Image Classification and Automated Machine Learning to Classify Lung Pathologies in Deceased Feedlot Cattle
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
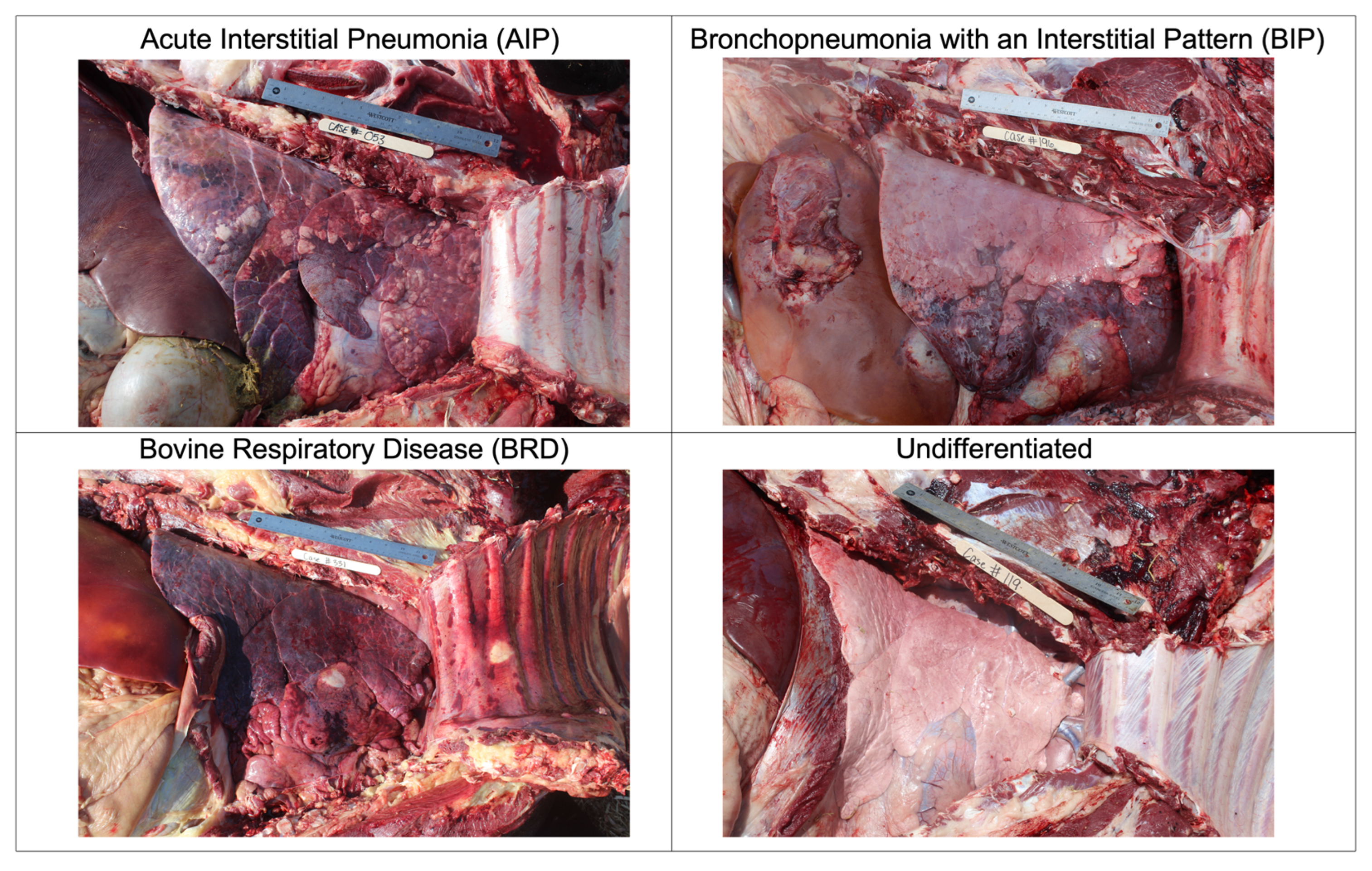
2.1. Images
2.2. Data Labeling
2.3. Modeling
3. Results
3.1. Descriptive Statistics
3.2. Image Classification Models Performance
3.2.1. Best Model Accuracies
3.2.2. Area under the Curve
3.2.3. Sensitivity, Specificity, and Accuracy
3.2.4. Positive and Negative Predictive Values
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- USDA APHIS-VS. National Animal Health Monitoring System Beef Feedlot Study 2011. In Types and Cost of Respiratory Disease Treatments in U.S. Feedlots; United States Department of Agriculture: Fort Collins, CO, USA, 2013. [Google Scholar]
- Haydock, L.A.J.; Fenton, R.K.; Sergejewich, L.; Squires, E.J.; Caswell, J.L. Acute Interstitial Pneumonia and the Biology of 3-Methylindole in Feedlot Cattle. Anim. Health Res. Rev. 2022, 23, 72–81. [Google Scholar] [CrossRef]
- Griffin, D. Economic Impact Associated with Respiratory Disease in Beef Cattle. Vet. Clin. North Am. Food Anim. Pract. 1997, 13, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Faber, R.; Hartwig, N. The Costs and Predictive Factors of Bovine Respiratory Disease in Standardized Steer Tests. Iowa State Univ. Anim. Ind. Rep. 2000. Available online: https://www.iastatedigitalpress.com/air/article/id/7342/ (accessed on 25 December 2022).
