol. Biol. 1993, 22, 83–90. [Google Scholar] [CrossRef]
- Nakagami, H.; Pitzschke, A.; Hirt, H. Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 2005, 10, 339–346. [Google Scholar] [CrossRef]
- Tatebayashi, K.; Takekawa, M.; Saito, H. A docking site determining specificity of Pbs2 MAPKK for Ssk2/Ssk22 MAPKKKs in the yeast HOG pathway. EMBO J. 2003, 22, 3624–3634. [Google Scholar] [CrossRef] [Green Version]
- Jonak, C.; Okresz, L.; Bogre, L.; Hirt, H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 2002, 5, 415–424. [Google Scholar] [CrossRef]
- Tena, G.; Asai, T.; Chiu, W.-L.; Sheen, J. Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 2001, 4, 392–400. [Google Scholar] [CrossRef]
- Bardwell, L. Mechanisms of MAPK Signalling Specificity; Portland Press Limited: London, UK, 2006. [Google Scholar]
- Lewis, T.S.; Shapiro, P.S.; Ahn, N.G. Signal transduction through MAP kinase cascades. In Advances in Cancer Research; Elsevier: Amsterdam, The Netherlands, 1998; Volume 74, pp. 49–139. [Google Scholar]
- Janitza, P.; Ullrich, K.K.; Quint, M. Toward a comprehensive phylogenetic reconstruction of the evolutionary history of mitogen-activated protein kinases in the plant kingdom. Front. Plant Sci. 2012, 3, 271. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C. Specificity of receptor tyrosine kinase signaling: Transient versus sustained extracellular signal-regulated kinase activation. Cell 1995, 80, 179–185. [Google Scholar] [CrossRef] [Green Version]
- Kissoudis, C.; van de Wiel, C.; Visser, R.G.; van der Linden, G. Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Front. Plant Sci. 2014, 5, 207. [Google Scholar] [CrossRef] [PubMed]
- Ranty, B.; Aldon, D.; Cotelle, V.; Galaud, J.-P.; Thuleau, P.; Mazars, C. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 2016, 7, 327. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Schikora, A.; Hirt, H. MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 2009, 12, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Müller-Xing, R.; Xing, Q.; Goodrich, J. Footprints of the sun: Memory of UV and light stress in plants. Front. Plant Sci. 2014, 5, 474. [Google Scholar]
- Davis, P.K.; Brachmann, R.K. Chromatin remodeling and cancer. Cancer Biol. Ther. 2003, 2, 23–30. [Google Scholar] [CrossRef]
- Rejeb, I.B.; Pastor, V.; Mauch-Mani, B. Plant responses to simultaneous biotic and abiotic stress: Molecular mechanisms. Plants 2014, 3, 458–475. [Google Scholar] [CrossRef]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef]
- Shao, Z.-Q.; Xue, J.-Y.; Wu, P.; Zhang, Y.-M.; Wu, Y.; Hang, Y.-Y.; Wang, B.; Chen, J.-Q. Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol. 2016. [Google Scholar] [CrossRef]
- Ramirez-Prado, J.S.; Abulfaraj, A.A.; Rayapuram, N.; Benhamed, M.; Hirt, H. Plant immunity: From signaling to epigenetic control of defense. Trends Plant Sci. 2018, 9, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.-L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Neupane, A.; Nepal, M.P.; Piya, S.; Subramanian, S.; Rohila, J.S.; Reese, R.N.; Benson, B.V. Identification, nomenclature, and evolutionary relationships of mitogen-activated protein kinase (MAPK) genes in soybean. Evol. Bioinform. 2013, 9, 363. [Google Scholar] [CrossRef] [PubMed]
