Beyond Harmful: Exploring Biofilm Formation by Enterococci Isolated from Portuguese Traditional Cheeses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbial Collection
2.2. Biofilm Formation
2.2.1. Bacterial Suspension
2.2.2. Biofilm Assays
Cell Viability
Biofilm Quantification
2.2.3. Data Analysis
3. Results and Discussion
3.1. Biofilm Formation: Preliminary Assays
3.2. Biofilm Formation at Different Temperatures
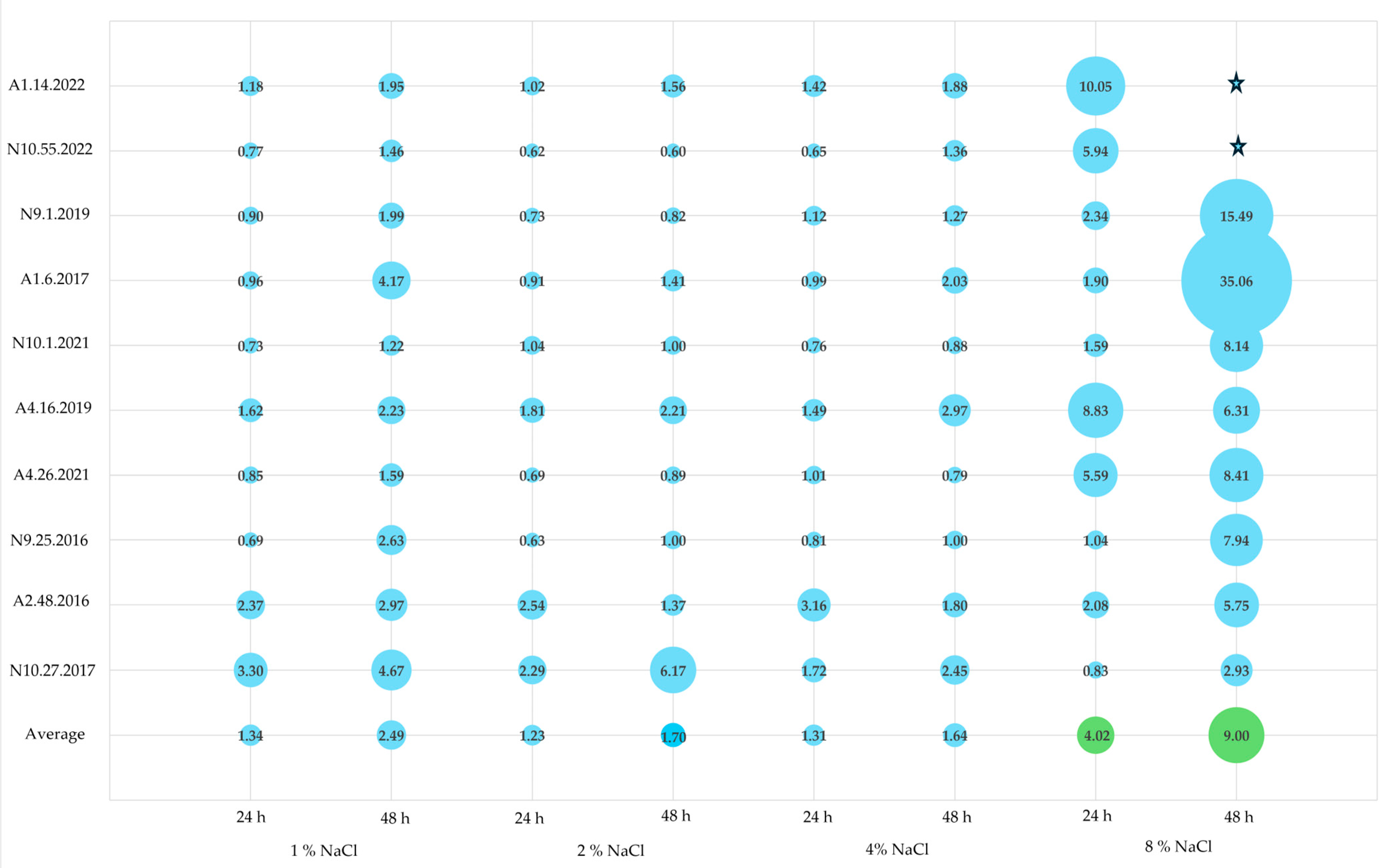
3.3. Biofilm Formation at Different NaCl Concentrations
3.4. Biofilm Formation at Different pH Values
3.5. Biofilm Quantification Versus Cell Viability
3.6. The Role of Biofilm Formation in Traditional Cheese Making
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health Benefits of Fermented Foods: Microbiota and Beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef]
- Zhang, M.; Dong, X.; Huang, Z.; Li, X.; Zhao, Y.; Wang, Y.; Zhu, H.; Fang, A.; Giovannucci, E.L. Cheese Consumption and Multiple Health Outcomes: An Umbrella Review and Updated Meta-Analysis of Prospective Studies. Adv. Nutr. 2023, 14, 1170–1186. [Google Scholar] [CrossRef] [PubMed]
- Kindstedt, P.S. The History of Cheese. In Global Cheesemaking Technology; Papademas, P., Bintsis, T., Eds.; Wiley: Hoboken, NJ, USA, 2017; pp. 1–19. [Google Scholar]
- Gajewska, J.; Chajęcka-Wierzchowska, W.; Byczkowska-Rostkowska, Z.; Saki, M. Biofilm Formation Capacity and Presence of Virulence Determinants among Enterococcus Species from Milk and Raw Milk Cheeses. Life 2023, 13, 495. [Google Scholar] [CrossRef] [PubMed]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C.G. Lactic Acid Bacteria in Raw-Milk Cheeses: From Starter Cultures to Probiotic Functions. Foods 2022, 11, 2276. [Google Scholar] [CrossRef] [PubMed]
- Freitas, C.; Malcata, F.X. Microbiology and Biochemistry of Cheeses with Appélation d’origine Protegée and Manufactured in the Iberian Peninsula from Ovine and Caprine Milks. J. Dairy Sci. 2000, 83, 584–602. [Google Scholar] [CrossRef]
- de Paula, J.C.J.; de Carvalho, A.F.; Furtado, M.M. Princípios Básicos de Fabricação de Queijo: Do Histórico à Saga. Rev. do Inst. Laticínios Cândido Tostes 2009, 64, 19–25. [Google Scholar]
- Guiné, R.P.F.; Florença, S.G. The Science behind Traditional Products: The Case of Portuguese Cheeses. ETP Int. J. Food Eng. 2020, 6, 45–51. [Google Scholar] [CrossRef]
- Serrano, S.; Morais, S.; Semedo-Lemsaddek, T. Tradition Unveiled: A Comprehensive Review of Microbiological Studies on Portuguese Traditional Cheeses, Merging Conventional and OMICs Analyses. Front. Ind. Microbiol. 2024, 2, 1420042. [Google Scholar] [CrossRef]
- EU Geographical Indications and Quality Schemes Explained. European Commission Website 2022. Available online: https://agriculture.ec.europa.eu/farming/geographical-indications-and-quality-schemes/geographical-indications-and-quality-schemes-explained_en (accessed on 5 June 2024).
