Enhanced Phenazine-1-Carboxamide Production in Pseudomonas chlororaphis H5△fleQ△relA through Fermentation Optimization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganism, Medium and Cultural Conditions
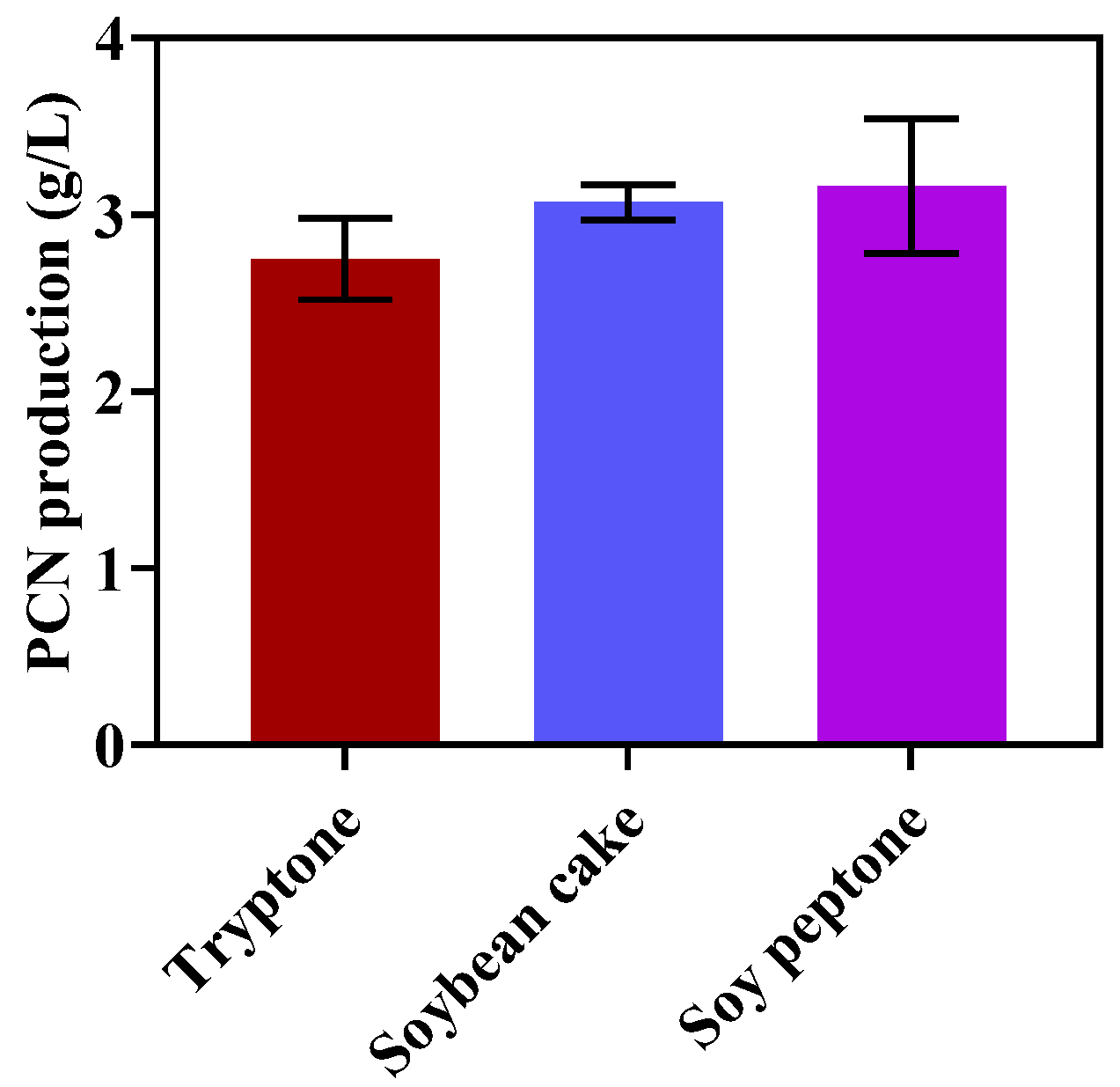
2.2. Optimization of Nitrogen Sources for PCN Production
2.3. Screening of Significant Factors for PCN Production by Plackett-Burman Design
2.4. Determined the Central Point of Central Composite Design by the Path of Steepest Ascent Experiment
2.5. Optimization of Significant Factors by Central Composite Design
2.6. Fermentation Optimization of the Strain H5△fleQ△relA in Bioreactor
2.7. The Measurement of Bacterial Density
2.8. The Extraction and Quantitative Analysis of PCN Content
2.9. Quantitative Real-Time PCR
2.10. UPLC-TOF MS Analyses of AHL Signal Molecule
2.11. The Detection of Glycerol Concentration
2.12. Statistical Analysis
3. Results
3.1. Optimization of Nitrogen Sources for PCN Production
3.2. Screening of Significant Factors for PCN Production by Plackett-Burman Design
3.3. Determined the Central Point of Central Composite Design by the Path of Steepest Ascent Experiment
3.4. Optimization of Significant Factors by Central Composite Design
3.5. Optimization pH in 1 L Bioreactor
3.6. Fed-Batch Fermentation in 1 L Bioreactor
3.7. The Scale-up Production of PCN in 30 L Bioreactor
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chandler, D.; Bailey, A.S.; Tatchell, G.M.; Davidson, G.; Greaves, J.; Grant, W.P. The development, regulation and use of biopesticides for integrated pest management. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 1987–1998. [Google Scholar] [CrossRef] [PubMed]
- Morán-Diez, M.E.; Glare, T.R. What are Microbial-based Biopesticides? Methods Mol. Biol. 2016, 1477, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, L.E.; Price-Whelan, A.; Petersen, A.; Whiteley, M.; Newman, D.K. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 2006, 61, 1308–1321. [Google Scholar] [CrossRef] [PubMed]
- Mavrodi, D.V.; Parejko, J.A.; Mavrodi, O.V.; Kwak, Y.S.; Weller, D.M.; Blankenfeldt, W.; Thomashow, L.S. Recent insights into the diversity, frequency and ecological roles of phenazines in fluorescent Pseudomonas spp. Environ. Microbiol. 2013, 15, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Zhou, L.; Jiang, H.; Sun, S.; Fang, Y.; Liu, J.; Zhang, X.; He, Y.W. Engineering the central biosynthetic and secondary metabolic pathways of Pseudomonas aeruginosa strain PA1201 to improve phenazine-1-carboxylic acid production. Metab. Eng. 2015, 32, 30–38. [Google Scholar] [CrossRef]