- Neupane, A.; Nepal, M.P.; Benson, B.V.; MacArthur, K.J.; Piya, S. Evolutionary history of mitogen-activated protein kinase (MAPK) genes in Lotus, Medicago, and Phaseolus. Plant Signal. Behav. 2013, 8, e27189. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Wang, W.; Li, Y.; Hou, X. Divergent evolutionary patterns of the MAPK cascade genes in Brassica rapa and plant phylogenetics. Hort. Res. 2017, 4, 17079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Wang, J.; Pan, C.; Guan, X.; Wang, Y.; Liu, S.; He, Y.; Chen, J.; Chen, L.; Lu, G. Genome-wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PLoS ONE 2014, 9, e103032. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, L.; Xue, C.; Fang, H.; Zhao, J.; Liu, M. Genome-wide identification and analysis of MAPK and MAPKK gene family in Chinese jujube (Ziziphus jujuba Mill.). BMC Genom. 2017, 18, 855. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Pan, J.; Zhang, D.; Jiang, S.; Cai, G.; Wang, L.; Li, D. Identification of mitogen-activated protein kinase kinase gene family and MKK–MAPK interaction network in maize. Biochem. Biophys. Res. Commun. 2013, 441, 964–969. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xu, R.; Luo, X.; Jiang, Z.; Shu, H. Genome-wide identification and expression analysis of MAPK and MAPKK gene family in Malus domestica. Gene 2013, 531, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hu, W.; Tan, S.; Wang, M.; Ma, Z.; Zhou, S.; Deng, X.; Zhang, Y.; Huang, C.; Yang, G. Genome-wide identification and analysis of MAPK and MAPKK gene families in Brachypodium distachyon. PLoS ONE 2012, 7, e46744. [Google Scholar] [CrossRef]
- Piao, Y.; Jin, K.; He, Y.; Liu, J.; Liu, S.; Li, X.; Piao, Z. Genome-wide identification and role of MKK and MPK gene families in clubroot resistance of Brassica rapa. PLoS ONE 2018, 13, e0191015. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.K.; Tulpan, D.; Chomistek, N.; Fundora, D.G.-P.; West, C.; Ellis, B.E.; Frick, M.; Laroche, A.; Foroud, N.A. Analysis of MAPK and MAPKK gene families in wheat and related Triticeae species. BMC Genom. 2018, 19, 178. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hu, W.; Tie, W.; Ding, Z.; Ding, X.; Liu, Y.; Yan, Y.; Wu, C.; Peng, M.; Xu, B. The MAPKKK and MAPKK gene families in banana: Identification, phylogeny and expression during development, ripening and abiotic stress. Sci. Rep. 2017, 7, 1159. [Google Scholar] [CrossRef] [PubMed]
- Cakir, B.; Kılıçkaya, O. Mitogen-activated protein kinase cascades in Vitis vinifera. Front. Plant Sci. 2015, 6, 556. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Mi, X.; Chen, C.; Wang, H.; Guo, W. Identification on mitogen-activated protein kinase signaling cascades by integrating protein interaction with transcriptional profiling analysis in cotton. Sci. Rep. 2018, 8, 8178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dóczi, R.; Ökrész, L.; Romero, A.E.; Paccanaro, A.; Bögre, L. Exploring the evolutionary path of plant MAPK networks. Trends Plant Sci. 2012, 17, 518–525. [Google Scholar] [CrossRef]
- Widmann, C.; Gibson, S.; Jarpe, M.B.; Johnson, G.L. Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol. Rev. 1999, 79, 143–180. [Google Scholar] [CrossRef]
- Iorizzo, M.; Ellison, S.; Senalik, D.; Zeng, P.; Satapoomin, P.; Huang, J.; Bowman, M.; Iovene, M.; Sanseverino, W.; Cavagnaro, P. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 2016, 48, 657. [Google Scholar] [CrossRef]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148. [Google Scholar] [CrossRef]