- Afshari, R.; Pillidge, C.J.; Dias, D.A.; Osborn, A.M.; Gill, H. Cheesomics: The Future Pathway to Understanding Cheese Flavour and Quality. Crit. Rev. Food Sci. Nutr. 2020, 60, 33–47. [Google Scholar] [CrossRef]
- Tavaria, F.K.; Dahl, S.; Carballo, F.J.; Malcata, F.X. Amino Acid Catabolism and Generation of Volatiles by Lactic Acid Bacteria. J. Dairy Sci. 2002, 85, 2462–2470. [Google Scholar] [CrossRef]
- Hatti-Kaul, R.; Chen, L.; Dishisha, T.; Enshasy, H. El Lactic Acid Bacteria: From Starter Cultures to Producers of Chemicals. FEMS Microbiol. Lett. 2018, 365, fny213. [Google Scholar] [CrossRef] [PubMed]
- Akoğlu, A. The Effect of Some Environmental Conditions on Planktonic Growth and Biofilm Formation by Some Lactic Acid Bacteria Isolated from a Local Cheese in Turkey. Biotechnol. Lett. 2020, 42, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Castellone, V.; Bancalari, E.; Rubert, J.; Gatti, M.; Neviani, E.; Bottari, B. Eating Fermented: Health Benefits of LAB-Fermented Foods. Foods 2021, 10, 2639. [Google Scholar] [CrossRef] [PubMed]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The Complex Microbiota of Raw Milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed]
- Muruzović, M.Ž.; Mladenović, K.G.; Žugić-Petrović, T.D.; Čomić, L.R. Characterization of Lactic Acid Bacteria Isolated from Traditionally Made Serbian Cheese and Evaluation of Their Antagonistic Potential against Enterobacteriaceae. J. Food Process. Preserv. 2018, 42, e13577. [Google Scholar] [CrossRef]
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; De Cesare, A.; Hilbert, F.; Lindqvist, R.; Nauta, M.; et al. Update of the List of Qualified Presumption of Safety (QPS) Recommended Microbiological Agents Intentionally Added to Food or Feed as Notified to EFSA 18: Suitability of Taxonomic Units Notified to EFSA until March 2023. EFSA J. 2023, 21, e08092. [Google Scholar] [CrossRef]
- Grujović, M.Ž.; Mladenović, K.G.; Semedo-Lemsaddek, T.; Laranjo, M.; Stefanović, O.D.; Kocić-Tanackov, S.D. Advantages and Disadvantages of Non-starter Lactic Acid Bacteria from Traditional Fermented Foods: Potential Use as Starters or Probiotics. Compr. Rev. Food Sci. Food Saf. 2022, 21, 1537–1567. [Google Scholar] [CrossRef]
- Grujović, M.Ž.; Marković, K.G.; Morais, S.; Semedo-Lemsaddek, T. Unveiling the Potential of Lactic Acid Bacteria from Serbian Goat Cheese. Foods 2024, 13, 2065. [Google Scholar] [CrossRef]
- Krawczyk, B.; Wityk, P.; Gałęcka, M.; Michalik, M. The Many Faces of Enterococcus spp.—Commensal, Probiotic and Opportunistic Pathogen. Microorganisms 2021, 9, 1900. [Google Scholar] [CrossRef]
- de Dapkevicius, M.L.E.; Sgardioli, B.; Câmara, S.P.A.; Poeta, P.; Malcata, F.X. Current Trends of Enterococci in Dairy Products: A Comprehensive Review of Their Multiple Roles. Foods 2021, 10, 821. [Google Scholar] [CrossRef]
- Bastião Rocha, P.A.; Monteiro Marques, J.M.; Barreto, A.S.; Semedo-Lemsaddek, T. Enterococcus spp. from Azeitão and Nisa PDO-Cheeses: Surveillance for Antimicrobial Drug Resistance. LWT 2022, 154, 112622. [Google Scholar] [CrossRef]
- Monteiro Marques, J.; Serrano, S.; Selmi, H.; Giesteira Cotovio, P.; Semedo-Lemsaddek, T. Antimicrobial and Antibiofilm Potential of Thymus Vulgaris and Cymbopogon Flexuosus Essential Oils against Pure and Mixed Cultures of Foodborne Bacteria. Antibiotics 2023, 12, 565. [Google Scholar] [CrossRef] [PubMed]
- Terzić-Vidojević, A.; Veljović, K.; Popović, N.; Tolinački, M.; Golić, N. Enterococci from Raw-Milk Cheeses: Current Knowledge on Safety, Technological, and Probiotic Concerns. Foods 2021, 10, 2753. [Google Scholar] [CrossRef]
- Ogier, J.C.; Serror, P. Safety Assessment of Dairy Microorganisms: The Enterococcus genus. Int. J. Food Microbiol. 2008, 126, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Cappitelli, F.; Polo, A.; Villa, F. Biofilm Formation in Food Processing Environments Is Still Poorly Understood and Controlled. Food Eng. Rev. 2014, 6, 29–42. [Google Scholar] [CrossRef]
- Carrascosa, C.; Raheem, D.; Ramos, F.; Saraiva, A.; Raposo, A. Microbial Biofilms in the Food Industry—A Comprehensive Review. Int. J. Environ. Res. Public Health 2021, 18, 2014. [Google Scholar] [CrossRef]
- Chitlapilly Dass, S.; Wang, R. Biofilm through the Looking Glass: A Microbial Food Safety Perspective. Pathogens 2022, 11, 346. [Google Scholar] [CrossRef]
- Gaillac, A.; Briandet, R.; Delahaye, E.; Deschamps, J.; Vigneau, E.; Courcoux, P.; Jaffrès, E.; Prévost, H. Exploring the Diversity of Biofilm Formation by the Food Spoiler Brochothrix Thermosphacta. Microorganisms 2022, 10, 2474. [Google Scholar] [CrossRef]
- Valentino, V.; De Filippis, F.; Sequino, G.; Ercolini, D. Psychrotrophic Bacteria Equipped with Virulence and Colonization Traits Populate the Ice Cream Manufacturing Environment. Appl. Environ. Microbiol. 2023, 89, e00765-23. [Google Scholar] [CrossRef]