- Yuan, L.L.; Li, Y.Q.; Wang, Y.; Zhang, X.H.; Xu, Y.Q. Optimization of critical medium components using response surface methodology for phenazine-1-carboxylic acid production by Pseudomonas sp. M-18Q. J. Biosci. Bioeng. 2008, 105, 232–237. [Google Scholar] [CrossRef]
- Pierson, L.S., 3rd; Pierson, E.A. Metabolism and function of phenazines in bacteria: Impacts on the behavior of bacteria in the environment and biotechnological processes. Appl. Microbiol. Biotechnol. 2010, 86, 1659–1670. [Google Scholar] [CrossRef] [Green Version]
- Chin-A-Woeng, T.F.; Thomas-Oates, J.E.; Lugtenberg, B.J.; Bloemberg, G.V. Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol. Plant-Microbe Interact. 2001, 14, 1006–1015. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Jiang, H.X.; Sun, S.; Yang, D.D.; Jin, K.M.; Zhang, W.; He, Y.W. Biotechnological potential of a rhizosphere Pseudomonas aeruginosa strain producing phenazine-1-carboxylic acid and phenazine-1-carboxamide. World J. Microbiol. Biotechnol. 2016, 32, 50. [Google Scholar] [CrossRef]
- Chin-A-Woeng, T.F.; Bloemberg, G.V.; Mulders, I.H.; Dekkers, L.C.; Lugtenberg, B.J. Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant-Microbe Interact. 2000, 13, 1340–1345. [Google Scholar] [CrossRef] [Green Version]
- Shanmugaiah, V.; Mathivanan, N.; Varghese, B. Purification, crystal structure and antimicrobial activity of phenazine-1-carboxamide produced by a growth-promoting biocontrol bacterium, Pseudomonas aeruginosa MML2212. J. Appl. Microbiol. 2010, 108, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Z.; Yao, W.; Zhang, X.; Wang, R.; Li, P.; Yang, K.; Wang, T.; Liu, K. Metabolic Engineering of Pseudomonas chlororaphis Qlu-1 for the enhanced production of phenazine-1-carboxamide. J. Agric. Food Chem. 2020, 68, 14832–14840. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Pan, K.; Peng, H.; Feng, L.; Hu, H.; Zhang, X. Engineering and systems-level analysis of Pseudomonas chlororaphis for production of phenazine-1-carboxamide using glycerol as the cost-effective carbon source. Biotechnol. Biofuels 2018, 11, 130. [Google Scholar] [CrossRef] [PubMed]
- Chin-A-Woeng, T.F.; van den Broek, D.; Lugtenberg, B.J.; Bloemberg, G.V. The Pseudomonas chlororaphis PCL1391 sigma regulator psrA represses the production of the antifungal metabolite phenazine-1-carboxamide. Mol. Plant-Microbe Interact. 2005, 18, 244–253. [Google Scholar] [CrossRef]
- Jin, Z.J.; Zhou, L.; Sun, S.; Cui, Y.; Song, K.; Zhang, X.; He, Y.W. Identification of a strong quorum sensing- and thermo-regulated promoter for the biosynthesis of a new metabolite pesticide phenazine-1-carboxamide in Pseudomonas strain PA1201. ACS Synth. Biol. 2020, 9, 1802–1812. [Google Scholar] [CrossRef]