- Consortium, T.G. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635. [Google Scholar] [CrossRef]
- Albert, V.A.; Barbazuk, W.B.; Der, J.P.; Leebens-Mack, J.; Ma, H.; Palmer, J.D.; Rounsley, S.; Sankoff, D.; Schuster, S.C.; Soltis, D.E. The Amborella genome and the evolution of flowering plants. Science 2013, 342, 1241089. [Google Scholar]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.; Schmutz, J.; Devos, N.; Shu, S.; Carrell, A.; Weston, D. The Sphagnum Genome Project: A new model for ecological and evolutionary genomics. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2016; Volume 78, pp. 167–187. [Google Scholar]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Kane, N.C.; Rieseberg, L.H.J.G. Selective sweeps reveal candidate genes for adaptation to drought and salt tolerance in common sunflower, Helianthus annuus. Genetics 2007, 175, 1823–1834. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2013, 42, W30–W38. [Google Scholar]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Prot. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. Evolution TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- He, S. Genome-wide identification and transcriptional expression analysis of mitogen-activated protein kinase and mitogen-activated protein kinase kinase genes in Capsicum annuum. Front. Plant Sci. 2015, 6, 780. [Google Scholar]
- Liang, W.; Yang, B.; Yu, B.-J.; Zhou, Z.; Li, C.; Jia, M.; Sun, Y.; Zhang, Y.; Wu, F.; Zhang, H. Identification and analysis of MKK and MPK gene families in canola (Brassica napus L.). BMC Genom. 2013, 14, 392. [Google Scholar] [CrossRef]
- Wang, J.; Pan, C.; Wang, Y.; Ye, L.; Wu, J.; Chen, L.; Zou, T.; Lu, G. Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genom. 2015, 16, 386. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, D.; Wang, L.; Li, D. Genome-Wide Analysis of Mitogen-Activated Protein Kinase Gene Family in Maize. Plant Mol. Biol. Rep. 2013, 31, 1446–1460. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Kong, Y.J.G. Btrim: A fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 2011, 98, 152–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grüning, B.; Dale, R.; Sjödin, A.; Chapman, B.A.; Rowe, J.; Tomkins-Tinch, C.H.; Valieris, R.; Köster, J.; Bioconda, T. Bioconda: Sustainable and comprehensive software distribution for the life sciences. Nat. Methods 2018, 15, 475–476. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X. iDEP: An integrated web application for differential expression and pathway analysis. bioRxiv 2017. [Google Scholar] [CrossRef]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef]
- Tajima, F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 1993, 135, 599–607. [Google Scholar]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2011, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L. The TIGR rice genome annotation resource: Improvements and new features. Nucleic Acids Res. 2006, 35, D883–D887. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.-M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463. [Google Scholar] [PubMed]
- Mohanta, T.K.; Park, Y.-H.; Bae, H. Novel genomic and evolutionary insight of WRKY transcription factors in plant lineage. Sci. Rep. 2016, 6, 37309. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, S. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015, 20, 56–64. [Google Scholar] [CrossRef] [PubMed]