- Furukawa, S. Studies on Formation, Control and Application of Biofilm Formed by Food Related Microorganisms. Biosci. Biotechnol. Biochem. 2015, 79, 1050–1056. [Google Scholar] [CrossRef]
- Percival, S.L.; Malic, S.; Cruz, H.; Williams, D.W. Introduction to Biofilms. In Biofilmes and Veterinary Medicine; Springer: Berlin/Heidelberg, Germany, 2011; pp. 41–68. [Google Scholar]
- Yuan, L.; Wang, H.; Liu, W.; Sadiq, F.A.; Zhao, Y. Editorial: Multi-Species Biofilms in the Food Industry. Front. Microbiol. 2022, 13, 1023428. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, M.H.; Idris, A.L.; Fan, X.; Guo, Y.; Yu, Y.; Jin, X.; Qiu, J.; Guan, X.; Huang, T. Beyond Risk: Bacterial Biofilms and Their Regulating Approaches. Front. Microbiol. 2020, 11, 928. [Google Scholar] [CrossRef]
- Jefferson, K. What Drives Bacteria to Produce a Biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173. [Google Scholar] [CrossRef]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hébraud, M.; Jaglic, Z.; et al. Critical Review on Biofilm Methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef]
- Hua, Z.; Korany, A.M.; El-Shinawy, S.H.; Zhu, M.J. Comparative Evaluation of Different Sanitizers Against Listeria monocytogenes Biofilms on Major Food-Contact Surfaces. Front. Microbiol. 2019, 10, 2462. [Google Scholar] [CrossRef] [PubMed]
- Wang, R. Biofilms and Meat Safety: A Mini-Review. J. Food Prot. 2019, 82, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Olanbiwoninu, A.A.; Popoola, B.M. Biofilms and Their Impact on the Food Industry. Saudi J. Biol. Sci. 2023, 30, 103523. [Google Scholar] [CrossRef]
- Pieniz, S.; de Moura, T.M.; Cassenego, A.P.V.; Andreazza, R.; Frazzon, A.P.G.; de Oliveira Camargo, F.A.; Brandelli, A. Evaluation of Resistance Genes and Virulence Factors in a Food Isolated Enterococcus durans with Potential Probiotic Effect. Food Control 2015, 51, 49–54. [Google Scholar] [CrossRef]
- Gómez, N.C.; Ramiro, J.M.P.; Quecan, B.X.V.; de Melo Franco, B.D.G. Use of Potential Probiotic Lactic Acid Bacteria (LAB) Biofilms for the Control of Listeria monocytogenes, Salmonella Typhimurium, and Escherichia coli O157:H7 Biofilms Formation. Front. Microbiol. 2016, 7, 863. [Google Scholar] [CrossRef]
- Kim, J.-H.; Lee, E.-S.; Song, K.-J.; Kim, B.-M.; Ham, J.-S.; Oh, M.-H. Development of Desiccation-Tolerant Probiotic Biofilms Inhibitory for Growth of Foodborne Pathogens on Stainless Steel Surfaces. Foods 2022, 11, 831. [Google Scholar] [CrossRef]
- El-Zamkan, M.A.; Mohamed, H.M.A. Antimicrobial Resistance, Virulence Genes and Biofilm Formation in Enterococcus Species Isolated from Milk of Sheep and Goat with Subclinical Mastitis. PLoS ONE 2021, 16, e0259584. [Google Scholar] [CrossRef] [PubMed]
- Muruzović, M.; Mladenović, K.G.; Čomić, L.R. In Vitro Evaluation of Resistance to Environmental Stress by Planktonic and Biofilm Form of Lactic Acid Bacteria Isolated from Traditionally Made Cheese from Serbia. Food Biosci. 2018, 23, 54–59. [Google Scholar] [CrossRef]
- Muruzović, M.Ž.; Mladenović, K.G.; Djilas, M.D.; Stefanović, O.D.; Čomić, L.R. In Vitro Evaluation of Antimicrobial Potential and Ability of Biofilm Formation of Autochthonous Lactobacillus Spp. and Lactococcus Spp. Isolated from Traditionally Made Cheese from Southeastern Serbia. J. Food Process. Preserv. 2018, 42, e13776. [Google Scholar] [CrossRef]
- Somers, E.B.; Johnson, M.E.; Wong, A.C.L. Biofilm Formation and Contamination of Cheese by Nonstarter Lactic Acid Bacteria in The Dairy Environment. J. Dairy Sci. 2001, 84, 1926–1936. [Google Scholar] [CrossRef]
- Popović, N.; Dinić, M.; Tolinački, M.; Mihajlović, S.; Terzić-Vidojević, A.; Bojić, S.; Djokić, J.; Golić, N.; Veljović, K. New Insight into Biofilm Formation Ability, the Presence of Virulence Genes and Probiotic Potential of Enterococcus sp. Dairy Isolates. Front. Microbiol. 2018, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the Food Industry: Health Aspects and Control Methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef] [PubMed]
- Ke, D.; Picard, F.J.; Martineau, F.; Ménard, C.; Roy, P.H.; Ouellette, M.; Bergeron, M.G. Development of a PCR Assay for Rapid Detection of Enterococci. J. Clin. Microbiol. 1999, 37, 3497–3503. [Google Scholar] [CrossRef]
- Jackson, C.R.; Fedorka-Cray, P.J.; Barrett, J.B. Use of a Genus- and Species-Specific Multiplex PCR for Identification of Enterococci. J. Clin. Microbiol. 2004, 42, 3558–3565. [Google Scholar] [CrossRef]
- Extremina, C.I.; Costa, L.; Aguiar, A.I.; Peixe, L.; Fonseca, A.P. Optimization of Processing Conditions for the Quantification of Enterococci Biofilms Using Microtitre-Plates. J. Microbiol. Methods 2011, 84, 167–173. [Google Scholar] [CrossRef]
- Aksoy, B.A.; Aksoy, P.; Hammerbacher, J. Resazurin Viability Assay for Human Primary T Cells in 96-Well Format. protocols.io 2018. Available online: https://www.protocols.io/view/resazurin-viability-assay-for-human-primary-t-cell-36wgqxo3lk57/v1 (accessed on 1 March 2024).
