The Emerging Role of the Gut Microbiome in Cardiovascular Disease: Current Knowledge and Perspectives
Abstract
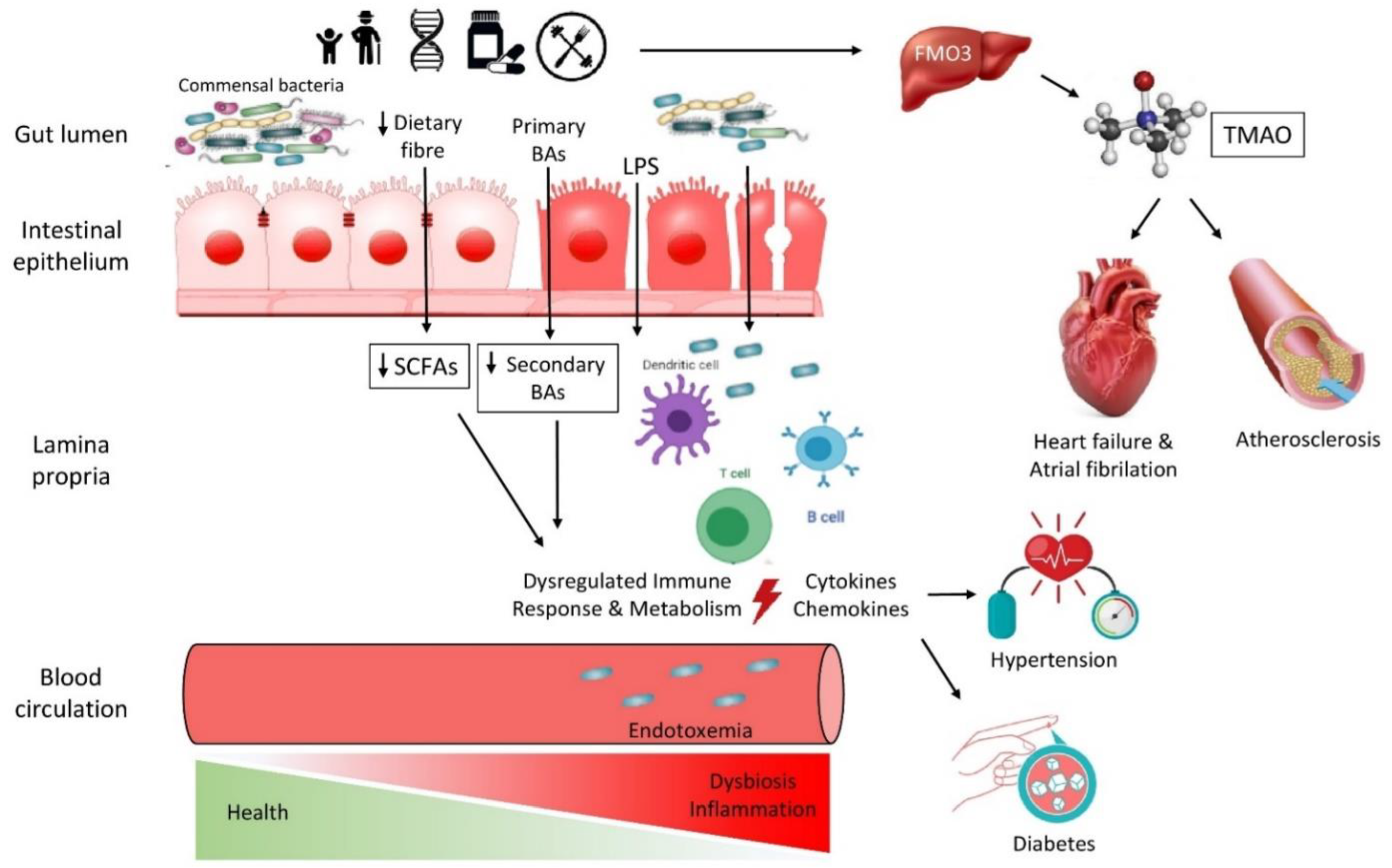
:1. Introduction
1.1. Indirect Association of Alterations in the Intestinal Microbiota and Significant Risk Factors for CVD
1.1.1. Diabetes Mellitus
1.1.2. Hypertension
1.1.3. Hypercholesterolemia
1.1.4. Obesity/Metabolic Syndrome and Lifestyle
1.1.5. Immune System Implication
1.1.6. Gut Metabolites
1.2. Direct Effects of Dysbiosis on Cardiovascular Health
1.2.1. Atheromatosis and CAD
1.2.2. Chronic Heart Failure
1.2.3. Atrial Fibrillation
2. Current Therapeutic Interventions and Future Perspectives in the Context of Personalized Medicine
2.1. Different Diets and Their Role in the Development of CVD
2.2. Pharmacological Therapy
2.3. FMT and Pre-/Probiotics
2.4. New Knowledge and Challenges
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Cardiovascular disease | CVD |
| Trimethylamine-N-oxide | TMAO |
| Blood pressure | BP |
| Bile salt hydrolase | BSH |
| G protein-coupled receptor | GPR43 |
| Low gene count | LGC |
| C-reactive protein | CRP |
| High-sensitivity C-reactive protein | CRP |
| Ankle brachial index | ABI |
| Coronary artery calcium | CAC |
| Tumor necrosis factor alpha | TNF-a |
| Lipopolysaccharides | LPS |
| Flavin-containing monooxygenase | FMO |
| Tissue factor | TF |
| P-selectin | CD62P |
| Short-chain fatty acids | SCFAs |
| Interleukin | IL |
| Hydroxy-methyl-glutaryl-coenzyme A | HMG-CoA |
References
- Methé, B.A.; Nelson, K.E.; Pop, M.; Creasy, H.H.; Giglio, M.G.; Huttenhower, C.; Gevers, D.; Petrosino, J.F.; Abubucker, S.; Mannon, P.J.; et al. A framework for human microbiome research. Nature 2012, 486, 215–221. [Google Scholar] [CrossRef] [Green Version]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canyelles, M.; Tondo, M.; Cedó, L.; Farràs, M.; Escolà-Gil, J.C.; Blanco-Vaca, F. Trimethylamine N-Oxide: A Link among Diet, Gut Microbiota, Gene Regulation of Liver and Intestine Cholesterol Homeostasis and HDL Function. Int. J. Mol. Sci. 2018, 19, 3228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [Green Version]
- Short Communications. J. Perin. Med. 1990, 18, 139–143. [CrossRef]
- Gabriel, I.; Olejek, A.; Stencel-Gabriel, K.; Wielgoś, M. The influence of maternal vaginal flora on the intestinal colonization in newborns and 3-month-old infants. J. Matern. Neonatal Med. 2017, 31, 1448–1453. [Google Scholar] [CrossRef]
- Mueller, N.T.; Whyatt, R.; Hoepner, L.; Oberfield, S.; Dominguez-Bello, M.G.; Widen, E.M.; Hassoun, A.; Perera, F.; Rundle, A. Prenatal exposure to antibiotics, cesarean section and risk of childhood obesity. Int. J. Obes. 2014, 39, 665–670. [Google Scholar] [CrossRef] [Green Version]
- Tamburini, S.; Shen, N.; Wu, H.C.; Clemente, J.C. The microbiome in early life: Implications for health outcomes. Nat. Med. 2016, 22, 713–722. [Google Scholar] [CrossRef] [Green Version]
- Leoni, C.; Ceci, O.; Manzari, C.; Fosso, B.; Volpicella, M.; Ferrari, A.; Fiorella, P.; Pesole, G.; Cicinelli, E.; Ceci, L.R.; et al. Human Endometrial Microbiota at Term of Normal Pregnancies. Genes 2019, 10, 971. [Google Scholar] [CrossRef] [Green Version]
- Bezirtzoglou, E.; Stavropoulou, E. Immunology and probiotic impact of the newborn and young children intestinal microflora. Anaerobe 2011, 17, 369–374. [Google Scholar] [CrossRef]
- Tsigalou, C.; Konstantinidis, T.; Stavropoulou, E.; Bezirtzoglou, E.E.; Tsakris, A. Potential Elimination of Human Gut Resistome by Exploiting the Benefits of Functional Foods. Front. Microbiol. 2020, 11, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- E Gustafsson, B. The physiological importance of the colonic microflora. Scand. J. Gastroenterol. Suppl. 1982, 77, 117–131. [Google Scholar]
- Adlerberth, I.; E Wold, A. Establishment of the gut microbiota in Western infants. Acta Paediatr. Int. J. Paediatr. 2009, 98, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Plovier, H.; Van Hul, M.; Geurts, L.; Delzenne, N.M.; Druart, C.; Everard, A. Endocannabinoids—At the crossroads between the gut microbiota and host metabolism. Nat. Rev. Endocrinol. 2016, 12, 133–143. [Google Scholar] [CrossRef]
- Cebra, J.J. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 1999, 69, 1046s–1051s. [Google Scholar] [CrossRef]
- Jama, H.A.; Kaye, D.M.; Marques, F.Z. The gut microbiota and blood pressure in experimental models. Curr. Opin. Nephrol Hypertens. 2019, 28, 97–104. [Google Scholar] [CrossRef]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef] [Green Version]
- Battson, M.L.; Lee, D.M.; Weir, T.L.; Gentile, C.L. The gut microbiota as a novel regulator of cardiovascular function and disease. J. Nutr. Biochem. 2018, 56, 1–15. [Google Scholar] [CrossRef]
- Mozaffarian, D. Dietary and Policy Priorities for Cardiovascular Disease, Diabetes, and Obesity: A Comprehensive Review. Circulation 2016, 133, 187–225. [Google Scholar] [CrossRef]
- NCD Risk Factor Collaboration (NCD-RisC). Worldwide trends in diabetes since 1980: A pooled analysis of 751 population-based studies with 4·4 million participants. Lancet 2016, 387, 1513–1530. [Google Scholar] [CrossRef] [Green Version]
- Velmurugan, G.; Ramprasath, T.; Gilles, M.; Swaminathan, K.; Ramasamy, S. Gut Microbiota, Endocrine-Disrupting Chemicals, and the Diabetes Epidemic. Trends Endocrinol. Metab. 2017, 28, 612–625. [Google Scholar] [CrossRef] [PubMed]
- Pflughoeft, K.J.; Versalovic, J. Human Microbiome in Health and Disease. Annu. Rev. Pathol. Mech. Dis. 2012, 7, 99–122. [Google Scholar] [CrossRef] [PubMed]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanna, S.; Van Zuydam, N.R.; Mahajan, A.; Kurilshikov, A.; Vila, A.V.; Võsa, U.; Mujagic, Z.; Masclee, A.A.M.; Jonkers, D.M.A.E.; Oosting, M.; et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 2019, 51, 600–605. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Li, Q.; Chang, Y.; Zhang, K.; Chen, H.; Tao, S.; Zhang, Z. Implication of the gut microbiome composition of type 2 diabetic patients from northern China. Sci. Rep. 2020, 10, 5450. [Google Scholar] [CrossRef]