- Ayroud, M.; Popp, J.D.; A VanderKop, M.; Yost, G.S.; Haines, D.M.; Majak, W.; Karren, D.; Yanke, L.J.; A McAllister, T. Characterization of Acute Interstitial Pneumonia in Cattle in Southern Alberta Feedyards. Can. Vet. J. 2000, 41, 547–554. [Google Scholar] [PubMed]
- Doster, A.R. Bovine Atypical Interstitial Pneumonia. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Wolfger, B.; Manns, B.J.; Barkema, H.W.; Schwartzkopf-Genswein, K.S.; Dorin, C.; Orsel, K. Evaluating the Cost Implications of a Radio Frequency Identification Feeding System for Early Detection of Bovine Respiratory Disease in Feedlot Cattle. Prev. Vet. Med. 2015, 118, 285–292. [Google Scholar] [CrossRef]
- Fiore, E.; Lisuzzo, A.; Beltrame, A.; Contiero, B.; Gianesella, M.; Schiavon, E.; Tessari, R.; Morgante, M.; Mazzotta, E. Lung Ultrasonography and Clinical Follow-Up Evaluations in Fattening Bulls Affected by Bovine Respiratory Disease (BRD) during the Restocking Period and after Tulathromycin and Ketoprofen Treatment. Animals 2022, 12, 994. [Google Scholar] [CrossRef]
- White, B.J.; Renter, D.G. Bayesian Estimation of the Performance of Using Clinical Observations and Harvest Lung Lesions for Diagnosing Bovine Respiratory Disease in Post-Weaned Beef Calves. J. Vet. Diagn. Investig. 2009, 21, 446–453. [Google Scholar] [CrossRef]
- Haydock, L.A.J.; Fenton, R.K.; Smerek, D.; Renaud, D.L.; Caswell, J.L. Bronchopneumonia with Interstitial Pneumonia in Feedlot Cattle: Epidemiologic Characteristics of Affected Animals. Vet. Pathol. 2023, 03009858221146096. [Google Scholar] [CrossRef]
- Griffin, D. Feedlot Euthanasia and Necropsy. Vet. Clin. N. Am. Food Anim. Pract. 2015, 31, 465–482. [Google Scholar] [CrossRef]
- Küker, S.; Faverjon, C.; Furrer, L.; Berezowski, J.; Posthaus, H.; Rinaldi, F.; Vial, F. The Value of Necropsy Reports for Animal Health Surveillance. BMC Vet. Res. 2018, 14, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panciera, R.J.; Confer, A.W. Pathogenesis and Pathology of Bovine Pneumonia. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 191–214. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.; Ferreras, M.D.C.; Giráldez, F.J.; Benavides, J.; Pérez, V. Production Significance of Bovine Respiratory Disease Lesions in Slaughtered Beef Cattle. Animals 2020, 10, 1770. [Google Scholar] [CrossRef]
- Wikse, S.E. Feedlot Cattle Pneumonias. Vet. Clin. N. Am. Food Anim. Pract. 1985, 1, 289–310. [Google Scholar] [CrossRef] [PubMed]
- Jensen, R.; E Pierson, R.; Braddy, P.M.; A Saari, D.; Lauerman, L.H.; England, J.J.; Benitez, A.; Horton, D.P.; E McChesney, A. Atypical Interstitial Pneumonia in Yearling Feedlot Cattle. J. Am. Vet. Med. Assoc. 1976, 169, 507–510. [Google Scholar]
- Woolums, A.R.; Mason, G.L.; Hawkins, L.L.; Brown, C.C.; Williams, S.M.; Gould, J.A.; Fox, J.J.; Sturgeon, S.D.; Anderson, J.L.; Duggan, F.E.; et al. Microbiologic Findings in Feedlot Cattle with Acute Interstitial Pneumonia. Am. J. Vet. Res. 2004, 65, 1525–1532. [Google Scholar] [CrossRef]
- Haydock, L.A.J.; Fenton, R.K.; Sergejewich, L.; Veldhuizen, R.A.W.; Smerek, D.; Ojkic, D.; Caswell, J.L. Bronchopneumonia with Interstitial Pneumonia in Beef Feedlot Cattle: Characterization and Laboratory Investigation. Vet. Pathol. 2023, 13, 030098582211460. [Google Scholar] [CrossRef]