- Gómez-Suárez, C.; Busscher, H.J.; Van Der Mei, H.C. Analysis of Bacterial Detachment from Substratum Surfaces by the Passage of Air-Liquid Interfaces. Appl. Environ. Microbiol. 2001, 67, 2531–2537. [Google Scholar] [CrossRef] [PubMed]
- Tasse, J.; Cara, A.; Saglio, M.; Villet, R.; Laurent, F. A Steam-Based Method to Investigate Biofilm. Sci. Rep. 2018, 8, 13040. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of Biofilm in Microtiter Plates: Overview of Testing Conditions and Practical Recommendations for Assessment of Biofilm Production by Staphylococci. Apmis 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Dassanayake, R.P.; Falkenberg, S.M.; Stasko, J.A.; Shircliff, A.L.; Lippolis, J.D.; Briggs, R.E. Identification of a Reliable Fixative Solution to Preserve the Complex Architecture of Bacterial Biofilms for Scanning Electron Microscopy Evaluation. PLoS ONE 2020, 15, e0233973. [Google Scholar] [CrossRef] [PubMed]
- Kırmusaoğlu, S. The Methods for Detection of Biofilm and Screening Antibiofilm Activity of Agents. In Antimicrobials, Antibiotic Resistance, Antibiofilm Strategies and Activity Methods; IntechOpen: London, UK, 2019; Volume 11, p. 18. ISBN 0000957720. [Google Scholar]
- Cruz, C.D.; Shah, S.; Tammela, P. Defining Conditions for Biofilm Inhibition and Eradication Assays for Gram-Positive Clinical Reference Strains. BMC Microbiol. 2018, 18, 173. [Google Scholar] [CrossRef] [PubMed]
- Mariscal, A.; Lopez-Gigosos, R.M.; Carnero-Varo, M.; Fernandez-Crehuet, J. Fluorescent Assay Based on Resazurin for Detection of Activity of Disinfectants against Bacterial Biofilm. Appl. Microbiol. Biotechnol. 2009, 82, 773–783. [Google Scholar] [CrossRef]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (Resazurin) Fluorescent Dye for the Assessment of Mammalian Cell Cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef]
- Goegan, P.; Johnson, G.; Vincent, R. Effects of Serum Protein and Colloid on the AlamarBlue Assay in Cell Cultures. Toxicol. In Vitro 1995, 9, 257–266. [Google Scholar] [CrossRef]
- Invitrogen AlamarBlue® Assay. U.S. Patent No 5501959, 16 July 2007. pp. 1–27.
- Mohamed, J.A.; Huang, D.B. Biofilm Formation by Enterococci. J. Med. Microbiol. 2007, 56, 1581–1588. [Google Scholar] [CrossRef]
- Nakazawa, M. fmsb: Functions for Medical Statistics Book with Some Demographic Data. R package Version 0.7.6. 2024. Available online: https://CRAN.R-project.org/package=fmsb (accessed on 15 July 2024).
- Rolf, F.J. Numerical Taxonomy and Multivariate Analysis System. Exet. Softw. 1993, 1, 80. [Google Scholar]
- Jollife, I.T.; Cadima, J. Principal Component Analysis: A Review and Recent Developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.I.; Prichula, J.; Santesteva, N.A.; d’Azevedo, P.A.; Motta, A.S.; Frazzon, A.P.G. Virulence Profiles in Enterococcus Spp. Isolated from Raw Buffalo’s Milk in South Brazil. Res. J. Microbiol. 2017, 12, 248–254. [Google Scholar] [CrossRef]
- Marinho, A.R.; Martins, P.D.; Ditmer, E.M.; d’Azevedo, P.A.; Frazzon, J.; Der Sand, S.T.V.; Frazzon, A.P.G. Biofilm Formation on Polystyrene under Different Temperatures by Antibiotic Resistant Enterococcus faecalis and Enterococcus faecium Isolated from Food. Brazilian J. Microbiol. 2013, 44, 423–426. [Google Scholar] [CrossRef]
- Chotinantakul, K.; Chansiw, N.; Okada, S. Biofilm Formation and Transfer of a Streptomycin Resistance Gene in Enterococci from Fermented Pork. J. Glob. Antimicrob. Resist. 2020, 22, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Maurya, A.; Kumar, R.; Singh, A.; Raj, A. Investigation on Biofilm Formation Activity of Enterococcus faecium under Various Physiological Conditions and Possible Application in Bioremediation of Tannery Effluent. Bioresour. Technol. 2021, 339, 125586. [Google Scholar] [CrossRef] [PubMed]
- Solheim, M.; La Rosa, S.L.; Mathisen, T.; Snipen, L.G.; Nes, I.F.; Brede, D.A. Transcriptomic and Functional Analysis of NaCl-Induced Stress in Enterococcus faecalis. PLoS ONE 2014, 9, e0094571. [Google Scholar] [CrossRef] [PubMed]
- Yilema, A.; Moges, F.; Tadele, S.; Endris, M.; Kassu, A.; Abebe, W.; Ayalew, G. Isolation of Enterococci, Their Antimicrobial Susceptibility Patterns and Associated Factors among Patients Attending at the University of Gondar Teaching Hospital. BMC Infect. Dis. 2017, 17, 276. [Google Scholar] [CrossRef]
- Hu, L.; Dong, Q.; Li, Z.; Ma, Y.; Aslam, M.Z.; Liu, Y. Modelling the Adhesion and Biofilm Formation Boundary of Listeria monocytogenes ST9. Foods 2022, 11, 1940. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Jeon, H.R.; Kwon, M.J.; Yoon, K.S. Control of Listeria Innocua Biofilms on Food Contact Surfaces with Slightly Acidic Electrolyzed Water and the Risk of Biofilm Cells Transfer to Duck Meat. J. Food Prot. 2018, 81, 582–592. [Google Scholar] [CrossRef]
- ASAE. What Is HACCP. Available online: https://www.asae.gov.pt/seguranca-food/haccp.aspx (accessed on 5 July 2024).