- Zhou, B.; Bentham, J.; Di Cesare, M.; Bixby, H.; Danaei, G.; Cowan, M.J.; Paciorek, C.J.; Singh, G.; Hajifathalian, K.; Bennett, J.E.; et al. Worldwide trends in blood pressure from 1975 to 2015: A pooled analysis of 1479 population-based measurement studies with 19·1 million participants. Lancet 2017, 389, 37–55. [Google Scholar] [CrossRef] [Green Version]
- Okamura, M.; Ueno, T.; Tanaka, S.; Murata, Y.; Kobayashi, H.; Miyamoto, A.; Abe, M.; Fukuda, N. Increased expression of acyl-CoA oxidase 2 in the kidney with plasma phytanic acid and altered gut microbiota in spontaneously hypertensive rats. Hypertens. Res. 2021, 651–661. [Google Scholar] [CrossRef]
- Yang, T.; Santisteban, M.M.; Rodriguez, V.; Li, E.; Ahmari, N.; Carvajal, J.M.; Zadeh, M.; Gong, M.; Qi, Y.; Zubcevic, J.; et al. Gut Dysbiosis Is Linked to Hypertension. Hypertension 2015, 65, 1331–1340. [Google Scholar] [CrossRef] [Green Version]
- Adnan, S.; Nelson, J.W.; Ajami, N.J.; Venna, V.R.; Petrosino, J.F.; Bryan, R.M.; Durgan, D.J. Alterations in the gut microbiota can elicit hypertension in rats. Physiol. Genom. 2017, 49, 96–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pluznick, J. A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes 2013, 5, 202–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Natarajan, N.; Hori, D.; Flavahan, S.; Steppan, J.; Flavahan, N.A.; Berkowitz, D.E.; Pluznick, J.L. Microbial short chain fatty acid metabolites lower blood pressure via endothelial G protein-coupled receptor 41. Physiol. Genom. 2016, 48, 826–834. [Google Scholar] [CrossRef]
- Silveira-Nunes, G.; Durso, D.F.; De Oliviera, L.R.A., Jr.; Cunha, E.H.M.; Maioli, T.U.; Vieira, A.T.; Speziali, E.; Corrêa-Oliveira, R.; Martins-Filho, O.A.; Teixeira-Carvalho, A.; et al. Hypertension Is Associated with Intestinal Microbiota Dysbiosis and Inflammation in a Brazilian Population. Front. Pharmacol. 2020, 11, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, C.R.; Kahn, S.; Kashyap, P.; Laine, L.; Rubin, D.; Atreja, A.; Moore, T.; Wu, G. Update on Fecal Microbiota Transplantation 2015: Indications, Methodologies, Mechanisms, and Outlook. Gastroenterology 2015, 149, 223–237. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017, 5, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Mailer, R.K.; Gisterå, A.; Polyzos, K.A.; Ketelhuth, D.F.; Hansson, G.K. Hypercholesterolemia Induces Differentiation of Regulatory T Cells in the Liver. Circ. Res. 2017, 120, 1740–1753. [Google Scholar] [CrossRef]
- Goldstein, J.L.; Brown, M.S. Regulation of the mevalonate pathway. Nat. 1990, 343, 425–430. [Google Scholar] [CrossRef]
- Yamamoto, H.; Yamanashi, Y.; Takada, T.; Mu, S.; Tanaka, Y.; Komine, T.; Suzuki, H. Hepatic Expression of Niemann-Pick C1-Like 1, a Cholesterol Reabsorber from Bile, Exacerbates Western Diet–Induced Atherosclerosis in LDL Receptor Mutant Mice. Mol. Pharmacol. 2019, 96, 47–55. [Google Scholar] [CrossRef] [Green Version]
- Ferrell, J.; Boehme, S.; Li, F.; Chiang, J.Y.L. Cholesterol 7α-hydroxylase-deficient mice are protected from high-fat/high-cholesterol diet-induced metabolic disorders. J. Lipid Res. 2016, 57, 1144–1154. [Google Scholar] [CrossRef] [Green Version]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.M.; Marchesi, J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.S.; Mason, M.R.; Brooker, M.R.; O’Brien, K. Pyrosequencing reveals unique microbial signatures associated with healthy and failing dental implants. J. Clin. Periodontol. 2012, 39, 425–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefebvre, P.; Cariou, B.; Lien, F.; Kuipers, F.; Staels, B. Role of Bile Acids and Bile Acid Receptors in Metabolic Regulation. Physiol. Rev. 2009, 89, 147–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoeler, M.; Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis, S.A.; Conceição, L.; Rosa, D.D.; Siqueira, N.P.; Peluzio, M.C.G. Mechanisms responsible for the hypocholesterolaemic effect of regular consumption of probiotics. Nutr. Res. Rev. 2016, 30, 36–49. [Google Scholar] [CrossRef]
- Kriaa, A.; Bourgin, M.; Potiron, A.; Mkaouar, H.; Jablaoui, A.; Gérard, P.; Maguin, E.; Rhimi, M. Microbial impact on cholesterol and bile acid metabolism: Current status and future prospects. J. Lipid Res. 2019, 60, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Kimura, I.; Ozawa, K.; Inoue, D.; Imamura, T.; Kimura, K.; Maeda, T.; Terasawa, K.; Kashihara, D.; Hirano, K.; Tani, T.; et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun. 2013, 4, 1829. [Google Scholar] [CrossRef] [Green Version]
- GBD 2015 Obesity Collaborators; Afshin, A.; Forouzanfar, M.H.; Reitsma, M.B.; Sur, P.; Estep, K.; Lee, A.; Marczak, L.; Mokdad, A.H.; Moradi-Lakeh, M.; et al. Health Effects of Overweight and Obesity in 195 Countries over 25 Years. N. Engl. J. Med. 2017, 377, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Saklayen, M.G. The Global Epidemic of the Metabolic Syndrome. Curr. Hypertens. Rep. 2018, 20, 12. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.K.; Chang, H.-W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [Green Version]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic Syndrome and Altered Gut Microbiota in Mice Lacking Toll-Like Receptor 5. Science 2010, 328, 228–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nirmalkar, K.; Murugesan, S.; Pizano-Zárate, M.L.; Villalobos-Flores, L.E.; García-González, C.; Morales-Hernández, R.M.; Nuñez-Hernández, J.A.; Hernández-Quiroz, F.; Romero-Figueroa, M.D.S.; Hernández-Guerrero, C.; et al. Gut Microbiota and Endothelial Dysfunction Markers in Obese Mexican Children and Adolescents. Nutrients 2018, 10, 2009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut Microbiota from Twins Discordant for Obesity Modulate Metabolism in Mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef] [Green Version]