- Wildman, B.K.; Schunicht, O.C.; Jim, G.K.; Guichon, P.T.; Booker, C.W.; A Tollens, R. The Use of Computer Imaging Technology to Facilitate the Capture of Feedlot Necropsy Information. Can. Vet. J. 2000, 41, 124–125. [Google Scholar]
- McEvoy, F.J.; Amigo, J.M. Using Machine Learning to Classify Image Features from Canine Pelvic Radiographs: Evaluation of Partial Least Squares Discriminant Analysis and Artificial Neural Network Models: Image Classification Using Machine Learning. Vet. Radiol. Ultrasound 2013, 54, 122–126. [Google Scholar] [CrossRef]
- Marschner, C.B.; Kokla, M.; Amigo, J.M.; Rozanski, E.A.; Wiinberg, B.; McEvoy, F.J. Texture analysis of pulmonary parenchymateous changes related to pulmonary thromboembolism in dogs—a novel approach using quantitative methods. BMC Vet. Res. 2017, 13, 219. [Google Scholar] [CrossRef]
- Arsomngern, P.; Numcharoenpinij, N.; Piriyataravet, J.; Teerapan, W.; Hinthong, W.; Phunchongharn, P. Computer-Aided Diagnosis for Lung Lesion in Companion Animals from X-Ray Images Using Deep Learning Techniques. In Proceedings of the 2019 IEEE 10th International Conference on Awareness Science and Technology (iCAST), Morioka, Japan, 23–25 October 2019; pp. 1–6. [Google Scholar]
- Biercher, A.; Meller, S.; Wendt, J.; Caspari, N.; Schmidt-Mosig, J.; De Decker, S.; Volk, H.A. Using Deep Learning to Detect Spinal Cord Diseases on Thoracolumbar Magnetic Resonance Images of Dogs. Front. Vet. Sci. 2021, 8, 721167. [Google Scholar] [CrossRef] [PubMed]
- Hennessey, E.; DiFazio, M.; Hennessey, R.; Cassel, N. Artificial Intelligence in Veterinary Diagnostic Imaging: A Literature Review. Vet. Radiol. Ultrasound 2022, 63, 851–870. [Google Scholar] [CrossRef] [PubMed]
- Pesapane, F.; Codari, M.; Sardanelli, F. Artificial Intelligence in Medical Imaging: Threat or Opportunity? Radiologists again at the Forefront of Innovation in Medicine. Eur. Radiol. Exp. 2018, 2, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garland, J.; Hu, M.; Kesha, K.; Glenn, C.; Morrow, P.; Stables, S.; Ondruschka, B.; Tse, R. Identifying Gross Post-Mortem Organ Images Using a Pre-Trained Convolutional Neural Network. J. Forensic Sci. 2021, 66, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Chien, R.C.; Sorensen, N.J.; Payton, M.E.; Confer, A.W. Comparative Histopathology of Bovine Acute Interstitial Pneumonia and Bovine Respiratory Syncytial Virus-Associated Interstitial Pneumonia. J. Comp. Pathol. 2022, 192, 23–32. [Google Scholar] [CrossRef]
- Woolums, A.R. Feedlot Acute Interstitial Pneumonia. Vet. Clin. N. Am. Food Anim. Pract. 2015, 31, 381–389. [Google Scholar] [CrossRef]
- Peng, H. Bioimage informatics: A new Area of Engineering Biology. Bioinformatics 2008, 24, 1827–1836. [Google Scholar] [CrossRef] [PubMed]
- Shamir, L.; Delaney, J.D.; Orlov, N.; Eckley, D.M.; Goldberg, I.G. Pattern Recognition Software and Techniques for Biological Image Analysis. PLoS Comput. Biol. 2010, 6, e1000974. [Google Scholar] [CrossRef]
