Sarcolipin Exhibits Abundant RNA Transcription and Minimal Protein Expression in Horse Gluteal Muscle
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequences
2.2. Animals and Samples
2.3. Muscle Fiber Type Composition
2.4. RNA Sequencing of the Horse Muscle Transcriptome
2.5. RNA-seq Quantitation of Gene Expression in Horse and Rabbit Muscle
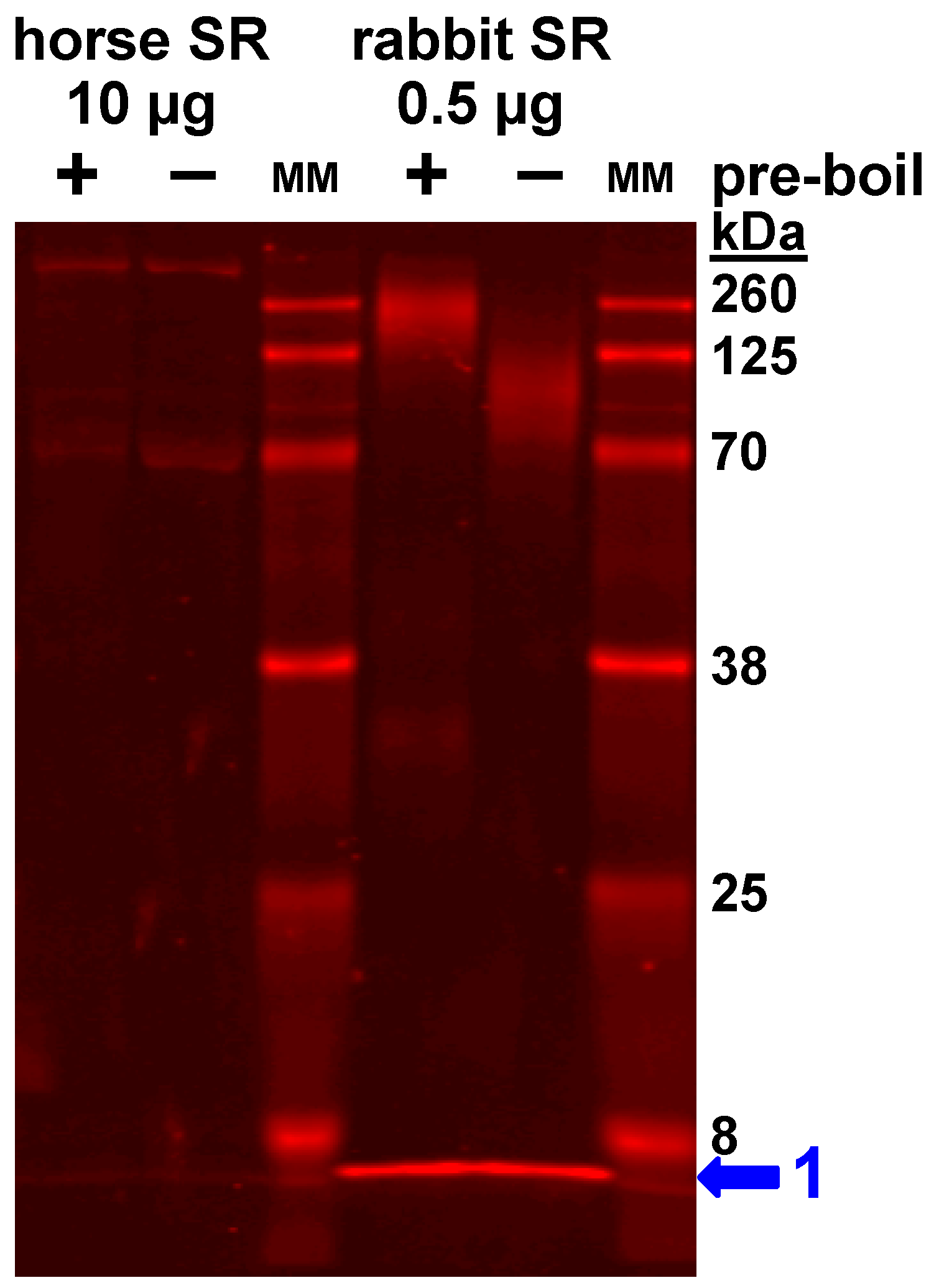
2.6. Purification of SR Vesicles from Horse Gluteal Muscle
2.7. Purification of SR Vesicles from Rabbit Skeletal Muscle
2.8. Synthesis of SLN Peptide Orthologs for Quantitative Immunoblotting
2.9. Quantitative Immunoblotting of Muscle Proteins
2.10. Secondary Immunolabeling and Target Quantitation Using Fluorescent Anti-IgG Antibodies
2.11. Custom Anti-Horse-SLN Polyclonal Antibody GS3379
2.12. Custom C-Terminal Anti-Rabbit/Mouse/Human-SLN Polyclonal Antibody PFD-1
2.13. Commercial Anti-SLN Polyclonal Antibodies
2.14. Anti-PLN Monoclonal Antibody 2D12
2.15. Experimental Design, Statistical Analysis, and Data Presentation
3. Results