- De Smet, I.; Voß, U.; Jürgens, G.; Beeckman, T. Receptor-like kinases shape the plant. Nat. Cell Biol. 2009, 11, 1166. [Google Scholar] [CrossRef]
- Li, J.; Tax, F.E. Receptor-like kinases: Key regulators of plant development and defense. J. Integr. Plant Biol. 2013, 55, 1184–1187. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Wang, S.; Sritubtim, S.; Chen, J.G.; Ellis, B.E. Arabidopsis mitogen-activated protein kinase MPK12 interacts with the MAPK phosphatase IBR5 and regulates auxin signaling. Plant J. 2009, 57, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Wang, H.; Li, B.; Huang, J.; Liu, X.; Zhou, Y.; Mou, Z.; Li, J. Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis. Plant Cell 2006, 18, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Park, S.-C.; Ji, C.Y.; Park, S.; Jeong, J.C.; Lee, H.-S.; Kwak, S.-S. Molecular characterization of biotic and abiotic stress-responsive MAP kinase genes, IbMPK3 and IbMPK6, in sweetpotato. Plant Physiol. Biochem. 2016, 108, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Abass, M.; Morris, P.C. The Hordeum vulgare signalling protein MAP kinase 4 is a regulator of biotic and abiotic stress responses. J. Plant Physiol. 2013, 170, 1353–1359. [Google Scholar] [CrossRef]
- Jalmi, S.K.; Sinha, A.K. ROS mediated MAPK signaling in abiotic and biotic stress-striking similarities and differences. Front. Plant Sci. 2015, 6, 769. [Google Scholar] [CrossRef] [PubMed]
- Jaggi, M.; Gupta, M.; Tuteja, N.; Sinha, A.K. Mitogen-Activated Protein Kinases in Abiotic Stress Tolerance in Crop Plants:“-Omics”Approaches. Improv. Crop Prod. Sustain. Agric. 2013, 107–132. [Google Scholar]
- Taj, G.; Agarwal, P.; Grant, M.; Kumar, A. MAPK machinery in plants: Recognition and response to different stresses through multiple signal transduction pathways. Plant Signal. Behav. 2010, 5, 1370–1378. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.S.; Tuteja, R.; Tuteja, N. Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 2006, 452, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.W.; Roux, M.; Petersen, M.; Mundy, J. MAP kinase cascades in Arabidopsis innate immunity. Front. Plant Sci. 2012, 3, 169. [Google Scholar] [CrossRef]
- Kültz, D. Evolution of osmosensory MAP kinase signaling pathways. Am. Zool. 2001, 41, 743–757. [Google Scholar] [CrossRef]
- Lee, D.; Redfern, O.; Orengo, C. Predicting protein function from sequence and structure. Nat. Rev. Mol. Cell Biol. 2007, 8, 995. [Google Scholar] [CrossRef]
- Kong, F.; Wang, J.; Cheng, L.; Liu, S.; Wu, J.; Peng, Z.; Lu, G. Genome-wide analysis of the mitogen-activated protein kinase gene family in Solanum lycopersicum. Gene 2012, 499, 108–120. [Google Scholar] [CrossRef]
- Liu, Q.; Xue, Q. Computational identification and phylogenetic analysis of the MAPK gene family in Oryza sativa. Plant Physiol. Biochem. 2007, 45, 6–14. [Google Scholar] [CrossRef]
- Barker, M.S.; Li, Z.; Kidder, T.I.; Reardon, C.R.; Lai, Z.; Oliveira, L.O.; Scascitelli, M.; Rieseberg, L.H. Most Compositae (Asteraceae) are descendants of a paleohexaploid and all share a paleotetraploid ancestor with the Calyceraceae. Am. J. Bot. 2016, 103, 1203–1211. [Google Scholar] [CrossRef] [Green Version]