- Fernandes, A.F.A.; Dórea, J.R.R.; Rosa, G.J.D.M. Image Analysis and Computer Vision Applications in Animal Sciences: An Overview. Front. Vet. Sci. 2020, 7, 551269. [Google Scholar] [CrossRef]
- Fernandes, A.F.; Turra, E.; de Alvarenga, R.; Passafaro, T.L.; Lopes, F.B.; Alves, G.F.; Singh, V.; Rosa, G.J. Deep Learning Image Segmentation for Extraction of Fish Body Measurements and Prediction of Body Weight and Carcass Traits in Nile Tilapia. Comput. Electron. Agric. 2020, 170, 105274. [Google Scholar] [CrossRef]
- Kashiha, M.; Bahr, C.; Ott, S.; Moons, C.P.; Niewold, T.A.; Ödberg, F.; Berckmans, D. Automatic Identification of Marked Pigs in a Pen Using Image Pattern Recognition. Comput. Electron. Agric. 2013, 93, 111–120. [Google Scholar] [CrossRef]



| Dataset | n 1 | BIP 2 | AIP 3 | BRD 4 | Undifferentiated 5 |
|---|---|---|---|---|---|
| Unaltered images gross diagnoses | 398 | 141 | 44 | 148 | 65 |
| Cropped images gross diagnoses | 318 | 120 | 41 | 113 | 44 |
| Unaltered images histopathology diagnoses | 167 | 67 | 21 | 57 | 22 |
| Cropped images histopathology diagnosis | 149 | 61 | 16 | 55 | 17 |
| Class | n | Se% 5 | Sp% 6 | PPV% 7 | NPV 8 | AUC 9 | Accuracy |
|---|---|---|---|---|---|---|---|
| Unaltered images gross diagnoses | |||||||
| AIP 1 | 14 | 14% | 98% | 67% | 85% | 0.68 | 83% |
| BIP 2 | 23 | 65% | 68% | 45% | 82% | 0.7 | 67% |
| BRD 3 | 29 | 69% | 52% | 45% | 75% | 0.65 | 58% |
| Undifferentiated 4 | 14 | 0% | 100% | 0% | 82% | 0.79 | 82% |
| Cropped images gross diagnoses | |||||||
| AIP | 8 | 0% | 100% | 0% | 76% | 0.44 | 76% |
| BIP | 10 | 100% | 13% | 32% | 100% | 0.61 | 38% |
| BRD | 10 | 20% | 100% | 100% | 75% | 0.71 | 76% |
| Undifferentiated | 6 | 17% | 100% | 100% | 84% | 0.76 | 85% |
| Unaltered images histopathology diagnoses | |||||||
| AIP | 3 | 23% | 98% | 75% | 83% | 0.46 | 83% |
| BIP | 30 | 72% | 61% | 50% | 81% | 0.71 | 65% |
| BRD | 23 | 31% | 80% | 40% | 73% | 0.63 | 65% |
| Undifferentiated | 8 | 70% | 89% | 54% | 94% | 0.91 | 86% |
| Cropped images histopathology diagnosis | |||||||
| AIP | 3 | 0% | 96% | 0% | 89% | 0.66 | 86% |
| BIP | 10 | 60% | 60% | 42% | 75% | 0.49 | 60% |
| BRD | 15 | 60% | 73% | 69% | 64% | 0.58 | 66% |
| Undifferentiated | 2 | 0% | 92% | 0% | 92% | 0.51 | 86% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bortoluzzi, E.M.; Schmidt, P.H.; Brown, R.E.; Jensen, M.; Mancke, M.R.; Larson, R.L.; Lancaster, P.A.; White, B.J. Image Classification and Automated Machine Learning to Classify Lung Pathologies in Deceased Feedlot Cattle. Vet. Sci. 2023, 10, 113. https://doi.org/10.3390/vetsci10020113
Bortoluzzi EM, Schmidt PH, Brown RE, Jensen M, Mancke MR, Larson RL, Lancaster PA, White BJ. Image Classification and Automated Machine Learning to Classify Lung Pathologies in Deceased Feedlot Cattle. Veterinary Sciences. 2023; 10(2):113. https://doi.org/10.3390/vetsci10020113
Chicago/Turabian StyleBortoluzzi, Eduarda M., Paige H. Schmidt, Rachel E. Brown, Makenna Jensen, Madeline R. Mancke, Robert L. Larson, Phillip A. Lancaster, and Brad J. White. 2023. "Image Classification and Automated Machine Learning to Classify Lung Pathologies in Deceased Feedlot Cattle" Veterinary Sciences 10, no. 2: 113. https://doi.org/10.3390/vetsci10020113