- Fei, N.; Zhao, L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2012, 7, 880–884. [Google Scholar] [CrossRef]
- Zhong, H.; Ren, H.; Lu, Y.; Fang, C.; Hou, G.; Yang, Z.; Chen, B.; Yang, F.; Zhao, Y.; Shi, Z.; et al. Distinct gut metagenomics and metaproteomics signatures in prediabetics and treatment-naïve type 2 diabetics. EBioMedicine 2019, 47, 373–383. [Google Scholar] [CrossRef]
- Makki, K.; Deehan, E.C.; Walter, J.; Bäckhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef] [Green Version]
- Bragina, T.; Elizarova, E.; Sheveleva, S. Intestinal microbiote of athletes. Vopr. Pitan. 2021, 90, 36–52. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Van den Munckhof, I.C.L.; Kurilshikov, A.; Ter Horst, R.; Riksen, N.P.; Joosten, L.A.B.; Zhernakova, A.; Fu, J.; Keating, S.T.; Netea, M.G.; De Graaf, J.; et al. Role of gut microbiota in chronic low-grade inflammation as potential driver for atherosclerotic cardiovascular disease: A systematic review of human studies. Obes. Rev. 2018, 19, 1719–1734. [Google Scholar] [CrossRef] [Green Version]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korporaal, S.J.; Van Eck, M.; Adelmeijer, J.; Ijsseldijk, M.; Out, R.; Lisman, T.; Lenting, P.J.; Van Berkel, T.J.; Akkerman, J.-W.N. Platelet Activation by Oxidized Low Density Lipoprotein Is Mediated by Cd36 and Scavenger Receptor-A. Arter. Thromb. Vasc. Biol. 2007, 27, 2476–2483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil-Cruz, C.; Perez-Shibayama, C.; De Martin, A.; Ronchi, F.; van der Borght, K.; Niederer, R.; Onder, L.; Lütge, M.; Novkovic, M.; Nindl, V.; et al. Microbiota-derived peptide mimics drive lethal inflammatory cardiomyopathy. Science 2019, 366, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ota, N.; Manzanillo, P.; Kates, L.; Zavala-Solorio, J.; Eidenschenk, C.; Zhang, J.; Lesch, J.; Lee, W.P.; Ross, J.; et al. Interleukin-22 alleviates metabolic disorders and restores mucosal immunity in diabetes. Nature 2014, 514, 237–241. [Google Scholar] [CrossRef]
- Sun, X.; Jiao, X.; Ma, Y.; Liu, Y.; Zhang, L.; He, Y.; Chen, Y. Trimethylamine N-oxide induces inflammation and endothelial dysfunction in human umbilical vein endothelial cells via activating ROS-TXNIP-NLRP3 inflammasome. Biochem. Biophys. Res. Commun. 2016, 481, 63–70. [Google Scholar] [CrossRef]
- Garber, K. Drugging the gut microbiome. Nat. Biotechnol. 2015, 33, 228–231. [Google Scholar] [CrossRef]
- Normén, L.; Lærke, H.N.; Jensen, B.B.; Langkilde, A.M.; Andersson, H. Small-bowel absorption of d-tagatose and related effects on carbohydrate digestibility: An ileostomy study. Am. J. Clin. Nutr. 2001, 73, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Sandek, A.; Bauditz, J.; Swidsinski, A.; Buhner, S.; Weber-Eibel, J.; von Haehling, S.; Schroedl, W.; Karhausen, T.; Doehner, W.; Rauchhaus, M.; et al. Altered Intestinal Function in Patients with Chronic Heart Failure. J. Am. Coll. Cardiol. 2007, 50, 1561–1569. [Google Scholar] [CrossRef] [Green Version]
- Sandek, A.; Bjarnason, I.; Volk, H.-D.; Crane, R.; Meddings, J.B.; Niebauer, J.; Kalra, P.R.; Buhner, S.; Herrmann, R.; Springer, J.; et al. Studies on bacterial endotoxin and intestinal absorption function in patients with chronic heart failure. Int. J. Cardiol. 2010, 157, 80–85. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.H.W.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal Microbial Metabolism of Phosphatidylcholine and Cardiovascular Risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; DuGar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.-M.; et al. Gut Flora Metabolism of Phosphatidylcholine Promotes Cardiovascular Disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, B.J.; de Aguiar Vallim, T.Q.; Wang, Z.; Shih, D.M.; Meng, Y.; Gregory, J.; Allayee, H.; Lee, R.; Graham, M.; Crooke, R.; et al. Trimethylamine-N-Oxide, a Metabolite Associated with Atherosclerosis, Exhibits Complex Genetic and Dietary Regulation. Cell Metab. 2013, 17, 49–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.; Chen, J.; Chen, J.; Tao, J.; Wu, S.; Xu, G.; Wang, Z.; Wei, D.; Yin, W. Trimethylamine N-oxide promotes apoE −/− mice atherosclerosis by inducing vascular endothelial cell pyroptosis via the SDHB/ROS pathway. J. Cell. Physiol. 2020, 235, 6582–6591. [Google Scholar] [CrossRef]
- Aldana-Hernández, P.; Leonard, K.-A.; Zhao, Y.-Y.; Curtis, J.M.; Field, C.J.; Jacobs, R.L. Dietary Choline or Trimethylamine N-oxide Supplementation Does Not Influence Atherosclerosis Development in Ldlr−/− and Apoe−/− Male Mice. J. Nutr. 2019, 150, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.; Srivatsav, V.; Rizwan, A.; Nashed, A.; Liu, R.; Shen, R.; Akhtar, M. Bridging the Gap between Gut Microbial Dysbiosis and Cardiovascular Diseases. Nutrients 2017, 9, 859. [Google Scholar] [CrossRef] [Green Version]
- Skye, S.M.; Zhu, W.; Romano, K.A.; Guo, C.-J.; Wang, Z.; Jia, X.; Kirsop, J.; Haag, B.; Lang, J.M.; DiDonato, J.A.; et al. Microbial Transplantation with Human Gut Commensals Containing CutC Is Sufficient to Transmit Enhanced Platelet Reactivity and Thrombosis Potential. Circ. Res. 2018, 123, 1164–1176. [Google Scholar] [CrossRef]
- Zhu, W.; Gregory, J.C.; Org, E.; Buffa, J.A.; Gupta, N.; Wang, Z.; Li, L.; Fu, X.; Wu, Y.; Mehrabian, M.; et al. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell 2016, 165, 111–124. [Google Scholar] [CrossRef] [Green Version]
- Shih, D.M.; Zhu, W.; Schugar, R.C.; Meng, Y.; Jia, X.; Miikeda, A.; Wang, Z.; Zieger, M.; Lee, R.; Graham, M.; et al. Genetic Deficiency of Flavin-Containing Monooxygenase 3 (Fmo3) Protects Against Thrombosis but Has Only a Minor Effect on Plasma Lipid Levels—Brief Report. Arter. Thromb. Vasc. Biol. 2019, 39, 1045–1054. [Google Scholar] [CrossRef] [Green Version]