- Gill, N.; Findley, S.; Walling, J.G.; Hans, C.; Ma, J.; Doyle, J.; Stacey, G.; Jackson, S.A. Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol. 2009, 151, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Soltis, D.E. The role of genetic and genomic attributes in the success of polyploids. Proc. Natl. Acad. Sci. USA 2000, 97, 7051–7057. [Google Scholar] [CrossRef] [Green Version]
- Gottlieb, L. Conservation and duplication of isozymes in plants. Science 1982, 216, 373–380. [Google Scholar] [CrossRef]
- Seiler, G. Wild annual Helianthus anomalus and H. deserticola for improving oil content and quality in sunflower. Ind. Crops Prod. 2007, 25, 95–100. [Google Scholar] [CrossRef]
- Colcombet, J.; Hirt, H. Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 2008, 413, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Nicole, M.-C.; Hamel, L.-P.; Morency, M.-J.; Beaudoin, N.; Ellis, B.E.; Séguin, A. MAP-ping genomic organization and organ-specific expression profiles of poplar MAP kinases and MAP kinase kinases. BMC Genom. 2006, 7, 223. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.P.; Richa, T.; Kumar, K.; Raghuram, B.; Sinha, A.K. In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Res. 2010, 17, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Chai, Z.; Xie, Y.; Gao, K.; Cui, M.; Jiang, Y.; Feng, J.J.P.o. Bioinformatics identification and transcript profile analysis of the mitogen-activated protein kinase gene family in the diploid woodland strawberry Fragaria vesca. PLoS ONE 2017, 12, e0178596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, Y.; Klessig, D.F. Multiple levels of tobacco WIPK activation during the induction of cell death by fungal elicitins. Plant J. 2000, 23, 339–347. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Ding, H.; Zhang, A.; Ma, F.; Cao, J.; Jiang, M. A novel mitogen-activated protein kinase gene in maize (Zea mays), ZmMPK3, is involved in response to diverse environmental cues. J. Integr. Plant Biol. 2010, 52, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, T.; Ichimura, K.; Mizoguchi, T.; Shinozaki, K. Oxidative stress activates ATMPK6, an Arabidopsis homologue of MAP kinase. Plant Cell Physiol. 2001, 42, 1012–1016. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, K.; Mizoguchi, T.; Yoshida, R.; Yuasa, T.; Shinozaki, K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 2000, 24, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Teige, M.; Scheikl, E.; Eulgem, T.; Dóczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Rohila, J.S.; Yang, Y. Rice mitogen-activated protein kinase gene family and its role in biotic and abiotic stress response. J. Integr. Plant Biol. 2007, 49, 751–759. [Google Scholar] [CrossRef]
- Danquah, A.; Zélicourt, A.; Boudsocq, M.; Neubauer, J.; Frei dit Frey, N.; Leonhardt, N.; Pateyron, S.; Gwinner, F.; Tamby, J.P.; Ortiz-Masia, D. Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 2015, 82, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Cheong, Y.H.; Moon, B.C.; Kim, J.K.; Kim, C.Y.; Kim, M.C.; Kim, I.H.; Park, C.Y.; Kim, J.C.; Park, B.O.; Koo, S.C. BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol. 2003, 132, 1961–1972. [Google Scholar] [CrossRef] [PubMed]
- Schoenbeck, M.A.; Samac, D.A.; Fedorova, M.; Gregerson, R.G.; Gantt, J.S.; Vance, C.P. The alfalfa (Medicago sativa) TDY1 gene encodes a mitogen-activated protein kinase homolog. Mol. Plant-Microbe Interact. 1999, 12, 882–893. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, T.; Irie, K.; Hirayama, T.; Hayashida, N.; Yamaguchi-Shinozaki, K.; Matsumoto, K.; Shinozaki, K. A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1996, 93, 765–769. [Google Scholar] [CrossRef] [PubMed]