- EUR-Lex Regulation (EC) No. 1935/2004 on Materials and Articles Intended to Come into Contact with Food and Repealing Directives 80/590/EEC and 89/109/EEC. Available online: http://data.europa.eu/eli/reg/2004/1935/2021-03-27 (accessed on 5 July 2024).
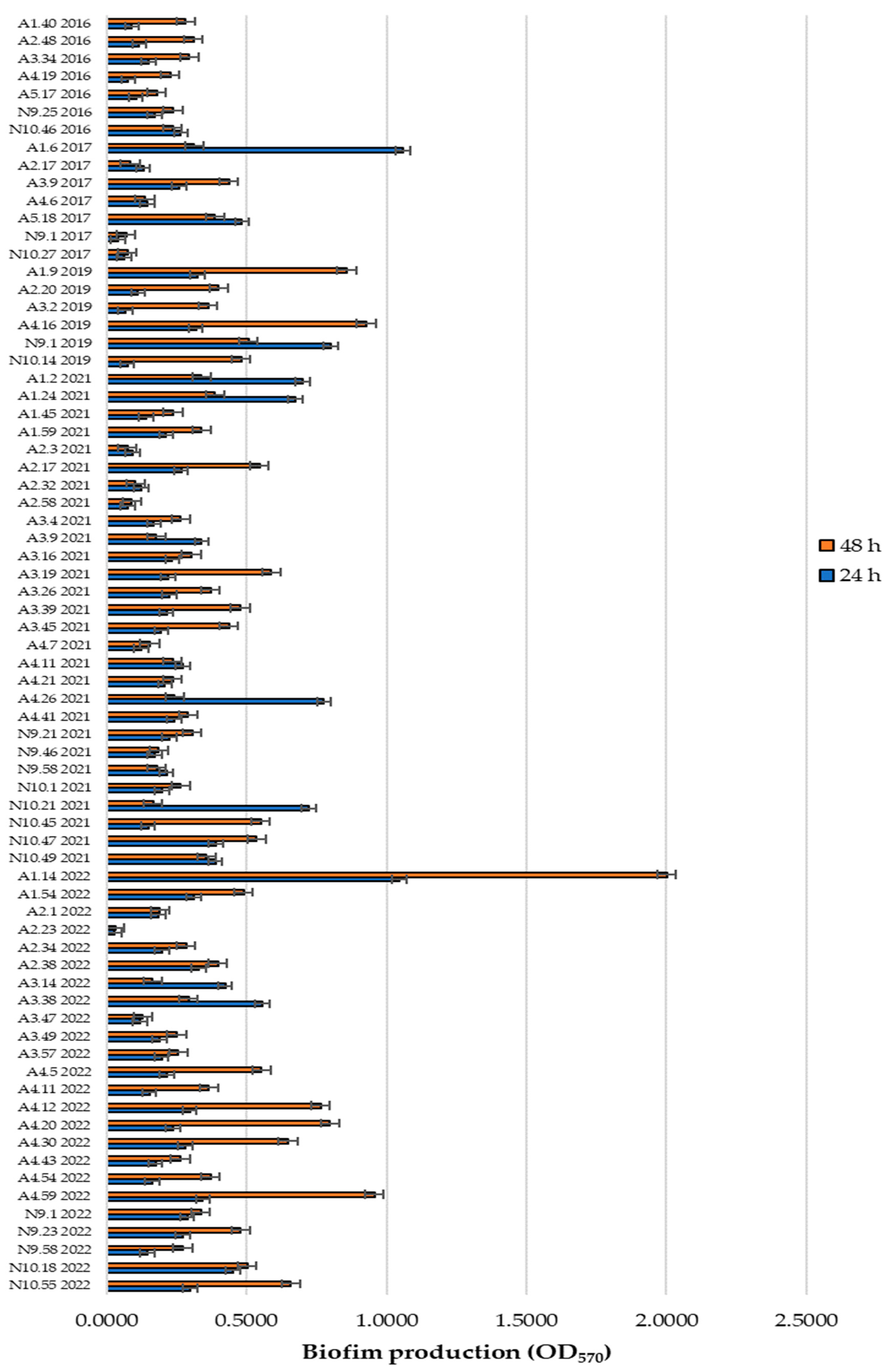
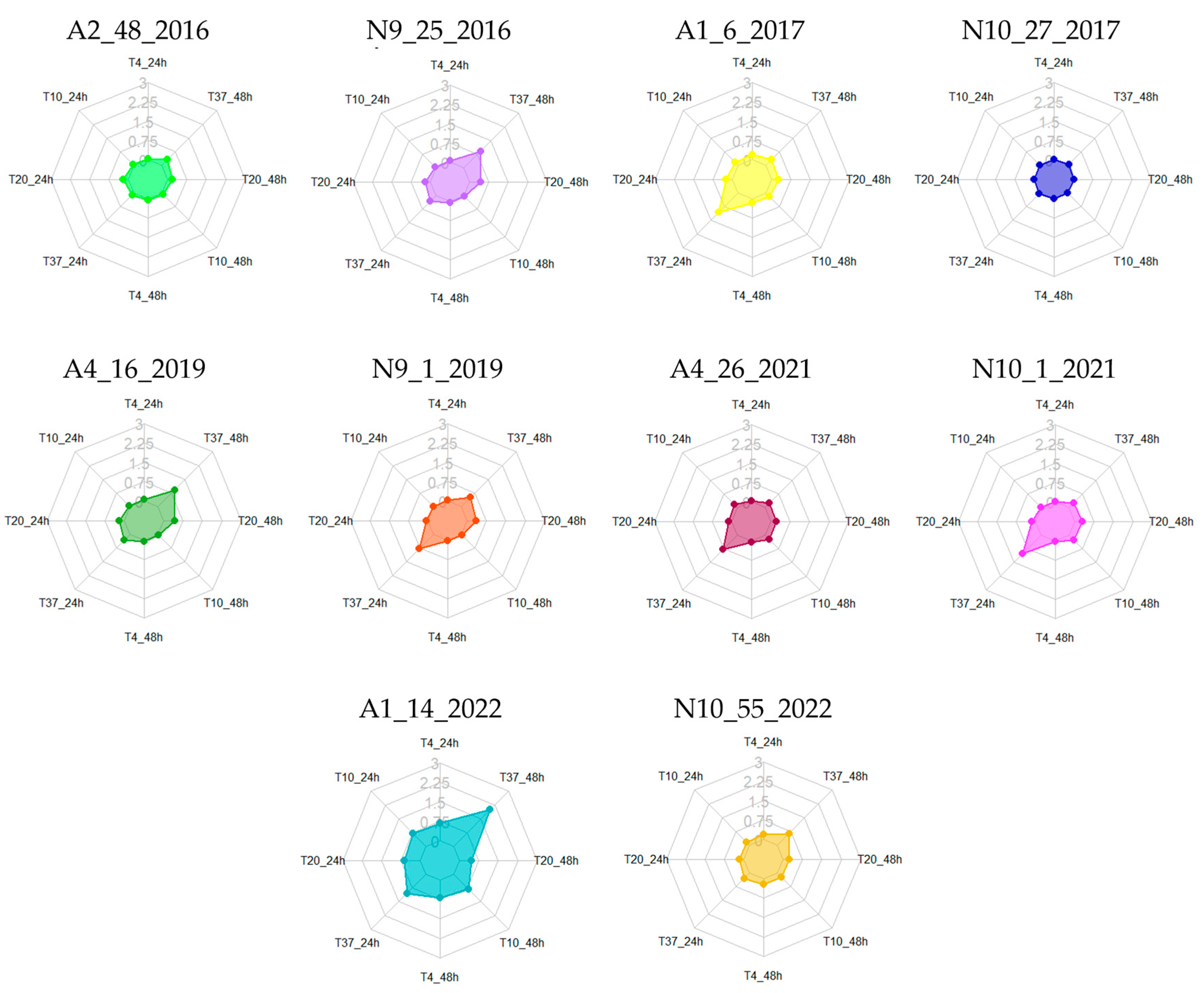
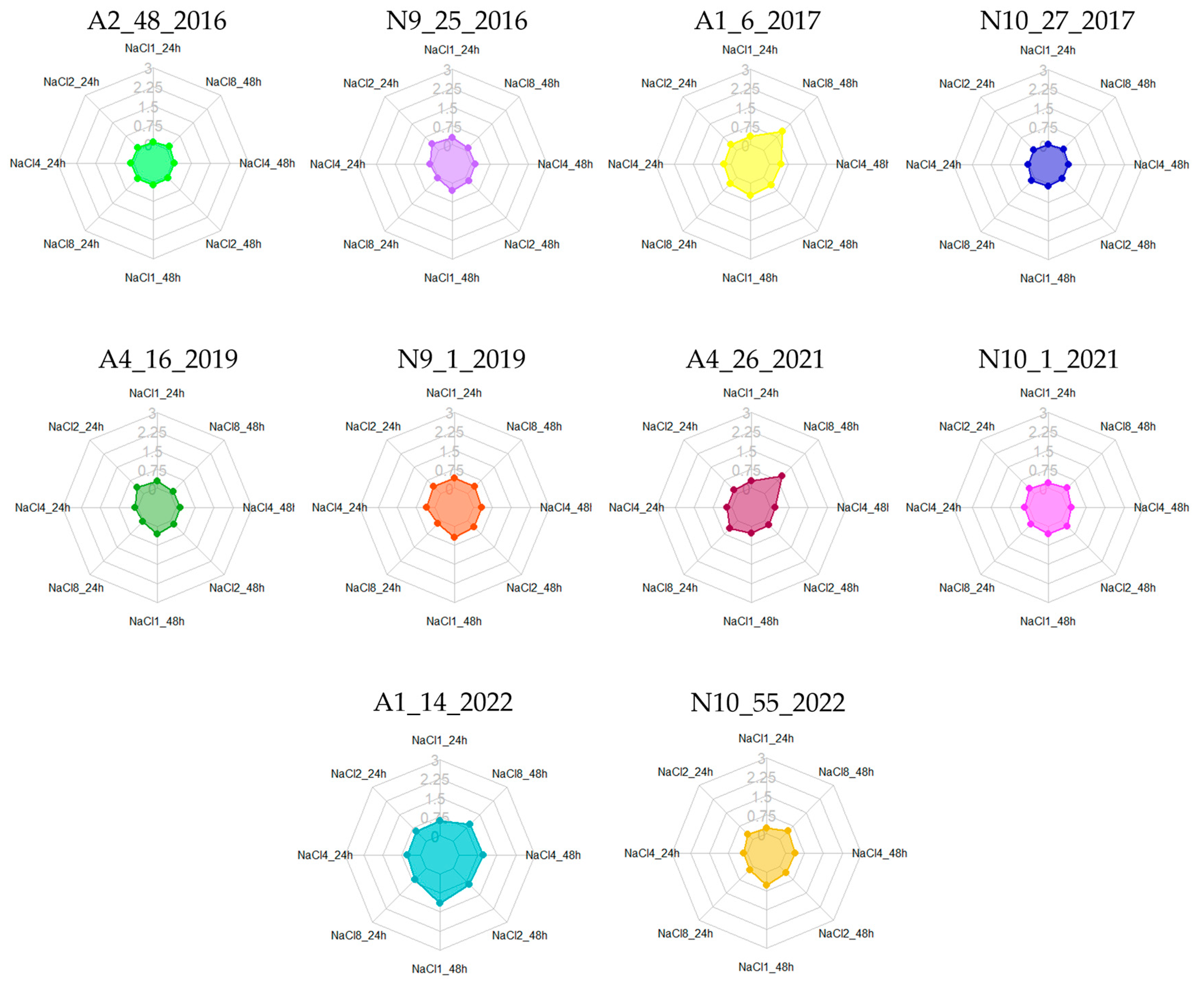

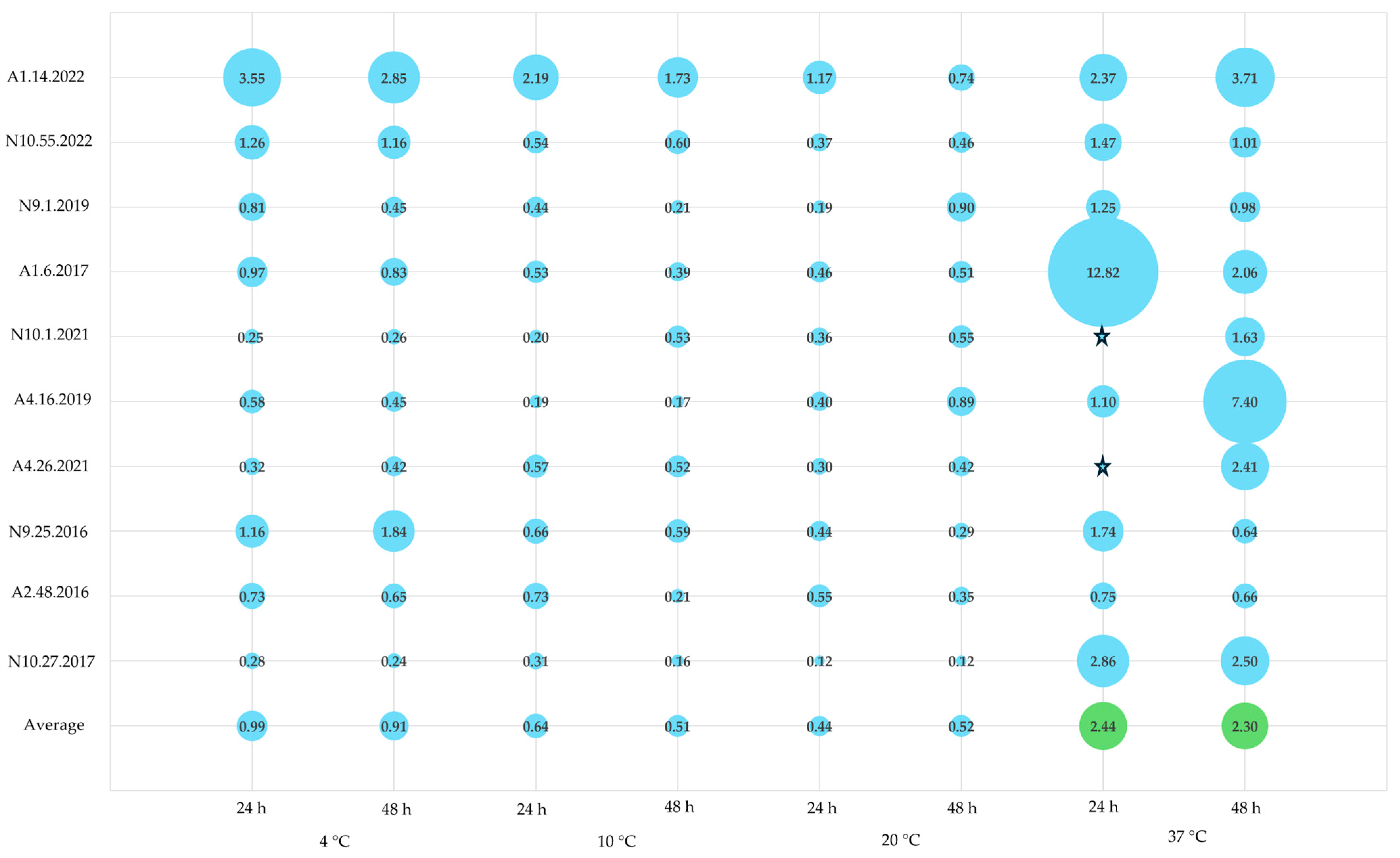


| Isolates | 24 h | 48 h | Isolates | 24 h | 48 h | Isolates | 24 h | 48 h | Isolates | 24 h | 48 h |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1.40 2016 | N.P. | W.P. | N9.1 2019 | M.P. | M.P. | A4.11 2021 | W.P. | N.P. | A3.14 2022 | W.P. | N.P. |
| A2.48 2016 | N.P. | W.P. | N10.14 2019 | N.P. | M.P. | A4.21 2021 | N.P. | N.P. | A3.38 2022 | M.P. | W.P. |
| A3.34 2016 | N.P. | W.P. | A1.2 2021 | M.P. | W.P. | A4.26 2021 | M.P. | W.P. | A3.47 2022 | N.P. | N.P. |
| A4.19 2016 | N.P. | N.P. | A1.24 2021 | M.P. | W.P. | A4.41 2021 | W.P. | W.P. | A3.49 2022 | N.P. | W.P. |
| A5.17 2016 | N.P. | N.P. | A1.45 2021 | N.P. | W.P. | N9.21 2021 | N.P. | W.P. | A3.57 2022 | N.P. | W.P. |
| N9.25 2016 | N.P. | W.P. | A1.59 2021 | N.P. | W.P. | N9.46 2021 | N.P. | N.P. | A4.5 2022 | N.P. | M.P. |
| N10.46 2016 | W.P. | N.P. | A2.3 2021 | N.P. | N.P. | N9.58 2021 | N.P. | N.P. | A4.11 2022 | N.P. | W.P. |
| A1.6 2017 | S.P. | W.P. | A2.17 2021 | W.P. | M.P. | N10.1 2021 | N.P. | W.P. | A4.12 2022 | W.P. | M.P. |
| A2.17 2017 | N.P. | N.P. | A2.32 2021 | N.P. | N.P. | N10.21 2021 | M.P. | N.P. | A4.20 2022 | W.P. | M.P. |
| A3.9 2017 | W.P. | W.P. | A2.58 2021 | N.P. | N.P. | N10.45 2021 | N.P. | M.P. | A4.30 2022 | W.P. | M.P. |