3.1. Muscle Fiber Type Composition
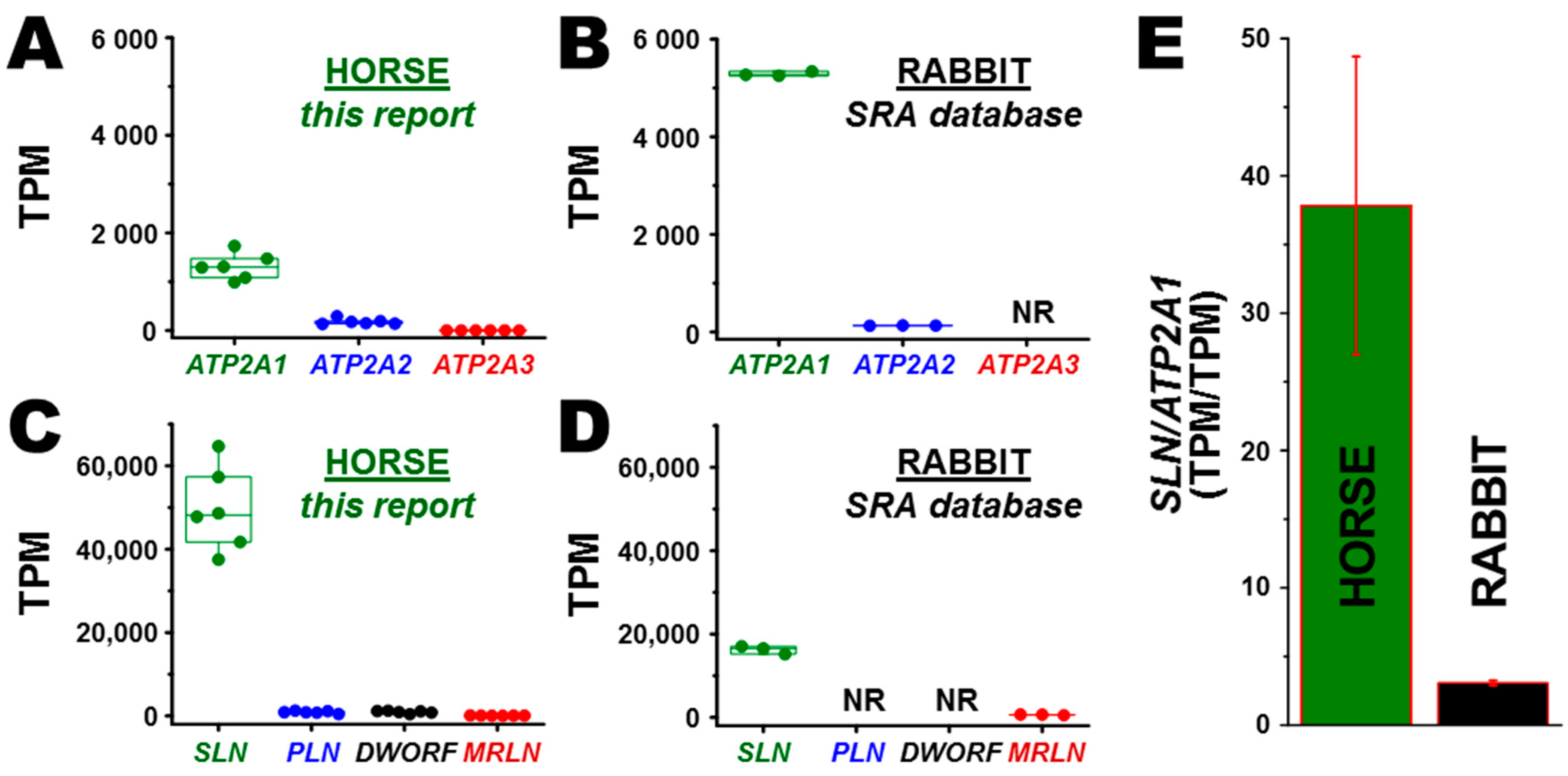
3.2. Transcription of ATP2A Genes Encoding SERCA Proteins
3.3. Transcription of SERCA Regulatory Peptide Genes: SLN, PLN, MRLN, and DWORF
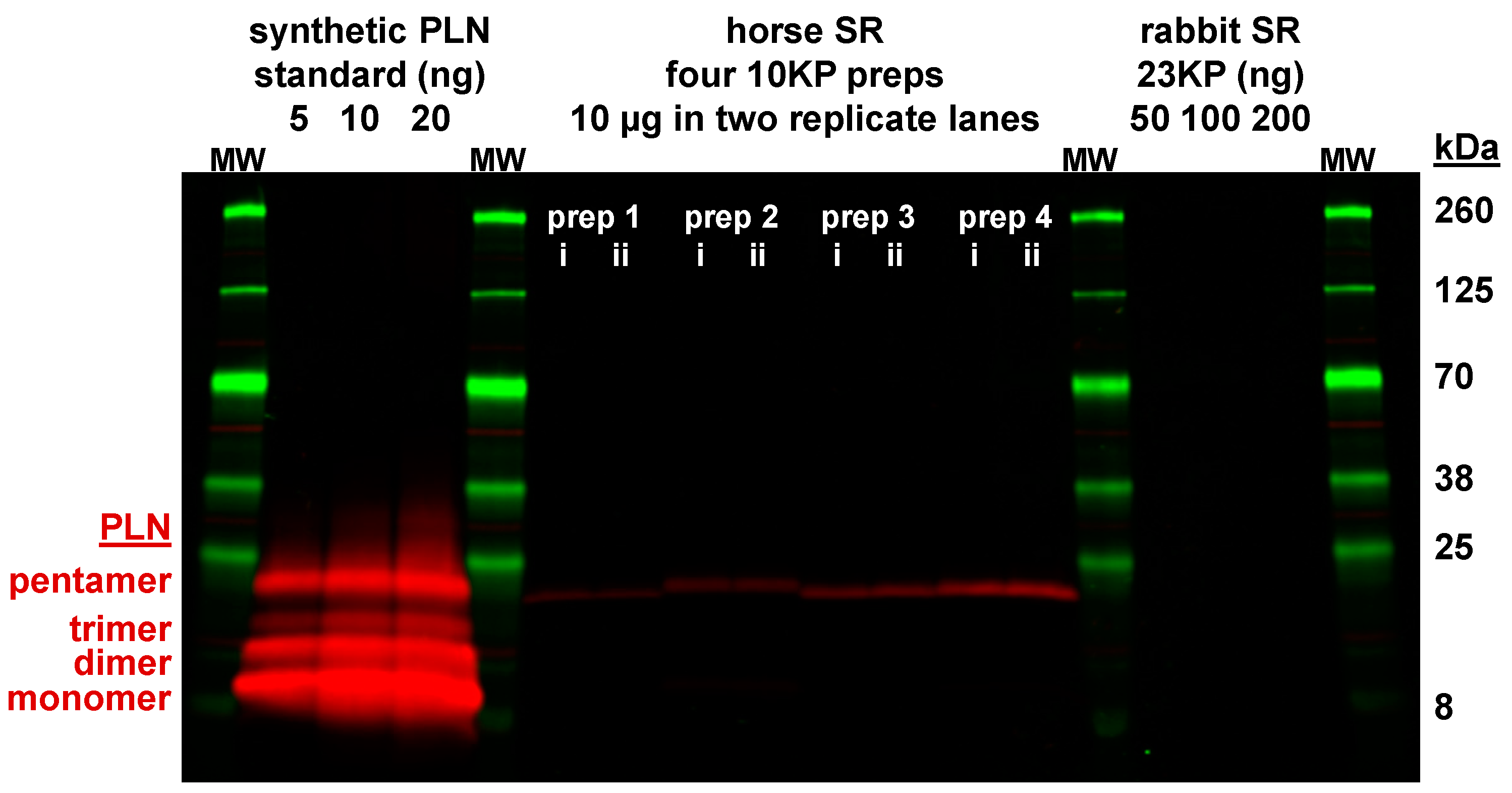
3.4. Expression of the SLN Peptide Relative to SERCA Protein in Horse SR vesicles, as Detected by Multiple Anti-SLN Antibodies
3.5. Correlation of Gene and Protein Expression of SLN in Horse and Rabbit Muscle
3.6. Horse Gluteus Expresses A Minimal Level of PLN Inhibitory Peptide Compared to SERCA Protein
4. Discussion
4.1. Gel analysis for Oligomerization of Horse and Rabbit SLN
4.2. Positive and Negative Correlations of Gene Transcription and Protein Expression of SR Ca2+ Transport Regulators in Horse Gluteal Muscle
4.3. Is There a Mammalian Striated Muscle Reported to Expresses SERCA in the Absence of Regulatory Transmembrane Peptide?
4.4. Does the SLN Gene Transcript Act as A Functional Long Non-Coding RNA for Controlling Contractility of Horse Gluteal Muscle?