- Senthong, V.; Li, X.S.; Hudec, T.; Coughlin, J.; Wu, Y.; Levison, B.; Wang, Z.; Hazen, S.L.; Tang, W.W. Plasma Trimethylamine N -Oxide, a Gut Microbe–Generated Phosphatidylcholine Metabolite, Is Associated with Atherosclerotic Burden. J. Am. Coll. Cardiol. 2016, 67, 2620–2628. [Google Scholar] [CrossRef]
- Haghikia, A.; Li, X.S.; Liman, T.G.; Bledau, N.; Schmidt, D.; Zimmermann, F.; Kränkel, N.; Widera, C.; Sonnenschein, K.; Haghikia, A.; et al. Gut Microbiota–Dependent Trimethylamine N -Oxide Predicts Risk of Cardiovascular Events in Patients with Stroke and Is Related to Proinflammatory Monocytes. Arter. Thromb. Vasc. Biol. 2018, 38, 2225–2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiattarella, G.; Sannino, A.; Toscano, E.; Giugliano, G.; Gargiulo, G.; Franzone, A.; Trimarco, B.; Esposito, G.; Perrino, C. Gut microbe-generated metabolite trimethylamine-N-oxide as cardiovascular risk biomarker: A systematic review and dose-response meta-analysis. Eur. Heart J. 2017, 38, 2948–2956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Kong, B.; Shuai, W.; Fu, H.; Jiang, X.; Huang, H. 3,3-Dimethyl-1-butanol attenuates cardiac remodeling in pressure-overload-induced heart failure mice. J. Nutr. Biochem. 2020, 78, 108341. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Ji, F.; Zuo, A.; Liu, H.; Qi, L.; He, Y.; Wang, Q.; Zhao, P. Destructive Role of TMAO in T-Tubule and Excitation-Contraction Coupling in the Adult Cardiomyocytes. Int. Heart J. 2020, 61, 355–363. [Google Scholar] [CrossRef] [Green Version]
- Senthong, V.; Wang, Z.; Fan, Y.; Wu, Y.; Hazen, S.L.; Tang, W.H.W. Trimethylamine N -Oxide and Mortality Risk in Patients with Peripheral Artery Disease. J. Am. Heart Assoc. 2016, 5. [Google Scholar] [CrossRef]
- Choucair, I.; Nemet, I.; Li, L.; Cole, M.A.; Skye, S.M.; Kirsop, J.D.; Fischbach, M.A.; Gogonea, V.; Brown, J.M.; Tang, W.H.W.; et al. Quantification of bile acids: A mass spectrometry platform for studying gut microbe connection to metabolic diseases. J. Lipid Res. 2020, 61, 159–177. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Harris, S.C.; Bhowmik, S.; Kang, D.-J.; Hylemon, P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016, 7, 22–39. [Google Scholar] [CrossRef] [Green Version]
- Mayerhofer, C.C.; Ueland, T.; Broch, K.; Vincent, R.; Cross, G.F.; Dahl, C.P.; Aukrust, P.; Gullestad, L.; Hov, J.R.; Trøseid, M. Increased Secondary/Primary Bile Acid Ratio in Chronic Heart Failure. J. Card. Fail. 2017, 23, 666–671. [Google Scholar] [CrossRef] [Green Version]
- Gryp, T.; Vanholder, R.; Vaneechoutte, M.; Glorieux, G. p-Cresyl Sulfate. Toxins 2017, 9, 52. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Arango, L.F.; Barrett, H.L.; McIntyre, H.D.; Callaway, L.K.; Morrison, M.; Nitert, M.D. Increased Systolic and Diastolic Blood Pressure Is Associated with Altered Gut Microbiota Composition and Butyrate Production in Early Pregnancy. Hypertension 2016, 68, 974–981. [Google Scholar] [CrossRef]
- Segain, J.-P.; De La Blétière, D.R.; Bourreille, A.; Leray, V.; Gervois, N.; Rosales, C.; Ferrier, L.; Bonnet, C.; Blottiere, H.; Galmiche, J.-P. Butyrate inhibits inflammatory responses through NFkappa B inhibition: Implications for Crohn’s disease. Gut 2000, 47, 397–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musso, G.; Gambino, R.; Cassader, M. Interactions Between Gut Microbiota and Host Metabolism Predisposing to Obesity and Diabetes. Annu. Rev. Med. 2011, 62, 361–380. [Google Scholar] [CrossRef] [PubMed]
- Koren, O.; Spor, A.; Felin, J.; Fåk, F.; Stombaugh, J.; Tremaroli, V.; Behre, C.J.; Knight, R.; Fagerberg, B.; Ley, R.E.; et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 2010, 108, 4592–4598. [Google Scholar] [CrossRef] [Green Version]
- Vajpeyee, A.; Chauhan, P.S.; Pandey, S.; Tiwari, S.; Yadav, L.B.; Shroti, A.K.; Vajpeyee, M. Metagenomics Analysis of Thrombus Samples Retrieved from Mechanical Thrombectomy. Neurointervention 2021, 16, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.K.; Brar, M.S.; Kirjavainen, P.V.; Chen, Y.; Peng, J.; Li, D.; Leung, F.C.-C.; El-Nezami, H. High fat diet induced atherosclerosis is accompanied with low colonic bacterial diversity and altered abundances that correlates with plaque size, plasma A-FABP and cholesterol: A pilot study of high fat diet and its intervention with Lactobacillus rhamnosus GG (LGG) or telmisartan in ApoE−/− mice. BMC Microbiol. 2016, 16, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Yin, J.; Liao, S.X.; He, Y.; Wang, S.; Xia, G.H.; Liu, F.T.; Zhu, J.J.; You, C.; Chen, Q.; Zhou, L.; et al. Dysbiosis of Gut Microbiota with Reduced Trimethylamine-N-Oxide Level in Patients with Large-Artery Atherosclerotic Stroke or Transient Ischemic Attack. J. Am. Heart Assoc. 2015, 4, e002699. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Dai, M. Trimethylamine N-Oxide Generated by the Gut Microbiota Is Associated with Vascular Inflammation: New Insights into Atherosclerosis. Mediat. Inflamm. 2020, 2020, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Pasini, E.; Aquilani, R.; Testa, C.; Baiardi, P.; Angioletti, S.; Boschi, F.; Verri, M.; Dioguardi, F.S. Pathogenic Gut Flora in Patients with Chronic Heart Failure. JACC Heart Fail. 2016, 4, 220–227. [Google Scholar] [CrossRef]
- Zhang, Q.; Xiao, X.; Li, M.; Yu, M.; Ping, F.; Zheng, J.; Wang, T.; Wang, X. Vildagliptin increases butyrate-producing bacteria in the gut of diabetic rats. PLoS ONE 2017, 12, e0184735. [Google Scholar] [CrossRef] [Green Version]
- Niebauer, J.; Volk, H.D.; Kemp, M.; Dominguez, M.; Schumann, R.R.; Rauchhaus, M.; Poole-Wilson, P.A.; Coats, A.J.; Anker, S.D. Endotoxin and immune activation in chronic heart failure: A prospective cohort study. Lancet 1999, 353, 1838–1842. [Google Scholar] [CrossRef]