| A4.6 2017 | N.P. | N.P. | A3.4 2021 | N.P. | W.P. | N10.47 2021 | W.P. | M.P. | A4.43 2022 | N.P. | W.P. |
| A5.18 2017 | M.P. | W.P. | A3.9 2021 | W.P. | N.P. | N10.49 2021 | W.P. | W.P. | A4.54 2022 | N.P. | W.P. |
| N9.1 2017 | N.P. | N.P. | A3.16 2021 | N.P. | W.P. | A1.14 2022 | S.P. | S.P. | A4.59 2022 | W.P. | W.P. |
| N10.27 2017 | N.P. | N.P. | A3.19 2021 | N.P. | M.P. | A1.54 2022 | W.P. | M.P. | N9.1 2022 | W.P. | W.P. |
| A1.9 2019 | W.P. | M.P. | A3.26 2021 | N.P. | W.P. | A2.1 2022 | N.P. | N.P. | N9.23 2022 | W.P. | M.P. |
| A2.20 2019 | N.P. | W.P. | A3.39 2021 | N.P. | M.P. | A2.23 2022 | N.P. | N.P. | N9.58 2022 | N.P. | W.P. |
| A3.2 2019 | N.P. | W.P. | A3.45 2021 | N.P. | W.P. | A2.34 2022 | N.P. | W.P. | N10.18 2022 | W.P. | M.P. |
| A4.16 2019 | W.P. | M.P. | A4.7 2021 | N.P. | N.P. | A2.38 2022 | W.P. | W.P. | N10.55 2022 | W.P. | M.P. |
| 48 h | N.P. | W.P. | M.P. | S.P. | |
|---|---|---|---|---|---|
| 24 h | |||||
| N.P. | N9.25.2016 N10.27.2017 | A2.48.2016 N10.1.2021 | |||
| W.P. | A4.16.2019 N10.55.2022 | ||||
| M.P. | A4.26.2021 | N9.1.2019 | |||
| S.P. | A1.6.2017 | A1.14.2022 | |||
| Temperature | ||||||||
|---|---|---|---|---|---|---|---|---|
| 4 °C | 10 °C | 20 °C | 37 °C | |||||
| Isolates | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h |
| A2.48.2016 | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | W.P. |
| N9.25.2016 | N.P. | W.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. |
| A1.6.2017 | N.P. | N.P. | N.P. | N.P. | W.P. | W.P. | W.P. | W.P. |
| N10.27.2017 | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. |