4.5. Potential Mechanisms to Control SLN Protein Expression and SERCA Regulation in Horse Gluteal Muscle
4.6. Study Limitations
5. Summary
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Acronyms and Abbreviations
References
- Nissen, P. Jens Christian Skou (1918–2018). Science 2018, 361, 133. [Google Scholar]
- Møller, J.V.; Olesen, C.; Winther, A.-M.L.; Nissen, P. The sarcoplasmic Ca2+-ATPase: Design of a perfect chemi-osmotic pump. Q. Rev. Biophys. 2010, 43, 501–566. [Google Scholar] [CrossRef] [PubMed]
- MacLennan, D.H. Isolation of proteins of the sarcoplasmic reticulum. Methods Enzymol. 1974, 32, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Tada, M.; Kirchberger, M.A.; Katz, A.M. Phosphorylation of a 22,000-dalton component of the cardiac sarcoplasmic reticulum by adenosine 3′:5′-monophosphate-dependent protein kinase. J. Biol. Chem. 1975, 250, 2640–2647. [Google Scholar] [PubMed]
- Wegener, A.D.; Simmerman, H.K.; Lindemann, J.P.; Jones, L.R. Phospholamban phosphorylation in intact ventricles. Phosphorylation of serine 16 and threonine 17 in response to beta-adrenergic stimulation. J. Biol. Chem. 1989, 264, 11468–11474. [Google Scholar] [PubMed]
- Bhupathy, P.; Babu, G.J.; Ito, M.; Periasamy, M. Threonine-5 at the N-terminus can modulate sarcolipin function in cardiac myocytes. J. Mol. Cell. Cardiol. 2009, 47, 723–729. [Google Scholar] [CrossRef] [Green Version]
- Montigny, C.; Decottignies, P.; Le Maréchal, P.; Capy, P.; Bublitz, M.; Olesen, C.; Møller, J.V.; Nissen, P.; Le Maire, M. S-Palmitoylation andS-Oleoylation of Rabbit and Pig Sarcolipin. J. Biol. Chem. 2014, 289, 33850–33861. [Google Scholar] [CrossRef] [Green Version]
- Sivakumaran, V.; Stanley, B.A.; Tocchetti, C.G.; Ballin, J.D.; Caceres, V.; Zhou, L.; Keceli, G.; Rainer, P.P.; Lee, D.I.; Huke, S.; et al. HNO Enhances SERCA2a Activity and Cardiomyocyte Function by Promoting Redox-Dependent Phospholamban Oligomerization. Antioxid. Redox Signal. 2013, 19, 1185–1197. [Google Scholar] [CrossRef] [Green Version]
- Zhou, T.; Li, J.; Zhao, P.; Liu, H.; Jia, D.; Jia, H.; He, L.; Cang, Y.; Boast, S.; Chen, Y.-H.; et al. Palmitoyl acyltransferase Aph2 in cardiac function and the development of cardiomyopathy. Proc. Natl. Acad. Sci. USA 2015, 112, 15666–15671. [Google Scholar] [CrossRef] [Green Version]
- Anderson, D.M.; Anderson, K.M.; Chang, C.-L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [Green Version]
- Desmond, P.F.; Muriel, J.; Markwardt, M.L.; Rizzo, M.A.; Bloch, R.J. Identification of Small Ankyrin 1 as a Novel Sarco(endo)plasmic Reticulum Ca2+-ATPase 1 (SERCA1) Regulatory Protein in Skeletal Muscle*. J. Biol. Chem. 2015, 290, 27854–27867. [Google Scholar] [CrossRef] [Green Version]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.; Reese, A.L.; McAnally, J.R.; Chen, X.; Kavalali, E.T.; et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016, 351, 271–275. [Google Scholar] [CrossRef] [Green Version]
- Anderson, D.M.; Makarewich, C.A.; Anderson, K.M.; Shelton, J.M.; Bezprozvannaya, S.; Bassel-Duby, R.; Olson, E.N. Widespread control of calcium signaling by a family of SERCA-inhibiting micropeptides. Sci. Signal. 2016, 9, ra119. [Google Scholar] [CrossRef] [Green Version]
- Makarewich, C.A.; Munir, A.Z.; Schiattarella, G.G.; Bezprozvannaya, S.; Raguimova, O.N.; Cho, E.E.; Vidal, A.H.; Robia, S.L.; Bassel-Duby, R.; Olson, E.N. The DWORF micropeptide enhances contractility and prevents heart failure in a mouse model of dilated cardiomyopathy. eLife 2018, 7, 7. [Google Scholar] [CrossRef]
- Vangheluwe, P.; Schuermans, M.; Zádor, E.; Waelkens, E.; Raeymaekers, L.; Wuytack, F. Sarcolipin and phospholamban mRNA and protein expression in cardiac and skeletal muscle of different species. Biochem. J. 2005, 389, 151–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fajardo, V.A.; Bombardier, E.; Vigna, C.; Devji, T.; Bloemberg, D.; Gamu, D.; Gramolini, A.O.; Quadrilatero, J.; Tupling, A.R. Co-Expression of SERCA Isoforms, Phospholamban and Sarcolipin in Human Skeletal Muscle Fibers. PLoS ONE 2013, 8, e84304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babu, G.J.; Bhupathy, P.; Carnes, C.A.; Billman, G.E.; Periasamy, M. Differential expression of sarcolipin protein during muscle development and cardiac pathophysiology. J. Mol. Cell. Cardiol. 2007, 43, 215–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odermatt, A.; Becker, S.; Khanna, V.K.; Kurzydlowski, K.; Leisner, E.; Pette, D.; MacLennan, D.H. Sarcolipin Regulates the Activity of SERCA1, the Fast-twitch Skeletal Muscle Sarcoplasmic Reticulum Ca2+-ATPase. J. Biol. Chem. 1998, 273, 12360–12369. [Google Scholar] [CrossRef] [Green Version]
- Smith, W.S. Sarcolipin uncouples hydrolysis of ATP from accumulation of Ca2+ by the Ca2+-ATPase of skeletal-muscle sarcoplasmic reticulum. Biochem. J. 2002, 361, 277–286. [Google Scholar] [CrossRef]
- Buck, B.; Zamoon, J.; Kirby, T.L.; DeSilva, T.M.; Karim, C.B.; Thomas, D.D.; Veglia, G. Overexpression, purification, and characterization of recombinant Ca-ATPase regulators for high-resolution solution and solid-state NMR studies. Protein Expr. Purif. 2003, 30, 253–261. [Google Scholar] [CrossRef]
- Espinoza-Fonseca, L.M.; Autry, J.M.; Thomas, D.D. Sarcolipin and phospholamban inhibit the calcium pump by populating a similar metal ion-free intermediate state. Biochem. Biophys. Res. Commun. 2015, 463, 37–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahoo, S.K.; Shaikh, S.A.; Sopariwala, D.H.; Bal, N.C.; Bruhn, D.S.; Kopec, W.; Khandelia, H.; Periasamy, M. The N Terminus of Sarcolipin Plays an Important Role in Uncoupling Sarco-endoplasmic Reticulum Ca2+-ATPase (SERCA) ATP Hydrolysis from Ca2+Transport. J. Biol. Chem. 2015, 290, 14057–14067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Autry, J.M.; Thomas, D.D.; Espinoza-Fonseca, L.M. Sarcolipin Promotes Uncoupling of the SERCA Ca2+ Pump by Inducing a Structural Rearrangement in the Energy-Transduction Domain. Biochemistry 2016, 55, 6083–6086. [Google Scholar] [CrossRef] [Green Version]