- Chugh, S.S.; Havmoeller, R.; Narayanan, K.; Singh, D.; Rienstra, M.; Benjamin, E.J.; Gillum, R.F.; Kim, Y.-H.; McAnulty, J.H., Jr.; Zheng, Z.-J.; et al. Worldwide Epidemiology of Atrial Fibrillation: A Global Burden of Disease 2010 Study. Circulation 2014, 129, 837–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, K.; Li, J.; Li, K.; Hu, C.; Gao, Y.; Chen, M.; Hu, R.; Liu, Y.; Chi, H.; Wang, H.; et al. Disordered gut microbiota and alterations in metabolic patterns are associated with atrial fibrillation. GigaScience 2019, 8, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, K.; Yin, X.; Li, K.; Zhang, J.; Wang, P.; Jiao, J.; Liu, Z.; Liu, X.; Liu, J.; Li, J.; et al. Different Types of Atrial Fibrillation Share Patterns of Gut Microbiota Dysbiosis. mSphere 2020, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Jin, M.; Liu, L.; Yu, Z.; Lu, X.; Zhang, H. Trimethylamine N-oxide and cardiovascular outcomes in patients with chronic heart failure after myocardial infarction. ESC Heart Fail. 2020, 7, 189–194. [Google Scholar] [CrossRef] [Green Version]
- Papandreou, C.; Bulló, M.; Hernández-Alonso, P.; Ruiz-Canela, M.; Li, J.; Guasch-Ferré, M.; Toledo, E.; Clish, C.; Corella, D.; Estruch, R.; et al. Choline Metabolism and Risk of Atrial Fibrillation and Heart Failure in the PREDIMED Study. Clin. Chem. 2020, 67, 288–297. [Google Scholar] [CrossRef]
- Wang, X.-H.; Li, Z.; Zang, M.-H.; Yao, T.-B.; Mao, J.-L.; Pu, J. Circulating primary bile acid is correlated with structural remodeling in atrial fibrillation. J. Interv. Card. Electrophysiol. 2019, 57, 371–377. [Google Scholar] [CrossRef]
- Dobrev, D.; Dudley, S.C. Oxidative stress: A bystander or a causal contributor to atrial remodelling and fibrillation? Cardiovasc. Res. 2021, 117, 2291–2293. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.; Li, B.; Luo, Y.; Gong, Y.; Jin, X.; Zhang, J.; Zhou, Y.; Zhuo, X.; Wang, Z.; et al. Gut microbiota dysbiosis promotes age-related atrial fibrillation by lipopolysaccharide and glucose-induced activation of NLRP3-inflammasome. Cardiovasc Res. 2022, 118, 785–797. [Google Scholar] [CrossRef]
- Scott, L., Jr.; Fender, A.C.; Saljic, A.; Li, L.; Chen, X.; Wang, X.; Linz, D.; Lang, J.; Hohl, M.; Twomey, D.; et al. NLRP3 inflammasome is a key driver of obesity-induced atrial arrhythmias. Cardiovasc. Res. 2021, 117, 1746–1759. [Google Scholar] [CrossRef]
- Liu, H.; Zhao, Y.; Xie, A.; Kim, T.-Y.; Terentyeva, R.; Liu, M.; Shi, G.; Feng, F.; Choi, B.-R.; Terentyev, D.; et al. Interleukin-1β, Oxidative Stress, and Abnormal Calcium Handling Mediate Diabetic Arrhythmic Risk. JACC Basic Transl. Sci. 2021, 6, 42–52. [Google Scholar] [CrossRef]
- Netto Candido, T.L.; Bressan, J.; de Alfenas, R.C.G. Dysbiosis and metabolic endotoxemia induced by high-fat diet. Nutr. Hosp. 2018, 35, 1432–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopp, W. How Western Diet And Lifestyle Drive The Pandemic Of Obesity And Civilization Diseases. Diabetes Metab. Syndr. Obes. Targets Ther. 2019, 12, 2221–2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-González, M.Á.; Corella, D.; Salas-Salvadó, J.; Ros, E.; Covas, M.I.; Fiol, M.; Wärnberg, J.; Arós, F.; Ruíz-Gutiérrez, V.; Lamuela-Raventós, R.M.; et al. Cohort Profile: Design and methods of the PREDIMED study. Int. J. Epidemiol. 2012, 41, 377–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Org, E.; Blum, Y.; Kasela, S.; Mehrabian, M.; Kuusisto, J.; Kangas, A.J.; Soininen, P.; Wang, Z.; Ala-Korpela, M.; Hazen, S.L.; et al. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 2017, 18, 1–14. [Google Scholar] [CrossRef]
- Berger, S.; Raman, G.; Vishwanathan, R.; Jacques, P.F.; Johnson, E.J. Dietary cholesterol and cardiovascular disease: A systematic review and meta-analysis. Am. J. Clin. Nutr. 2015, 102, 276–294. [Google Scholar] [CrossRef] [Green Version]
- Wilck, N.; Matus, M.G.; Kearney, S.M.; Olesen, S.W.; Forslund, K.; Bartolomaeus, H.; Haase, S.; Mähler, A.; Balogh, A.; Markó, L.; et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 2017, 551, 585–589. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [Green Version]
- Simopoulos, A.P. The Mediterranean Diets: What Is So Special about the Diet of Greece? The Scientific Evidence. J. Nutr. 2001, 131, 3065S–3073S. [Google Scholar] [CrossRef]
- Krznarić, Ž.; Bender, D.V.; Meštrović, T. The Mediterranean diet and its association with selected gut bacteria. Curr. Opin. Clin. Nutr. Metab. Care 2019, 22, 401–406. [Google Scholar] [CrossRef]
- Garcia-Mantrana, I.; Selma-Royo, M.; Alcantara, C.; Collado, M.C. Shifts on Gut Microbiota Associated to Mediterranean Diet Adherence and Specific Dietary Intakes on General Adult Population. Front. Microbiol. 2018, 9, 890. [Google Scholar] [CrossRef]
- Bagga, D.; Wang, L.; Farias-Eisner, R.; Glaspy, J.A.; Reddy, S.T. Differential effects of prostaglandin derived from ω-6 and ω-3 polyunsaturated fatty acids on COX-2 expression and IL-6 secretion. Proc. Natl. Acad. Sci. USA 2003, 100, 1751–1756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Angelis, M.; Garruti, G.; Minervini, F.; Bonfrate, L.; Portincasac, P.; Gobbetti, M. The Food-gut Human Axis: The Effects of Diet on Gut Microbiota and Metabolome. Curr. Med. Chem. 2019, 26, 3567–3583. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Zhou, J.; Li, Z.; Yu, Z.; Ouyang, D. The Association between Trimethylamine N-Oxide and Its Predecessors Choline, L-Carnitine, and Betaine with Coronary Artery Disease and Artery Stenosis. Cardiol. Res. Pr. 2020, 2020, 5854919. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Wang, Z.; Lai, H.; Otto, M.D.O.; Lemaitre, R.; Fretts, A.; Sotoodehnia, N.; Budoff, M.; DiDonato, J.; McKnight, B.; et al. Longitudinal Measures of Trimethylamine N-oxide and Incident Atherosclerotic Cardiovascular Disease Events in Older Adults: The Cardiovascular Health Study. Curr. Dev. Nutr. 2020, 4, 1434. [Google Scholar] [CrossRef]