| A4.16.2019 | N.P. | N.P. | N.P. | N.P. | N.P. | W.P. | W.P. | M.P. |
| N9.1.2019 | N.P. | N.P. | N.P. | N.P. | N.P. | W.P. | M.P. | M.P. |
| A4.26.2021 | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | M.P. | W.P. |
| N10.1.2021 | N.P. | N.P. | N.P. | W.P. | N.P. | W.P. | N.P. | W.P. |
| A1.14.2022 | M.P. | M.P. | M.P. | M.P. | M.P. | W.P. | S.P. | S.P. |
| N10.55.2022 | N.P. | N.P. | N.P. | N.P. | N.P. | W.P. | W.P. | M.P. |
| NaCl Concentration | ||||||||
|---|---|---|---|---|---|---|---|---|
| 1% | 2% | 4% | 8% | |||||
| Isolates | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h |
| A2.48.2016 | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. |
| N9.25.2016 | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | W.P. | W.P. |
| A1.6.2017 | W.P. | M.P. | W.P. | W.P. | W.P. | W.P. | W.P. | S.P. |
| N10.27.2017 | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. |
| A4.16.2019 | W.P. | W.P. | W.P. | N.P. | N.P. | N.P. | N.P. | N.P. |
| N9.1.2019 | W.P. | W.P. | W.P. | W.P. | W.P. | W.P. | N.P. | W.P. |
| A4.26.2021 | W.P. | W.P. | N.P. | N.P. | N.P. | N.P. | W.P. | S.P. |
| N10.1.2021 | N.P. | W.P. | W.P. | W.P. | N.P. | N.P. | N.P. | W.P. |
| A1.14.2022 | M.P. | S.P. | M.P. | M.P. | M.P. | S.P. | M.P. | M.P. |
| N10.55.2022 | N.P. | M.P. | W.P. | W.P. | N.P. | W.P. | N.P. | M.P. |
| pH Values | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pH 5 | pH 6 | pH 7 | pH 8 | pH 9 | ||||||
| Isolates | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h |
| A2.48.2016 | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | W.P. | N.P. | N.P. |
| N9.25.2016 | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | M.P. | N.P. | W.P. |