- Espinoza-Fonseca, L.M.; Autry, J.M.; Ramírez-Salinas, G.L.; Thomas, D.D. Atomic-Level Mechanisms for Phospholamban Regulation of the Calcium Pump. Biophys. J. 2015, 108, 1697–1708. [Google Scholar] [CrossRef] [Green Version]
- Gortari, E.F.-D.; Espinoza-Fonseca, L.M. Structural basis for relief of phospholamban-mediated inhibition of the sarcoplasmic reticulum Ca2+-ATPase at saturating Ca2+conditions. J. Biol. Chem. 2018, 293, 12405–12414. [Google Scholar] [CrossRef] [Green Version]
- Campbell, K.L.; Dicke, A.A. Sarcolipin Makes Heat, but Is It Adaptive Thermogenesis? Front. Physiol. 2018, 9, 714. [Google Scholar] [CrossRef] [Green Version]
- Gramolini, A.O.; Trivieri, M.G.; Oudit, G.Y.; Kislinger, T.; Li, W.; Patel, M.M.; Emili, A.; Kranias, E.G.; Backx, P.H.; MacLennan, D.H. Cardiac-specific overexpression of sarcolipin in phospholamban null mice impairs myocyte function that is restored by phosphorylation. Proc. Natl. Acad. Sci. USA 2006, 103, 2446–2451. [Google Scholar] [CrossRef] [Green Version]
- Barbot, T.; Montigny, C.; Decottignies, P.; Le Maire, M.; Jaxel, C.; Jamin, N.; Beswick, V. Functional and Structural Insights into Sarcolipin, a Regulator of the Sarco-Endoplasmic Reticulum Ca2+-ATPases; Chakraborti, S., Dhalla, N.S., Eds.; Springer: Cham, Switzerland, 2016; pp. 153–186. [Google Scholar]
- Johnson, C.N.; Pattanayek, R.; Potet, F.; Rebbeck, R.T.; Blackwell, D.J.; Nikolaienko, R.; Sequeira, V.; Le Meur, R.; Radwański, P.B.; Davis, J.P.; et al. The CaMKII inhibitor KN93-calmodulin interaction and implications for calmodulin tuning of NaV1.5 and RyR2 function. Cell Calcium 2019, 82, 102063. [Google Scholar] [CrossRef]
- McCarthy, M.R.; Savich, Y.; Cornea, R.L.; Thomas, D.D. Resolved Structural States of Calmodulin in Regulation of Skeletal Muscle Calcium Release. Biophys. J. 2020, 118, 1090–1100. [Google Scholar] [CrossRef]
- Cala, S.E.; Scott, B.T.; Jones, L.R. Intralumenal sarcoplasmic reticulum Ca(2+)-binding proteins. Semin. Cell Biol. 1990, 1, 265–275. [Google Scholar] [PubMed]
- Valberg, S.J.; Soave, K.; Williams, Z.J.; Perumbakkam, S.; Schott, M.; Finno, C.J.; Petersen, J.L.; Fenger, C.; Autry, J.M.; Thomas, D.D. Coding sequences of sarcoplasmic reticulum calcium ATPase regulatory peptides and expression of calcium regulatory genes in recurrent exertional rhabdomyolysis. J. Vet. Intern. Med. 2019, 33, 933–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaudry, M.J.; Jastroch, M.; Treberg, J.R.; Hofreiter, M.; Paijmans, J.L.A.; Starrett, J.; Wales, N.; Signore, A.V.; Springer, M.S.; Campbell, K.L. Inactivation of thermogenic UCP1 as a historical contingency in multiple placental mammal clades. Sci. Adv. 2017, 3, e1602878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorski, P.A.; Glaves, J.P.; Vangheluwe, P.; Young, H.S. Sarco(endo)plasmic Reticulum Calcium ATPase (SERCA) Inhibition by Sarcolipin Is Encoded in Its Luminal Tail. J. Biol. Chem. 2013, 288, 8456–8467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gramolini, A.O.; Kislinger, T.; Asahi, M.; Li, W.; Emili, A.; MacLennan, D.H. Sarcolipin retention in the endoplasmic reticulum depends on its C-terminal RSYQY sequence and its interaction with sarco(endo)plasmic Ca2+-ATPases. Proc. Natl. Acad. Sci. USA 2004, 101, 16807–16812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, J.; Lee, A.G.; Wilson, D.I.; Spalluto, C.; Hanley, N.; East, J.M. Phospholamban and sarcolipin are maintained in the endoplasmic reticulum by retrieval from the ER-Golgi intermediate compartment. Cardiovasc. Res. 2007, 74, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Martín, Y.; Martín-Romero, F.J.; Iñesta-Vaquera, F.A.; Gutiérrez-Merino, C.; Henao, F. Modulation of sarcoplasmic reticulum Ca2+-ATPase by chronic and acute exposure to peroxynitrite. JBIC J. Biol. Inorg. Chem. 2004, 271, 2647–2657. [Google Scholar] [CrossRef]
- Knyushko, T.V.; Sharov, V.S.; Williams, T.D.; Schöneich, C.; Bigelow, D.J. 3-Nitrotyrosine Modification of SERCA2a in the Aging Heart: A Distinct Signature of the Cellular Redox Environment†. Biochemistry 2005, 44, 13071–13081. [Google Scholar] [CrossRef]
- MacLeay, J.M.; Sorum, S.A.; Valberg, S.J.; Marsh, W.E.; Sorum, M.D. Epidemiologic analysis of factors influencing exertional rhabdomyolysis in Thoroughbreds. Am. J. Vet. Res. 1999, 60, 1562–1566. [Google Scholar]
- Cole, F.L.; Mellor, D.J.; Hodgson, D.R.; Reid, S.W.J. Prevalence and demographic characteristics of exertional rhabdomyolysis in horses in Australia. Vet. Rec. 2004, 155, 625–630. [Google Scholar] [CrossRef]
- Wilberger, M.S.; McKenzie, E.; Payton, M.E.; Rigas, J.D.; Valberg, S.J. Prevalence of exertional rhabdomyolysis in endurance horses in the Pacific Northwestern United States. Equine Vet. J. 2015, 47, 165–170. [Google Scholar] [CrossRef]
- Cheng, A.J.; Andersson, D.; Lanner, J.T. Can’t live with or without it: Calcium and its role in Duchenne muscular dystrophy-induced muscle weakness. Focus on “SERCA1 overexpression minimizes skeletal muscle damage in dystrophic mouse models”. Am. J. Physiol. Physiol. 2015, 308, C697–C698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fruen, B.R.; Mickelson, J.R.; Louis, C.F. Dantrolene Inhibition of Sarcoplasmic Reticulum Ca2+Release by Direct and Specific Action at Skeletal Muscle Ryanodine Receptors. J. Biol. Chem. 1997, 272, 26965–26971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, J.G.T.; Newtont, J.R.; Ramzan, P.H.L.; Pilsworth, R.C.; Shepherd, M.C. The efficacy of dantrolene sodium in controlling exertional rhabdomyolysis in the Thoroughbred racehorse. Equine Vet. J. 2003, 35, 707–711. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, E.C.; Valberg, S.J.; Godden, S.M.; Finno, C.J.; Murphy, M.J. Effect of oral administration of dantrolene sodium on serum creatine kinase activity after exercise in horses with recurrent exertional rhabdomyolysis. Am. J. Vet. Res. 2004, 65, 74–79. [Google Scholar] [CrossRef]