- Boini, K.M.; Puchchakayala, G.; Zhang, Y.; Koka, S. TMAO Activates Carotid Endothelial Inflammasomes Leading to Enhanced Neointimal Formation in Nlrp3 Mice. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Tsigalou, C.; Paraschaki, A.; Karvelas, A.; Kantartzi, K.; Gagali, K.; Tsairidis, D.; Bezirtzoglou, E. Gut microbiome and Mediterranean diet in the context of obesity. Current knowledge, perspectives and potential therapeutic targets. Metab. Open 2021, 9, 100081. [Google Scholar] [CrossRef]
- Ali, K.M.; Wonnerth, A.; Huber, K.; Wojta, J. Cardiovascular disease risk reduction by raising HDL cholesterol—Current therapies and future opportunities. J. Cereb. Blood Flow Metab. 2012, 167, 1177–1194. [Google Scholar] [CrossRef] [Green Version]
- Endo, A. The Discovery and Development of HMG-CoA Reductase Inhibitors. J. Lipid Res. 1992, 33, 1569–1582. [Google Scholar] [CrossRef]
- Tuteja, S.; Ferguson, J.F. Gut Microbiome and Response to Cardiovascular Drugs. Circ. Genom. Precis. Med. 2019, 12, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Khan, T.J.; Ahmed, Y.M.; Zamzami, M.A.; Siddiqui, A.M.; Khan, I.; Baothman, O.A.S.; Mehanna, M.G.; Kuerban, A.; Kaleemuddin, M.; Yasir, M. Atorvastatin Treatment Modulates the Gut Microbiota of the Hypercholesterolemic Patients. OMICS A J. Integr. Biol. 2018, 22, 154–163. [Google Scholar] [CrossRef]
- Vieira-Silva, S.; Falony, G.; Belda, E.; Nielsen, T.; Aron-Wisnewsky, J.; Chakaroun, R.; Forslund, S.K.; Assmann, K.; Valles-Colomer, M.; Nguyen, T.T.D.; et al. Statin Therapy Is Associated with Lower Prevalence of Gut Microbiota Dysbiosis. Nature 2020, 581, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.D.; Panza, G.; Zaleski, A.; Taylor, B. Statin-Associated Side Effects. J. Am. Coll. Cardiol. 2016, 67, 2395–2410. [Google Scholar] [CrossRef]
- Repa, J.; Dietschy, J.M.; Turley, S.D. Inhibition of cholesterol absorption by SCH 58053 in the mouse is not mediated via changes in the expression of mRNA for ABCA1, ABCG5, or ABCG8 in the enterocyte. J. Lipid Res. 2002, 43, 1864–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudhop, T.; Lütjohann, D.; Kodal, A.; Igel, M.; Tribble, D.L.; Shah, S.; Perevozskaya, I.; von Bergmann, K. Inhibition of Intestinal Cholesterol Absorption by Ezetimibe in Humans. Circulation 2002, 106, 1943–1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakulj, L.; van Dijk, T.H.; de Boer, J.F.; Kootte, R.S.; Schonewille, M.; Paalvast, Y.; Boer, T.; Bloks, V.W.; Boverhof, R.; Nieuwdorp, M.; et al. Transintestinal Cholesterol Transport Is Active in Mice and Humans and Controls Ezetimibe-Induced Fecal Neutral Sterol Excretion. Cell Metab. 2016, 24, 783–794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.H.; Portincasa, P.; Mendez–Sanchez, N.; Uribe, M.; Wang, D.Q. Effect of Ezetimibe on the Prevention and Dissolution of Cholesterol Gallstones. Gastroenterology 2008, 134, 2101–2110. [Google Scholar] [CrossRef] [Green Version]
- O’Connor, C.M.; Dunne, M.W.; Pfeffer, M.A.; Muhlestein, J.B.; Yao, L.; Gupta, S.; Benner, R.J.; Fisher, M.R.; Cook, T.D.; for the Investigators in the WIZARD Study. Azithromycin for the Secondary Prevention of Coronary Heart Disease Events. JAMA 2003, 290, 1459–1466. [Google Scholar] [CrossRef] [Green Version]
- Andraws, R.; Berger, J.; Brown, D.L. Effects of Antibiotic Therapy on Outcomes of Patients with Coronary Artery Disease. JAMA 2005, 293, 2641–2647. [Google Scholar] [CrossRef]
- Algeri, A.; Tassi, F.; Ferrero, I.; Puglisi, P.P. Different phenotypes for the lactose utilization system inKluyveromyces andSaccharomyces species. Antonie van Leeuwenhoek 1978, 44, 177–182. [Google Scholar] [CrossRef]
- Santisteban, M.; Qi, Y.; Zubcevic, J.; Kim, S.; Yang, T.; Shenoy, V.; Cole-Jeffrey, C.T.; Lobaton, G.; Stewart, D.C.; Rubiano, A.; et al. Hypertension-Linked Pathophysiological Alterations in the Gut. Circ. Res. 2017, 120, 312–323. [Google Scholar] [CrossRef]
- Yan, X.; Jin, J.; Su, X.; Yin, X.; Gao, J.; Wang, X.; Zhang, S.; Bu, P.; Wang, M.; Zhang, Y.; et al. Intestinal Flora Modulates Blood Pressure by Regulating the Synthesis of Intestinal-Derived Corticosterone in High Salt-Induced Hypertension. Circ. Res. 2020, 126, 839–853. [Google Scholar] [CrossRef] [PubMed]
- Najmanová, I.; Pourová, J.; Vopršalová, M.; Pilařová, V.; Semecký, V.; Nováková, L.; Mladěnka, P. Flavonoid metabolite 3-(3-hydroxyphenyl)propionic acid formed by human microflora decreases arterial blood pressure in rats. Mol. Nutr. Food Res. 2016, 60, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.A.M.; Aronoff, D.M. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin. Microbiol. Infect. 2016, 22, 178.e1–178.e9. Available online: http://www.ncbi.nlm.nih.gov/pubmed/26482265 (accessed on 19 January 2019). [CrossRef] [Green Version]
- Bjarnason, I.; Scarpignato, C.; Holmgren, E.; Olszewski, M.; Rainsford, K.D.; Lanas, A. Mechanisms of Damage to the Gastrointestinal Tract from Nonsteroidal Anti-Inflammatory Drugs. Gastroenterology 2018, 154, 500–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foretz, M.; Guigas, B.; Viollet, B. Understanding the glucoregulatory mechanisms of metformin in type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2019, 15, 569–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rena, G.; Hardie, D.G.; Pearson, E.R. The mechanisms of action of metformin. Diabetologia 2017, 60, 1577–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, E.W.; Gao, L.; Stastka, P.; Cheney, M.C.; Mahabamunuge, J.; Soto, M.T.; Ford, C.B.; Bryant, J.A.; Henn, M.R.; Hohmann, E.L. Fecal microbiota transplantation for the improvement of metabolism in obesity: The FMT-TRIM double-blind placebo-controlled pilot trial. PLoS Med. 2020, 17, e1003051. [Google Scholar] [CrossRef]