| A1.6.2017 | W.P. | W.P. | W.P. | W.P. | W.P. | W.P. | W.P. | W.P. | W.P. | W.P. |
| N10.27.2017 | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. | N.P. |
| A4.16.2019 | N.P. | W.P. | W.P. | W.P. | W.P. | W.P. | W.P. | M.P. | W.P. | M.P. |
| N9.1.2019 | M.P. | M.P. | W.P. | W.P. | S.P. | M.P. | W.P. | W.P. | W.P. | W.P. |
| A4.26.2021 | N.P. | W.P. | N.P. | N.P. | W.P. | N.P. | N.P. | W.P. | W.P. | N.P. |
| N10.1.2021 | W.P. | M.P. | W.P. | W.P. | W.P. | W.P. | W.P. | W.P. | W.P. | W.P. |
| A1.14.2022 | W.P. | M.P. | M.P. | M.P. | M.P. | S.P. | M.P. | S.P. | S.P. | S.P. |
| N10.55.2022 | N.P. | W.P. | N.P. | W.P. | W.P. | W.P. | N.P. | M.P. | M.P. | S.P. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serrano, S.; Ferreira, M.V.; Alves-Barroco, C.; Morais, S.; Barreto-Crespo, M.T.; Tenreiro, R.; Semedo-Lemsaddek, T. Beyond Harmful: Exploring Biofilm Formation by Enterococci Isolated from Portuguese Traditional Cheeses. Foods 2024, 13, 3067. https://doi.org/10.3390/foods13193067
Serrano S, Ferreira MV, Alves-Barroco C, Morais S, Barreto-Crespo MT, Tenreiro R, Semedo-Lemsaddek T. Beyond Harmful: Exploring Biofilm Formation by Enterococci Isolated from Portuguese Traditional Cheeses. Foods. 2024; 13(19):3067. https://doi.org/10.3390/foods13193067
Chicago/Turabian StyleSerrano, Susana, Maria Vitória Ferreira, Cinthia Alves-Barroco, Susana Morais, Maria Teresa Barreto-Crespo, Rogério Tenreiro, and Teresa Semedo-Lemsaddek. 2024. "Beyond Harmful: Exploring Biofilm Formation by Enterococci Isolated from Portuguese Traditional Cheeses" Foods 13, no. 19: 3067. https://doi.org/10.3390/foods13193067