- McDonald, A.G.; Boyce, S.; Tipton, K.F. ExplorEnz: The primary source of the IUBMB enzyme list. Nucleic Acids Res. 2009, 37, D593–D597. [Google Scholar] [CrossRef]
- Strausberg, R.L. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA 2002, 99, 16899–16903. [Google Scholar]
- Zhang, Y.; Fujii, J.; Phillips, M.S.; Chen, H.-S.; Karpati, G.; Yee, W.-C.; Schrank, B.; Cornblath, D.R.; Boylan, K.B.; MacLennan, D.H. Characterization of cDNA and Genomic DNA Encoding SERCA1, the Ca2+-ATPase of Human Fast-Twitch Skeletal Muscle Sarcoplasmic Reticulum, and Its Elimination as a Candidate Gene for Brody Disease. Genomics 1995, 30, 415–424. [Google Scholar] [CrossRef]
- Odermatt, A.; Taschner, P.E.M.; Scherer, S.W.; Beatty, B.; Khanna, V.K.; Cornblath, D.R.; Chaudhry, V.; Yee, W.-C.; Schrank, B.; Karpati, G.; et al. Characterization of the Gene Encoding Human Sarcolipin (SLN), a Proteolipid Associated with SERCA1: Absence of Structural Mutations in Five Patients with Brody Disease. Genomics 1997, 45, 541–553. [Google Scholar] [CrossRef] [Green Version]
- Fujii, J.; Lytton, J.; Tada, M.; MacLennan, D.H. Rabbit cardiac and slow-twitch muscle express the same phospholamban gene. FEBS Lett. 1988, 227, 51–55. [Google Scholar] [CrossRef] [Green Version]
- Fujii, J.; Zarain-Herzberg, A.; Willard, H.F.; Tada, M.; MacLennan, D.H. Structure of the rabbit phospholamban gene, cloning of the human cDNA, and assignment of the gene to human chromosome 6. J. Biol. Chem. 1991, 266, 11669–11675. [Google Scholar]
- Fujii, J.; Ueno, A.; Kitano, K.; Tanaka, S.; Kadoma, M.; Tada, M. Complete complementary DNA-derived amino acid sequence of canine cardiac phospholamban. J. Clin. Investig. 1987, 79, 301–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2019, 48, D84–D86. [Google Scholar]
- Lindholm, A.; Piehl, K. Fibre composition, enzyme activity and concentrations of metabolites and electrolytes in muscles of standardbred horses. Acta Vet. Scand. 1974, 15, 287–309. [Google Scholar] [PubMed]
- Ablorh, N.-A.D.; Dong, X.; James, Z.M.; Xiong, Q.; Zhang, J.; Thomas, D.D.; Karim, C.B. Synthetic Phosphopeptides Enable Quantitation of the Content and Function of the Four Phosphorylation States of Phospholamban in Cardiac Muscle. J. Biol. Chem. 2014, 289, 29397–29405. [Google Scholar] [CrossRef] [Green Version]
- Ablorh, N.-A.; Miller, T.; Nitu, F.; Gruber, S.J.; Karim, C.B.; Thomas, D.D. Accurate quantitation of phospholamban phosphorylation by immunoblot. Anal. Biochem. 2012, 425, 68–75. [Google Scholar] [CrossRef] [Green Version]
- Autry, J.M.; Jones, L.R. High-level coexpression of the canine cardiac calcium pump and phospholamban in Sf21 insect cells. Ann. New York Acad. Sci. 1998, 853, 92–102. [Google Scholar] [CrossRef]
- Autry, J.M.; Karim, C.B.; Cocco, M.; Carlson, S.F.; Thomas, D.D.; Valberg, S.J. Purification of sarcoplasmic reticulum vesicles from horse gluteal muscle. Anal. Biochem. 2020, 610, 113965. [Google Scholar] [CrossRef]
- Valberg, S.J.; Perumbakkam, S.; McKenzie, E.C.; Finno, C.J. Proteome and transcriptome profiling of equine myofibrillar myopathy identifies diminished peroxiredoxin 6 and altered cysteine metabolic pathways. Physiol. Genom. 2018, 50, 1036–1050. [Google Scholar] [CrossRef]
- Leberer, E.; Pette, D. Immunochemical quantification of sarcoplasmic reticulum Ca-ATPase, of calsequestrin and of parvalbumin in rabbit skeletal muscles of defined fiber composition. Jbic J. Biol. Inorg. Chem. 1986, 156, 489–496. [Google Scholar] [CrossRef]
- Yates, A.D. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, M.I.L.R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikemoto, N.; Sreter, F.; Gergely, J. Structural features of the surface of the vesicles of FSR—Lack of functional role in Ca2+ uptake and ATPase activity. Arch. Biochem. Biophys. 1971, 147, 571–582. [Google Scholar] [CrossRef]
- Karim, C.B.; Zhang, Z.; Thomas, D.D. Synthesis of TOAC spin-labeled proteins and reconstitution in lipid membranes. Nat. Protoc. 2007, 2, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; James, Z.M.; Dong, X.; Karim, C.B.; Thomas, D.D. Structural and Functional Dynamics of an Integral Membrane Protein Complex Modulated by Lipid Headgroup Charge. J. Mol. Biol. 2012, 418, 379–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Autry, J.M.; Jones, L.R. Functional Co-expression of the canine cardiac Ca2+ pump and phospholamban in Spodoptera frugiperda (Sf21) cells reveals new insights on ATPase regulation. J. Biol. Chem. 1997, 272, 15872–15880. [Google Scholar] [CrossRef] [Green Version]
- Mahaney, J.E.; Autry, J.M.; Jones, L.R. Kinetics Studies of the Cardiac Ca-ATPase Expressed in Sf21 Cells: New Insights on Ca-ATPase Regulation by Phospholamban. Biophys. J. 2000, 78, 1306–1323. [Google Scholar] [CrossRef] [Green Version]
- Guerrero-Hernandez, A.; Sánchez-Vázquez, V.H.; Martínez-Martínez, E.; Sandoval-Vázquez, L.; Perez-Rosas, N.C.; Lopez-Farias, R.; Dagnino-Acosta, A. Sarco-Endoplasmic Reticulum Calcium Release Model Based on Changes in the Luminal Calcium Content. Atherosclerosis 2020, 1131, 337–370. [Google Scholar]
- Desmond, P.F.; Labuza, A.; Muriel, J.; Markwardt, M.L.; Mancini, A.E.; Rizzo, M.A.; Bloch, R.J. Interactions between small ankyrin 1 and sarcolipin coordinately regulate activity of the sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA1). J. Biol. Chem. 2017, 292, 10961–10972. [Google Scholar] [CrossRef] [Green Version]
- Zaman, J.A.; Harling, L.; Ashrafian, H.; Darzi, A.; Gooderham, N.; Athanasiou, T.; Peters, N.S. Post-operative atrial fibrillation is associated with a pre-existing structural and electrical substrate in human right atrial myocardium. Int. J. Cardiol. 2016, 220, 580–588. [Google Scholar] [CrossRef] [Green Version]