- Im, J.H.; Lee, H.; Kim, J.; Kim, H.B.; Seyoung, K.; Kim, B.M.; An, C.S. A salt stress-activated mitogen-activated protein kinase in soybean is regulated by phosphatidic acid in early stages of the stress response. J. Plant Biol. 2012, 55, 303–309. [Google Scholar] [CrossRef]
- Zong, X.-j.; Li, D.-p.; Gu, L.-k.; Li, D.-q.; Liu, L.-x.; Hu, X.-l. Abscisic acid and hydrogen peroxide induce a novel maize group C MAP kinase gene, ZmMPK7, which is responsible for the removal of reactive oxygen species. Planta 2009, 229, 485. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, X.; Yu, Y.; Chen, C.; Wang, J.; Cai, C.; Guo, W.J.S.r. Integration analysis of MKK and MAPK family members highlights potential MAPK signaling modules in cotton. Sci. Rep. 2016, 6, 29781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lespinet, O.; Wolf, Y.I.; Koonin, E.V.; Aravind, L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002, 12, 1048–1059. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, D.; Nanmori, T.; Sato, K.i.; Fukami, Y.; Kikkawa, U.; Yasuda, T. Activation of AtMEK1, an Arabidopsis mitogen-activated protein kinase kinase, in vitro and in vivo: Analysis of active mutants expressed in E. coli and generation of the active form in stress response in seedlings. Plant J. 2002, 29, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Yang, H.; Zhang, S. Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 2002, 277, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Hwa, C.-M.; Yang, X.-C. The AtMKK3 pathway mediates ABA and salt signaling in Arabidopsis. Acta Physiol. Planta 2008, 30, 277–286. [Google Scholar] [CrossRef]
- Liu, Y. Roles of mitogen-activated protein kinase cascades in ABA signaling. Plant Cell Rep. 2012, 31, 1–12. [Google Scholar] [CrossRef]
- Zhong, B.; Xi, Z.; Goremykin, V.V.; Fong, R.; Mclenachan, P.A.; Novis, P.M.; Davis, C.C.; Penny, D. Streptophyte algae and the origin of land plants revisited using heterogeneous models with three new algal chloroplast genomes. Mol. Biol. Evol. 2013, 31, 177–183. [Google Scholar] [CrossRef]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; Chumley, T.W.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [Green Version]
- Stevens, P.F.; Davis, H. Angiosperm Phylogeny Website; Missouri Botanical Garden: St. Louis, MO, USA, 2001. [Google Scholar]
- Mockaitis, K.; Howell, S.H.J.T.P.J. Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 2000, 24, 785–796. [Google Scholar] [CrossRef]
- Seo, S.; Katou, S.; Seto, H.; Gomi, K.; Ohashi, Y. The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic and salicylic acids in wounded tobacco plants. Plant J. 2007, 49, 899–909. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Cai, Z.; Guo, Y.; Gan, S. An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 2009, 150, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Kong, X.P.; Zong, X.J.; Li, D.P.; Li, D.Q. Expression analysis of five maize MAP kinase genes in response to various abiotic stresses and signal molecules. Mol. Biol. Rep. 2011, 38, 3967–3975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zou, D.; Li, Y.; Sun, X.; Wang, N.-N.; Gong, S.-Y.; Zheng, Y.; Li, X.B. GhMPK17, a cotton mitogen-activated protein kinase, is involved in plant response to high salinity and osmotic stresses and ABA signaling. PLoS ONE 2014, 9, e95642. [Google Scholar]