- De Groot, P.; Scheithauer, T.; Bakker, G.J.; Prodan, A.; Levin, E.; Khan, M.T.; Herrema, H.; Ackermans, M.; Serlie, M.J.M.; De Brauw, M.; et al. Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut 2020, 69, 502–512. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Lu, Y.; Yan, Y.; Tian, S.; Zheng, D.; Leng, D.; Wang, C.; Jiao, J.; Wang, Z.; Bai, Y. Promising Treatment for Type 2 Diabetes: Fecal Microbiota Transplantation Reverses Insulin Resistance and Impaired Islets. Front. Cell. Infect. Microbiol. 2020, 9, 455. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.-F.; Zhang, W.-Y.; Wen, Q.; Chen, W.-J.; Wang, Z.-M.; Chen, J.; Zhu, F.; Liu, K.; Cheng, L.-X.; Yang, J.; et al. Fecal microbiota transplantation alleviates myocardial damage in myocarditis by restoring the microbiota composition. Pharmacol. Res. 2019, 139, 412–421. [Google Scholar] [CrossRef]
- Toral, M.; Robles-Vera, I.; de la Visitación, N.; Romero, M.; Yang, T.; Sánchez, M.; Gómez-Guzmán, M.; Jiménez, R.; Raizada, M.K.; Duarte, J. Critical Role of the Interaction Gut Microbiota—Sympathetic Nervous System in the Regulation of Blood Pressure. Front. Physiol. 2019, 10, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smits, L.P.; Kootte, R.S.; Levin, E.; Prodan, A.; Fuentes, S.; Zoetendal, E.G.; Wang, Z.; Levison, B.S.; Cleophas, M.C.P.; Kemper, E.M.; et al. Effect of Vegan Fecal Microbiota Transplantation on Carnitine- and Choline-Derived Trimethylamine-N-Oxide Production and Vascular Inflammation in Patients with Metabolic Syndrome. J. Am. Heart Assoc. 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Khalili, L.; Alipour, B.; Jafarabadi, M.A.; Hassanalilou, T.; Abbasi, M.M.; Faraji, I. Probiotic assisted weight management as a main factor for glycemic control in patients with type 2 diabetes: A randomized controlled trial. Diabetol. Metab. Syndr. 2019, 11, 5. [Google Scholar] [CrossRef]
- Zeng, Z.; Yuan, Q.; Yu, R.; Zhang, J.; Ma, H.; Chen, S. Ameliorative Effects of Probiotic Lactobacillus paracasei NL41 on Insulin Sensitivity, Oxidative Stress, and Beta-Cell Function in a Type 2 Diabetes Mellitus Rat Model. Mol. Nutr. Food Res. 2019, 63, e1900457. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, T.V.; Succurro, E.; Marini, M.A.; Pedace, E.; Andreozzi, F.; Perticone, M.; Sciacqua, A.; Perticone, F.; Sesti, G. HDL cholesterol is an independent predictor of β-cell function decline and incident type 2 diabetes: A longitudinal study. Diabetes Metabolism Res. Rev. 2020, 36, e3289. [Google Scholar] [CrossRef]
- Bock, P.M.; Telo, G.H.; Ramalho, R.; Sbaraini, M.; Leivas, G.; Martins, A.F.; Schaan, B.D. The effect of probiotics, prebiotics or synbiotics on metabolic outcomes in individuals with diabetes: A systematic review and meta-analysis. Diabetologia 2020, 64, 26–41. [Google Scholar] [CrossRef]
- Gómez-Guzmán, M.; Toral, M.; Romero, M.; Jiménez, R.; Galindo, P.; Sánchez, M.; Zarzuelo, M.J.; Olivares, M.; Gálvez, J.; Duarte, J. Antihypertensive effects of probioticsLactobacillusstrains in spontaneously hypertensive rats. Mol. Nutr. Food Res. 2015, 59, 2326–2336. [Google Scholar] [CrossRef]
- Moludi, J.; Khedmatgozar, H.; Nachvak, S.M.; Abdollahzad, H.; Moradinazar, M.; Tabaei, A.S. The effects of co-administration of probiotics and prebiotics on chronic inflammation, and depression symptoms in patients with coronary artery diseases: A randomized clinical trial. Nutr. Neurosci. 2021, 1–10. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Vlasov, A.A.; Shperling, M.I.; Terkin, D.A.; Bystrova, O.V.; Osipov, G.A.; Salikova, S.P.; Grinevich, V.B. Effect of Prebiotic Complex on Gut Microbiota and Endotoxemia in Female Rats with Modeled Heart Failure. Bull. Exp. Biol. Med. 2020, 168, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, G.; Li, X.; Zheng, F.; Zeng, X. Yeast β-glucan, a potential prebiotic, showed a similar probiotic activity to inulin. Food Funct. 2020, 11, 10386–10396. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.Y.; Rutka, J.T.; Chan, W.C. Nanomedicine. N. Engl. J. Med. 2010, 363, 2434–2443. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Hou, Y.; Chen, X.; Ding, X.; Liu, Y.; Shen, X.; Cai, K. Recyclable heparin and chitosan conjugated magnetic nanocomposites for selective removal of low-density lipoprotein from plasma. J. Mater. Sci. Mater. Med. 2014, 25, 1055–1064. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, L.; Zhao, W.; Dou, Y.; An, H.; Tao, H.; Xu, X.; Jia, Y.; Lu, S.; Zhang, J.; et al. Targeted Therapy of Atherosclerosis by a Broad-Spectrum Reactive Oxygen Species Scavenging Nanoparticle with Intrinsic Anti-inflammatory Activity. ACS Nano 2018, 12, 8943–8960. [Google Scholar] [CrossRef] [PubMed]
- Gan, J.; Dou, Y.; Li, Y.; Wang, Z.; Wang, L.; Liu, S.; Li, Q.; Yu, H.; Liu, C.; Han, C.; et al. Producing anti-inflammatory macrophages by nanoparticle-triggered clustering of mannose receptors. Biomaterials 2018, 178, 95–108. [Google Scholar] [CrossRef]
- Papi, M.; Caracciolo, G. Principal component analysis of personalized biomolecular corona data for early disease detection. Nano Today 2018, 21, 14–17. [Google Scholar] [CrossRef]
- Caracciolo, G.; Safavi-Sohi, R.; Malekzadeh, R.; Poustchi, H.; Vasighi, M.; Chiozzi, R.Z.; Capriotti, A.L.; Laganà, A.; Hajipour, M.; Di Domenico, M.; et al. Disease-specific protein corona sensor arrays may have disease detection capacity. Nanoscale Horiz. 2019, 4, 1063–1076. [Google Scholar] [CrossRef]
- Ashkarran, A.A.; Olfatbakhsh, T.; Ramezankhani, M.; Crist, R.C.; Berrettini, W.H.; Milani, A.S.; Pakpour, S.; Mahmoudi, M. Evolving Magnetically Levitated Plasma Proteins Detects Opioid Use Disorder as a Model Disease. Adv. Healthc. Mater. 2020, 9, e1901608. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, M.; Pakpour, S.; Perry, G. Drug-Abuse Nanotechnology: Opportunities and Challenges. ACS Chem. Neurosci. 2018, 9, 2288–2298. [Google Scholar] [CrossRef] [PubMed]
- Guthrie, L.; Kelly, L. Bringing microbiome-drug interaction research into the clinic. EBioMedicine 2019, 44, 708–715. [Google Scholar] [CrossRef] [Green Version]
- Roberts, A.; Gu, X.; Buffa, J.A.; Hurd, A.G.; Wang, Z.; Zhu, W.; Gupta, N.; Skye, S.M.; Cody, D.B.; Levison, B.S.; et al. Development of a gut microbe–targeted nonlethal therapeutic to inhibit thrombosis potential. Nat. Med. 2018, 24, 1407–1417. [Google Scholar] [CrossRef]