- Rowland, L.A.; Maurya, S.K.; Bal, N.C.; Kozak, L.; Periasamy, M. Sarcolipin and uncoupling protein 1 play distinct roles in diet-induced thermogenesis and do not compensate for one another. Obesity 2016, 24, 1430–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.S.; Dagnino-Acosta, A.; Yarotskyy, V.; Hanna, A.; Lyfenko, A.; Knoblauch, M.; Georgiou, D.K.; Poché, R.A.; Swank, M.W.; Long, C.; et al. Ca(2+) permeation and/or binding to CaV1.1 fine-tunes skeletal muscle Ca(2+) signaling to sustain muscle function. Skelet. Muscle 2015, 5, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sham, J.S.; Jones, L.R.; Morad, M. Phospholamban mediates the beta-adrenergic-enhanced Ca2+ uptake in mammalian ventricular myocytes. Am. J. Physiol. Circ. Physiol. 1991, 261, H1344–H1349. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.R.; Cornea, R.L.; Chen, Z. Close Proximity between Residue 30 of Phospholamban and Cysteine 318 of the Cardiac Ca2+Pump Revealed by Intermolecular Thiol Cross-linking. J. Biol. Chem. 2002, 277, 28319–28329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacLennan, D.H.; Rice, W.J.; Green, N.M. The Mechanism of Ca2+Transport by Sarco(Endo)plasmic Reticulum Ca2+-ATPases. J. Biol. Chem. 1997, 272, 28815–28818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, J.; Boknik, P.; DePaoli-Roach, A.A.; Field, L.J.; Rockman, H.A.; Kobayashi, Y.M.; Kelley, J.S.; Jones, L.R. Targeted Overexpression of Phospholamban to Mouse Atrium Depresses Ca2+Transport and Contractility. J. Mol. Cell. Cardiol. 1998, 30, 1991–2002. [Google Scholar] [CrossRef]
- Shutova, A.N.; Storey, K.B.; Lopina, O.D.; Rubtsov, A.M. Comparative characteristics of sarcoplasmic reticulum preparations from skeletal muscles of the ground squirrel Spermophilus undulatus, rats, and rabbits. Biochemistry (Moscow) 1999, 64, 1250–1257. [Google Scholar] [PubMed]
- Kumar, S.; Li, C.; Montigny, C.; Le Maire, M.; Barth, A. Conformational changes of recombinant Ca2+-ATPase studied by reaction-induced infrared difference spectroscopy. FEBS J. 2013, 280, 5398–5407. [Google Scholar] [CrossRef] [PubMed]
- Autry, J.M.; Rubin, J.E.; Svensson, B.; Li, J.; Thomas, D.D. Nucleotide Activation of the Ca-ATPase. J. Biol. Chem. 2012, 287, 39070–39082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Autry, J.M.; Rubin, J.E.; Pietrini, S.D.; Winters, D.L.; Robia, S.L.; Thomas, D.D. Oligomeric Interactions of Sarcolipin and the Ca-ATPase. J. Biol. Chem. 2011, 286, 31697–31706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, D.R.; Dalton, M.P.; Cho, E.E.; Pribadi, M.P.; Zak, T.J.; Šeflová, J.; Makarewich, C.A.; Olson, E.N.; Robia, S.L. Newly Discovered Micropeptide Regulators of SERCA Form Oligomers but Bind to the Pump as Monomers. J. Mol. Biol. 2019, 431, 4429–4443. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, C.; Iwasawa, S.; Ogawa, H.; Hirata, A.; Tsueda, J.; Inesi, G. Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state. Nat. Cell Biol. 2013, 495, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.; Smyth, N.; Broadbridge, R.; Council, C.E.; Lee, A.G.; Stocker, C.J.; Hislop, D.C.; Arch, J.R.; Cawthorne, M.A.; East, J.M. The effects of sarcolipin over-expression in mouse skeletal muscle on metabolic activity. Arch. Biochem. Biophys. 2015, 569, 26–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohnoki, S.; Martonosi, A. Purification and characterization of the proteolipid of rabbit sarcoplasmic reticulum. Biochim. Et Biophys. Acta (Bba) Protein Struct. 1980, 626, 170–178. [Google Scholar] [CrossRef]
- Hellstern, S.; Pegoraro, S.; Karim, C.B.; Lustig, A.; Thomas, D.D.; Moroder, L.; Engel, J. Sarcolipin, the Shorter Homologue of Phospholamban, Forms Oligomeric Structures in Detergent Micelles and in Liposomes. J. Biol. Chem. 2001, 276, 30845–30852. [Google Scholar] [CrossRef] [Green Version]
- Glaves, J.P. Cryo-Electron Microscopy of SERCA Interacting with Oligomeric Phospholamban and Oligomeric Sarcolipin; University of Alberta: Edmonton, AB, Canada, 2011. [Google Scholar]
- Movsesian, M.A.; Karimi, M.; Green, K.; Jones, L.R. Ca(2+)-transporting ATPase, phospholamban, and calsequestrin levels in nonfailing and failing human myocardium. Circulation 1994, 90, 653–657. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Akin, B.L.; Jones, L.R. Mechanism of Reversal of Phospholamban Inhibition of the Cardiac Ca2+-ATPase by Protein Kinase A and by Anti-phospholamban Monoclonal Antibody 2D12. J. Biol. Chem. 2007, 282, 20968–20976. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Jones, P.P.; Guo, J.; Zhong, X.; Clark, R.B.; Zhou, Q.; Wang, R.; Vallmitjana, A.; Benítez, R.; Hove-Madsen, L.; et al. Phospholamban knockout breaks arrhythmogenic Ca2+ waves and suppresses catecholaminergic polymorphic ventricular tachycardia in mice. Circ. Res. 2013, 113, 517–526. [Google Scholar] [CrossRef] [Green Version]
- Jones, L.R.; Field, L.J. Residues 2-25 of phospholamban are insufficient to inhibit Ca2+ transport ATPase of cardiac sarcoplasmic reticulum. J. Biol. Chem. 1993, 268, 11486–11488. [Google Scholar]
- Kelly, E.M.; Hou, Z.; Bossuyt, J.; Bers, D.M.; Robia, S.L. Phospholamban Oligomerization, Quaternary Structure, and Sarco(endo)plasmic Reticulum Calcium ATPase Binding Measured by Fluorescence Resonance Energy Transfer in Living Cells. J. Biol. Chem. 2008, 283, 12202–12211. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Wu, X.; Yang, R.; Wang, X.; Sun, H.; Lee, I. Self-assembling Study of Sarcolipin and Its Mutants in Multiple Molecular Dynamic Simulations. Proteins: Struct. Funct. Bioinform. 2017, 85, 1065–1077. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wu, X.; Wang, X.; Sun, H.; Lee, I. Transmembrane dynamics of the Thr-5 phosphorylated sarcolipin pentameric channel. Arch. Biochem. Biophys. 2016, 604, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Kurzydlowski, K.; Tada, M.; MacLennan, D.H. Phospholamban Inhibitory Function Is Activated by Depolymerization. J. Biol. Chem. 1997, 272, 15061–15064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornea, R.L.; Autry, J.M.; Chen, Z.; Jones, L.R. Reexamination of the Role of the Leucine/Isoleucine Zipper Residues of Phospholamban in Inhibition of the Ca2+Pump of Cardiac Sarcoplasmic Reticulum. J. Biol. Chem. 2000, 275, 41487–41494. [Google Scholar] [CrossRef] [Green Version]