- Chakraborty, C.; Sharma, A.R.; Patra, B.C.; Bhattacharya, M.; Sharma, G.; Lee, S.-S. MicroRNAs mediated regulation of MAPK signaling pathways in chronic myeloid leukemia. Oncotarget 2016, 7, 42683. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, J.; Gao, J.; Li, Y. MicroRNA-4728 mediated regulation of MAPK oncogenic signaling in papillary thyroid carcinoma. Saudi J. Biol. Sci. 2018, 25, 986–990. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Li, S.; Yu, B. miRNA limits MAP kinase-mediated immunity: Optimization of plant fitness. J. Exp. Bot. 2017, 68, 5685. [Google Scholar] [CrossRef]
- Raghuram, B.; Sheikh, A.H.; Sinha, A.K. Regulation of MAP kinase signaling cascade by microRNAs in Oryza sativa. Plant Signal. Behav. 2014, 9, e972130. [Google Scholar] [CrossRef]





| Plant Species | Ploidy | Size of Genome (Mbs) γ | No. of loci γ | MPK | MKK |
|---|---|---|---|---|---|
| Amborella trichopoda ‡ | Diploid | 706 | 26846 | 8 | 7 |
| Aquilegia coerulea ‡ | Diploid | 302 | 24823 | 11 | 5 |
| Arabidopsis thaliana | Diploid | 135 | 27416 | 20 a | 10 a |
| Chlamydomonas reinhardtii ‡ | Haploid | 111.1 | 17741 | 6 | 1 |
| Daucus carota ‡ | Diploid | 421 | 32,113 | 17 | 5 |
| Glycine max | Tetraploid | 975 | 56044 | 38 b | 11 b |
| Helianthus annuus ‡ | Diploid | 3600 | 52243 | 28 | 8 |
| Oryza sativa | Diploid | 372 | 39049 | 16 c | 8 c |
| Solanum lycopersicum ‡ | Diploid | 900 | 34727 | 15 | 5 |
| Sphagnum fallax ‡ | Haploid/Diploid | 395 | 26939 | 11 | 6 |
| Vitis vinifera | Diploid | 487 | 26346 | 14 d | 5 d |
| Name | Gene ID | Chr | Str | Start | End | PL | Exo | Int | Sl | pI | Mw |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MPK | |||||||||||
| HaMPK6-1 | HanXRQChr01g0023391 | Ha1 | - | 130301686 | 130292965 | 359 | 6 | 5 | - | 5.85 | 41581.61 |
| HaMPK16-1 | HanXRQChr03g0071491 | Ha3 | - | 77378137 | 77372246 | 564 | 10 | 9 | - | 9.17 | 64059.43 |
| HaMPK7 | HanXRQChr03g0074811 | Ha3 | + | 102410161 | 102406169 | 353 | 3 | 2 | - | 7.62 | 40274.83 |
| HaMPK23-1 | HanXRQChr03g0081221 | Ha3 | + | 129978443 | 129973452 | 453 | 15 | 14 | - | 9.65 | 50392.28 |
| HaMPK23-3 | HanXRQChr03g0081391 | Ha3 | + | 130506162 | 130500013 | 423 | 16 | 15 | - | 8.91 | 47648.79 |
| HaMPK22 | HanXRQChr04g0108301 | Ha4 | - | 77321727 | 77315970 | 432 | 18 | 17 | - | 5.46 | 49633.87 |
| HaMPK11-1 | HanXRQChr04g0121371 | Ha4 | + | 158781451 | 158778221 | 358 | 6 | 5 | M | 6.42 | 41228.21 |
| HaMPK3-1 | HanXRQChr05g0133161 | Ha5 | + | 21064225 | 21061089 | 358 | 6 | 5 | - | 5.68 | 41323.35 |
| HaMPK8 | HanXRQChr05g0143371 | Ha5 | - | 116774638 | 116767923 | 505 | 11 | 10 | - | 6.8 | 57051.89 |
| HaMPK2 | HanXRQChr05g0151241 | Ha5 | - | 169574750 | 169571609 | 349 | 3 | 2 | - | 6.54 | 40295.67 |
| HaMPK11-2 | HanXRQChr06g0167011 | Ha6 | - | 7104659 | 7099870 | 359 | 6 | 5 | M | 6.25 | 41336.17 |
| HaMPK4 | HanXRQChr06g0170261 | Ha6 | + | 16894292 | 16893100 | 157 | 2 | 1 | M | 8.36 | 17702.54 |
| HaMPK13-1 | HanXRQChr06g0175501 | Ha6 | - | 34635251 | 34631528 | 363 | 7 | 6 | - | 5.22 | 41353.31 |
| HaMPK9-1 | HanXRQChr06g0183531 | Ha6 | + | 90706107 | 90699312 | 478 | 11 | 10 | - | 6.53 | 54442.91 |
| HaMPK23-4 | HanXRQChr08g0226701 | Ha8 | + | 84318787 | 84308381 | 442 | 18 | 17 | - | 9.52 | 49480.06 |