- Costea, P.I.; Zeller, G.; Sunagawa, S.; Pelletier, E.; Alberti, A.; Levenez, F.; Tramontano, M.; Driessen, M.; Hercog, R.; Jung, F.-E.; et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 2017, 35, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Badal, V.D.; Wright, D.; Katsis, Y.; Kim, H.-C.; Swafford, A.D.; Knight, R.; Hsu, C.-N. Challenges in the construction of knowledge bases for human microbiome-disease associations. Microbiome 2019, 7, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahrens, A.P.; Culpepper, T.; Saldivar, B.; Anton, S.; Stoll, S.; Handberg, E.M.; Xu, K.; Pepine, C.; Triplett, E.W.; Aggarwal, M. A Six-Day, Lifestyle-Based Immersion Program Mitigates Cardiovascular Risk Factors and Induces Shifts in Gut Microbiota, Specifically Lachnospiraceae, Ruminococcaceae, Faecalibacterium prausnitzii: A Pilot Study. Nutrients 2021, 13, 3459. [Google Scholar] [CrossRef]
- Zhou, X.; Li, J.; Guo, J.; Geng, B.; Ji, W.; Zhao, Q.; Li, J.; Liu, X.; Liu, J.; Guo, Z.; et al. Gut-dependent microbial translocation induces inflammation and cardiovascular events after ST-elevation myocardial infarction. Microbiome 2018, 6, 66. [Google Scholar] [CrossRef]
- Ivashkin, V.T.; Kashukh, Y.A. Impact of L-carnitine and phosphatidylcholine containing products on the proatherogenic metabolite TMAO production and gut microbiome changes in patients with coronary artery disease. Voprosy Pitaniia 2019, 88, 25–33. (In Russian) [Google Scholar] [CrossRef]
- Liu, H.; Zhu, C.-G.; Cui, C.-J.; Cao, Y.; Jin, J.; Wu, N.-Q.; Guo, Y.-L.; Gao, Y.; Sun, J.; Dong, Q.; et al. Abstract 15197: Lipopolysaccharide-Nuclear Factor-kappa B Pathway and Lipoprotein Apheresis Effects in Patients with Familial Hypercholesterolemia and Coronary Artery Disease. Circulation 2020, 142, A15197. [Google Scholar] [CrossRef]
- Schuett, K.; Kleber, M.E.; Scharnagl, H.; Lorkowski, S.; März, W.; Niessner, A.; Marx, N.; Meinitzer, A. Trimethylamine-N-oxide and Heart Failure with Reduced Versus Preserved Ejection Fraction. J. Am. Coll. Cardiol. 2017, 70, 3202–3204. [Google Scholar] [CrossRef] [PubMed]
- Alhmoud, T.; Kumar, A.; Lo, C.-C.; Al-Sadi, R.; Clegg, S.; Alomari, I.; Zmeili, T.; Gleasne, C.D.; Mcmurry, K.; Dichosa, A.E.K.; et al. Investigating intestinal permeability and gut microbiota roles in acute coronary syndrome patients. Hum. Microbiome J. 2019, 13, 100059. [Google Scholar] [CrossRef]
- Zuo, K.; Li, J.; Wang, P.; Liu, Y.; Liu, Z.; Yin, X.; Liu, X.; Yang, X. Duration of Persistent Atrial Fibrillation Is Associated with Alterations in Human Gut Microbiota and Metabolic Phenotypes. mSystems 2019, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastori, D.; Carnevale, R.; Nocella, C.; Novo, M.; Santulli, M.; Cammisotto, V.; Menichelli, D.; Pignatelli, P.; Violi, F. Gut-Derived Serum Lipopolysaccharide is Associated with Enhanced Risk of Major Adverse Cardiovascular Events in Atrial Fibrillation: Effect of Adherence to Mediterranean Diet. J. Am. Heart Assoc. 2017, 6, e005784. [Google Scholar] [CrossRef]
- Gong, D.; Zhang, L.; Zhang, Y.; Wang, F.; Zhao, Z.; Zhou, X. Gut Microbial Metabolite Trimethylamine N-Oxide Is Related to Thrombus Formation in Atrial Fibrillation Patients. Am. J. Med. Sci. 2019, 358, 422–428. [Google Scholar] [CrossRef] [PubMed]
| Clinical Trials | |||
|---|---|---|---|
| Diseases | Sample (Controls excluded) | Gut Microbiota-related effect | Method |
| Atheromatosis | 100 STEMI patients [178] | Translocation of gut bacteria to the bloodstream | 16S rRNA PCR (V4 region) |
| 4144 older adults [124] | Increased TMAO levels | Stable isotope dilution LC/MS/MS | |
| CAD | 29 CAD patients [179] | Increased L-carnitine and TMAO levels | Electrospray ionization LC/MS-8060 |
| 63 non-FH CAD patients [180] | Increased LPS levels and inflammatory cytokines | LBP, ELISA | |
| Chronic Heart Failure | 428 HFrEF patients 395 HFpEF patients [181] | Increased TMAO levels—increased HF susceptibility | LC/MS/MS |
| Heart attack | 19 ACS patients [182] | Increased gut leakiness and endotoxemia | L/M ratio, LC/MS, 16S sequencing |
| Atrial fibrillation | 20 psAF patients [183] | Distinctive and progressive alterations in gut microbiome and metabolic structure | LC/MS (+/− ion mode), metagenomic sequencing |
| 912 AF patients [184] | Increased LPS levels and platelet hyperreactivity | ELISA | |
| 20 psAF—30 PAF patients [182] | Distinctive and progressive alterations in gut microbiome | LC/MS, Whole-metagenome shotgun sequnecing | |
| 117 rheumatic heart disease patients with AF [185] | Increased TMAO levels and thrombus development | Light transmittance aggregometry, LC/MS | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadopoulos, P.D.; Tsigalou, C.; Valsamaki, P.N.; Konstantinidis, T.G.; Voidarou, C.; Bezirtzoglou, E. The Emerging Role of the Gut Microbiome in Cardiovascular Disease: Current Knowledge and Perspectives. Biomedicines 2022, 10, 948. https://doi.org/10.3390/biomedicines10050948
Papadopoulos PD, Tsigalou C, Valsamaki PN, Konstantinidis TG, Voidarou C, Bezirtzoglou E. The Emerging Role of the Gut Microbiome in Cardiovascular Disease: Current Knowledge and Perspectives. Biomedicines. 2022; 10(5):948. https://doi.org/10.3390/biomedicines10050948
Chicago/Turabian StylePapadopoulos, Panagiotis D., Christina Tsigalou, Pipitsa N. Valsamaki, Theocharis G. Konstantinidis, Chrysoula Voidarou, and Eugenia Bezirtzoglou. 2022. "The Emerging Role of the Gut Microbiome in Cardiovascular Disease: Current Knowledge and Perspectives" Biomedicines 10, no. 5: 948. https://doi.org/10.3390/biomedicines10050948
APA StylePapadopoulos, P. D., Tsigalou, C., Valsamaki, P. N., Konstantinidis, T. G., Voidarou, C., & Bezirtzoglou, E. (2022). The Emerging Role of the Gut Microbiome in Cardiovascular Disease: Current Knowledge and Perspectives. Biomedicines, 10(5), 948. https://doi.org/10.3390/biomedicines10050948