- Glaves, J.P. Interaction of a Sarcolipin Pentamer and Monomer with the Sarcoplasmic Reticulum Calcium Pump, SERCA. Biophys. J. 2020, 118, 518–531. [Google Scholar] [CrossRef]
- Lu, P.; Vogel, C.; Wang, R.; Yao, X.; Marcotte, E.M. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 2007, 25, 117–124. [Google Scholar] [CrossRef]
- Eichenberger, R.M.; Ramakrishnan, C.; Russo, G.; Deplazes, P.; Hehl, A.B. Genome-wide analysis of gene expression and protein secretion of Babesia canis during virulent infection identifies potential pathogenicity factors. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Kislinger, T.; Cox, B.; Kannan, A.; Chung, C.; Hu, P.; Ignatchenko, A.; Scott, M.S.; Gramolini, A.O.; Morris, Q.; Hallett, M.T.; et al. Global Survey of Organ and Organelle Protein Expression in Mouse: Combined Proteomic and Transcriptomic Profiling. Cell 2006, 125, 173–186. [Google Scholar] [CrossRef] [Green Version]
- Pant, M.; Bal, N.C.; Periasamy, M. Cold adaptation overrides developmental regulation of sarcolipin expression in mice skeletal muscle: SOS for muscle-based thermogenesis? J. Exp. Biol. 2015, 218, 2321–2325. [Google Scholar] [CrossRef] [Green Version]
- Primeau, J.O.; Armanious, G.P.; Fisher, M.E.; Young, H.S. The SarcoEndoplasmic Reticulum Calcium ATPase. Subcell. Biochem. 2018, 87, 229–258. [Google Scholar] [PubMed]
- Nef, H.M.; Möllmann, H.; Troidl, C.; Kostin, S.; Voss, S.; Hilpert, P.; Behrens, C.B.; Rolf, A.; Rixe, J.; Weber, M.; et al. Abnormalities in intracellular Ca2+ regulation contribute to the pathomechanism of Tako-Tsubo cardiomyopathy. Eur. Hear. J. 2009, 30, 2155–2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asahi, M.; Kurzydlowski, K.; Tada, M.; MacLennan, D.H.; Čabart, P.; Murphy, S. Sarcolipin Inhibits Polymerization of Phospholamban to Induce Superinhibition of Sarco(endo)plasmic Reticulum Ca2+-ATPases (SERCAs). J. Biol. Chem. 2002, 277, 26725–26728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asahi, M.; Sugita, Y.; Kurzydlowski, K.; De Leon, S.; Tada, M.; Toyoshima, C.; MacLennan, D.H. Sarcolipin regulates sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) by binding to transmembrane helices alone or in association with phospholamban. Proc. Natl. Acad. Sci. USA 2003, 100, 5040–5045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorski, P.A.; Ceholski, D.K.; Young, H.S. Structure-Function Relationship of the SERCA Pump and Its Regulation by Phospholamban and Sarcolipin. Adv. Exp. Med. Biol. 2017, 981, 77–119. [Google Scholar] [PubMed]
- Young, H.S.; Lemieux, M.J. Regulating the regulator: Intramembrane proteolysis of vesicular trafficking proteins and the SERCA regulator phospholamban. Embo Rep. 2019, 20. [Google Scholar] [CrossRef]
- Morita, T.; Hussain, D.; Asahi, M.; Tsuda, T.; Kurzydlowski, K.; Toyoshima, C.; MacLennan, D.H. Interaction sites among phospholamban, sarcolipin, and the sarco(endo)plasmic reticulum Ca2+-ATPase. Biochem. Biophys. Res. Commun. 2008, 369, 188–194. [Google Scholar] [CrossRef]
- Tupling, A.R.; Bombardier, E.; Gupta, S.C.; Hussain, D.; Vigna, C.; Bloemberg, D.; Quadrilatero, J.; Trivieri, M.G.; Babu, G.J.; Backx, P.H.; et al. Enhanced Ca2+ transport and muscle relaxation in skeletal muscle from sarcolipin-null mice. Am. J. Physiol. Physiol. 2011, 301, C841–C849. [Google Scholar] [CrossRef] [Green Version]
- Shanmugam, M.; Molina, C.E.; Gao, S.; Severac-Bastide, R.; Fischmeister, R.; Babu, G.J. Decreased sarcolipin protein expression and enhanced sarco(endo)plasmic reticulum Ca2+ uptake in human atrial fibrillation. Biochem. Biophys. Res. Commun. 2011, 410, 97–101. [Google Scholar] [CrossRef] [Green Version]
- Molina, C.E.; Abu-Taha, I.H.; Wang, Q.; Roselló-Díez, E.; Kamler, M.; Nattel, S.; Ravens, U.; Wehrens, X.H.T.; Hove-Madsen, L.; Heijman, J.; et al. Profibrotic, Electrical, and Calcium-Handling Remodeling of the Atria in Heart Failure Patients With and Without Atrial Fibrillation. Front. Physiol. 2018, 9, 1383. [Google Scholar] [CrossRef] [Green Version]
- Kozak, M. An analysis of vertebrate mRNA sequences: Intimations of translational control. J. Cell Biol. 1991, 115, 887–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Zhang, Y.; Li, T.; Ma, Z.; Jia, H.; Chen, Q.; Zhao, Y.; Zhai, L.; Zhong, R.; Li, C.; et al. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 2017, 8, 14016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Cao, F.; Yu, X.; Chen, C.; Meng, J.; Zhong, R.; Zhang, Y.; Zhu, D. Linc-RAM is required for FGF2 function in regulating myogenic cell differentiation. Rna Biol. 2018, 15, 404–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sathish, V.; Thompson, M.A.; Bailey, J.P.; Pabelick, C.M.; Prakash, Y.S.; Sieck, G.C. Effect of proinflammatory cytokines on regulation of sarcoplasmic reticulum Ca2+ reuptake in human airway smooth muscle. Am. J. Physiol. Cell. Mol. Physiol. 2009, 297, L26–L34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Jiao, L.; Sun, L.-H.; Li, Y.; Gao, Y.; Xu, C.; Shao, Y.; Li, M.; Li, C.; Lu, Y.; et al. LncRNA ZFAS1 as a SERCA2a Inhibitor to Cause Intracellular Ca 2+ Overload and Contractile Dysfunction in a Mouse Model of Myocardial Infarction. Circ. Res. 2018, 122, 1354–1368. [Google Scholar] [CrossRef]