| HaMPK15 | HanXRQChr08g0227231 | Ha8 | + | 87599490 | 87591577 | 501 | 11 | 10 | - | 8.53 | 57073.07 |
| HaMPK3-2 | HanXRQChr08g0229941 | Ha8 | - | 101013127 | 101009864 | 358 | 6 | 5 | - | 5.58 | 41298.31 |
| HaMPK13-2 | HanXRQChr08g0230171 | Ha8 | - | 102808229 | 102804252 | 362 | 6 | 5 | - | 5.85 | 41552.83 |
| HaMPK14 | HanXRQChr09g0243011 | Ha9 | + | 34673154 | 34669292 | 362 | 3 | 2 | - | 5.57 | 41423.42 |
| HaMPK16-2 | HanXRQChr09g0248301 | Ha9 | + | 76212398 | 76202758 | 559 | 10 | 9 | - | 9.07 | 63370.4 |
| HaMPK1 | HanXRQChr09g0269211 | Ha9 | - | 185086347 | 185083825 | 361 | 3 | 2 | - | 6.64 | 41831.44 |
| HaMPK19-2 | HanXRQChr11g0330461 | Ha11 | + | 43791321 | 43784989 | 574 | 9 | 8 | - | 9.33 | 65344.85 |
| HaMPK6-2 | HanXRQChr11g0343001 | Ha11 | - | 125967866 | 125963374 | 359 | 6 | 5 | - | 5.8 | 41553.72 |
| HaMPK19-1 | HanXRQChr13g0389781 | Ha13 | - | 19048315 | 19044532 | 588 | 10 | 9 | - | 9.06 | 66613.36 |
| HaMPK23-2 | HanXRQChr13g0411961 | Ha13 | - | 142634442 | 142625511 | 459 | 18 | 17 | - | 9.63 | 50984.95 |
| HaMPK9-2 | HanXRQChr14g0432771 | Ha14 | - | 49683290 | 49679650 | 484 | 10 | 9 | - | 6.57 | 55530.13 |
| HaMPK17 | HanXRQChr15g0484561 | Ha15 | - | 84424855 | 84420653 | 429 | 11 | 10 | - | 6.24 | 49909.6 |
| HaMPK18 | HanXRQChr15g0495321 | Ha15 | - | 160155012 | 160149273 | 563 | 9 | 8 | - | 9.47 | 64374.62 |
| MKK | |||||||||||
| HaMKK9 | HanXRQChr03g0087071 | Ha3 | - | 148424902 | 148425825 | 308 | 1 | 0 | M | 6.75 | 34332.34 |
| HaMKK4 | HanXRQChr04g0094171 | Ha4 | + | 471743 | 472816 | 351 | 1 | 0 | C | 9.04 | 38917.18 |
| HaMKK6-1 | HanXRQChr09g0238861 | Ha9 | + | 9311933 | 9322916 | 357 | 8 | 7 | - | 6.76 | 39934.36 |
| HaMKK5 | HanXRQChr10g0311571 | Ha10 | + | 219604899 | 219606004 | 355 | 1 | 0 | C | 9.25 | 39840.46 |
| HaMKK6-2 | HanXRQChr10g0318871 | Ha10 | + | 244056044 | 244064185 | 355 | 8 | 7 | - | 7.13 | 39751.09 |
| HaMKK2 | HanXRQChr10g0319531 | Ha10 | - | 245318274 | 245324118 | 371 | 9 | 8 | - | 5.43 | 40967.01 |
| HaMKK1 | HanXRQChr12g0354521 | Ha12 | - | 1236278 | 1243005 | 358 | 10 | 9 | - | 5.77 | 39199.81 |
| HaMKK3 | HanXRQChr14g0450561 | Ha14 | - | 141579116 | 141587170 | 520 | 12 | 11 | M | 5.79 | 68568.6 |
| Clades | Consensus Common Docking Sites |
|---|---|
| Clade A | K-M-L-V-F-D-P-N-K-R-I-V-E-E-A-L |
| Clade B | K-M-L-V-F-D-P-S-K-R-I-S-V-T-E-A-L |
| Clade C | S-L-C-S-W-D-P-C-K-R-P-T-A-E-E-A-L |
| Clade D | R-L-L-A-F-D-P-K-D-R-P-T-A-E-E-A-L |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neupane, S.; Schweitzer, S.E.; Neupane, A.; Andersen, E.J.; Fennell, A.; Zhou, R.; Nepal, M.P. Identification and Characterization of Mitogen-Activated Protein Kinase (MAPK) Genes in Sunflower (Helianthus annuus L.). Plants 2019, 8, 28. https://doi.org/10.3390/plants8020028
Neupane S, Schweitzer SE, Neupane A, Andersen EJ, Fennell A, Zhou R, Nepal MP. Identification and Characterization of Mitogen-Activated Protein Kinase (MAPK) Genes in Sunflower (Helianthus annuus L.). Plants. 2019; 8(2):28. https://doi.org/10.3390/plants8020028
Chicago/Turabian StyleNeupane, Surendra, Sarah E. Schweitzer, Achal Neupane, Ethan J. Andersen, Anne Fennell, Ruanbao Zhou, and Madhav P. Nepal. 2019. "Identification and Characterization of Mitogen-Activated Protein Kinase (MAPK) Genes in Sunflower (Helianthus annuus L.)" Plants 8, no. 2: 28. https://doi.org/10.3390/plants8020028