- Soller, K.J.; Verardi, R.; Jing, M.; Abrol, N.; Yang, J.; Walsh, N.; Vostrikov, V.V.; Robia, S.L.; Bowser, M.T.; Veglia, G. Rheostatic Regulation of the SERCA/Phospholamban Membrane Protein Complex Using Non-Coding RNA and Single-Stranded DNA oligonucleotides. Sci. Rep. 2015, 5, 13000. [Google Scholar] [CrossRef] [Green Version]
- Soller, K.J.; Yang, J.; Veglia, G.; Bowser, M.T. Reversal of Phospholamban Inhibition of the Sarco(endo)plasmic Reticulum Ca2+-ATPase (SERCA) Using Short, Protein-interacting RNAs and Oligonucleotide Analogs. J. Biol. Chem. 2016, 291, 21510–21518. [Google Scholar] [CrossRef] [Green Version]
- Van Heesch, S.; Witte, F.; Schneider-Lunitz, V.; Schulz, J.F.; Adami, E.; Faber, A.B.; Kirchner, M.; Maatz, H.; Blachut, S.; Sandmann, C.-L.; et al. The Translational Landscape of the Human Heart. Cell 2019, 178, 242–260.e29. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, M.A.; Ziman, A.P.; Strong, J.; Zhang, Y.; Hartford, A.K.; Ward, C.W.; Randall, W.R.; Kontrogianni-Konstantopoulos, A.; Bloch, R.J. Integrity of the network sarcoplasmic reticulum in skeletal muscle requires small ankyrin 1. J. Cell Sci. 2011, 124, 3619–3630. [Google Scholar] [CrossRef] [Green Version]
- Solarewicz, J.; Manly, A.; Kokoszka, S.; Sleiman, N.; Leff, T.; Cala, S.E. Adiponectin secretion from cardiomyocytes produces canonical multimers and partial co-localization with calsequestrin in junctional SR. Mol. Cell. Biochem. 2019, 457, 201–214. [Google Scholar] [CrossRef]
- He, W.; Huang, D.; Guo, S.; Wang, D.; Guo, J.; Cala, S.E.; Chen, Z. Association with SERCA2a directs phospholamban trafficking to sarcoplasmic reticulum from a nuclear envelope pool. J. Mol. Cell. Cardiol. 2020, 143, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.; Watson, H.R.; Lee, A.G.; Schuppe, H.-J.; East, J.M. Retrieval from the ER-golgi intermediate compartment is key to the targeting of c-terminally anchored ER-resident proteins. J. Cell. Biochem. 2011, 112, 3543–3548. [Google Scholar] [CrossRef] [PubMed]
- Teng, A.C.T.; Miyake, T.; Yokoe, S.; Zhang, L.; Rezende, J.L.M.; Sharma, P.; MacLennan, D.H.; Liu, P.P.; Gramolini, A.O. Metformin increases degradation of phospholamban via autophagy in cardiomyocytes. Proc. Natl. Acad. Sci. USA 2015, 112, 7165–7170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, T.; Yokoe, S.; Asahi, M. Phospholamban degradation is induced by phosphorylation-mediated ubiquitination and inhibited by interaction with cardiac type Sarco(endo)plasmic reticulum Ca2+-ATPase. Biochem. Biophys. Res. Commun. 2016, 472, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, J.; Mentrup, T.; Heidasch, R.; Müller, S.A.; Biswas, U.; Meyer, R.; Papadopoulou, A.A.; Dederer, V.; Haug-Kröper, M.; Adamski, V.; et al. The intramembrane protease SPPL 2c promotes male germ cell development by cleaving phospholamban. Embo Rep. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Willis, I.M.; Moir, R.D. Signaling to and from the RNA Polymerase III Transcription and Processing Machinery. Annu. Rev. Biochem. 2018, 87, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Subbaiah, K.C.V.; Hedaya, O.; Wu, J.; Jiang, F.; Yao, P. Mammalian RNA switches: Molecular rheostats in gene regulation, disease, and medicine. Comput. Struct. Biotechnol. J. 2019, 17, 1326–1338. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Cai, B.; Zhang, Y.; Zhao, Y.; Wang, J.; Li, T.; Zhang, Y.; Jiang, Y.; Jin, X.; Xue, G.; Li, P.; et al. Long Noncoding RNA–DACH1 (Dachshund Homolog 1) Regulates Cardiac Function by Inhibiting SERCA2a (Sarcoplasmic Reticulum Calcium ATPase 2a). Hypertension 2019, 74, 833–842. [Google Scholar] [CrossRef]
- Lentz, L.R.; Valberg, S.J.; Balog, E.M.; Mickelson, J.R.; Gallant, E.M. Abnormal regulation of muscle contraction in horses with recurrent exertional rhabdomyolysis. Am. J. Vet. Res. 1999, 60, 992–999. [Google Scholar]
- Voit, A.; Patel, V.; Pachon, R.; Shah, V.; Bakhutma, M.; Kohlbrenner, E.; McArdle, J.J.; Dell’Italia, L.J.; Mendell, J.R.; Xie, L.-H.; et al. Reducing sarcolipin expression mitigates Duchenne muscular dystrophy and associated cardiomyopathy in mice. Nat. Commun. 2017, 8, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fajardo, V.A.; Chambers, P.J.; Juracic, E.S.; Rietze, B.A.; Gamu, D.; Bellissimo, C.; Kwon, F.; Quadrilatero, J.; Tupling, A.R. Sarcolipin deletion in mdx mice impairs calcineurin signalling and worsens dystrophic pathology. Hum. Mol. Genet. 2018, 27, 4094–4102. [Google Scholar] [CrossRef] [PubMed]
- Niranjan, N.; Mareedu, S.; Tian, Y.; Kodippili, K.; Fefelova, N.; Voit, A.; Xie, L.-H.; Duan, N.; Babu, G.J. Sarcolipin overexpression impairs myogenic differentiation in Duchenne muscular dystrophy. Am. J. Physiol. Physiol. 2019, 317, C813–C824. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Autry, J.M.; Karim, C.B.; Perumbakkam, S.; Finno, C.J.; McKenzie, E.C.; Thomas, D.D.; Valberg, S.J. Sarcolipin Exhibits Abundant RNA Transcription and Minimal Protein Expression in Horse Gluteal Muscle. Vet. Sci. 2020, 7, 178. https://doi.org/10.3390/vetsci7040178
Autry JM, Karim CB, Perumbakkam S, Finno CJ, McKenzie EC, Thomas DD, Valberg SJ. Sarcolipin Exhibits Abundant RNA Transcription and Minimal Protein Expression in Horse Gluteal Muscle. Veterinary Sciences. 2020; 7(4):178. https://doi.org/10.3390/vetsci7040178
Chicago/Turabian StyleAutry, Joseph M., Christine B. Karim, Sudeep Perumbakkam, Carrie J. Finno, Erica C. McKenzie, David D. Thomas, and Stephanie J. Valberg. 2020. "Sarcolipin Exhibits Abundant RNA Transcription and Minimal Protein Expression in Horse Gluteal Muscle" Veterinary Sciences 7, no. 4: 178. https://doi.org/10.3390/vetsci7040178
APA StyleAutry, J. M., Karim, C. B., Perumbakkam, S., Finno, C. J., McKenzie, E. C., Thomas, D. D., & Valberg, S. J. (2020). Sarcolipin Exhibits Abundant RNA Transcription and Minimal Protein Expression in Horse Gluteal Muscle. Veterinary Sciences, 7(4), 178. https://doi.org/10.3390/vetsci7040178