- Van Rij, E.T.; Wesselink, M.; Chin-A-Woeng, T.F.; Bloemberg, G.V.; Lugtenberg, B.J. Influence of environmental conditions on the production of phenazine-1-carboxamide by Pseudomonas chlororaphis PCL1391. Mol. Plant-Microbe Interact. 2004, 17, 557–566. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.; Ouyang, Y.; Bilal, M.; Wang, W.; Hu, H.; Zhang, X. Identification, synthesis and regulatory function of the N-acylated homoserine lactone signals produced by Pseudomonas chlororaphis HT66. Microb. Cell Fact. 2018, 17, 9. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Shen, X.; Peng, H.; Hu, H.; Wang, W.; Zhang, X. Comparative genomic analysis and phenazine production of Pseudomonas chlororaphis, a plant growth-promoting rhizobacterium. Genom. Data 2015, 4, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Li, H.L.; Deng, R.X.; Wang, W.; Liu, K.Q.; Hu, H.B.; Huang, X.Q.; Zhang, X.H. Biosynthesis and characterization of medium-chain-length polyhydroxyalkanoate with an enriched 3-Hydroxydodecanoate monomer from a Pseudomonas chlororaphis cell factory. J. Agric. Food Chem. 2021, 69, 3895–3903. [Google Scholar] [CrossRef]
- Jin, X.J.; Peng, H.S.; Hu, H.B.; Huang, X.Q.; Wang, W.; Zhang, X.H. iTRAQ-based quantitative proteomic analysis reveals potential factors associated with the enhancement of phenazine-1-carboxamide production in Pseudomonas chlororaphis P3. Sci. Rep. 2016, 6, 27393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.A.; Mavrodi, D.V.; Thomashow, L.S.; Roessle, M.; Blankenfeldt, W. Ligand binding induces an ammonia channel in 2-amino-2-desoxyisochorismate (ADIC) synthase PhzE. J. Biol. Chem. 2011, 286, 18213–18221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.H.; Yue, S.J.; Feng, T.T.; Li, S.; Huang, P.; Hu, H.B.; Wang, W.; Zhang, X.H. Characterization and engineering of Pseudomonas chlororaphis LX24 with high production of 2-Hydroxyphenazine. J. Agric. Food Chem. 2021, 69, 4778–4784. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lee, S.H.; Seeve, C.; Yu, J.M.; Pierson, L.S., 3rd; Pierson, E.A. Roles of the Gac-Rsm pathway in the regulation of phenazine biosynthesis in Pseudomonas chlororaphis 30-84. Microbiologyopen 2013, 2, 505–524. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.T.; Maeda, T.; Wood, T.K. Metabolic engineering of Escherichia coli to enhance hydrogen production from glycerol. Appl. Microbiol. Biotechnol. 2014, 98, 4757–4770. [Google Scholar] [CrossRef] [PubMed]
- Kohlstaedt, M.; Buschmann, S.; Langer, J.D.; Xie, H.; Michel, H. Subunit CcoQ is involved in the assembly of the Cbb3-type cytochrome c oxidases from Pseudomonas stutzeri ZoBell but not required for their activity. Biochim. Biophys. Acta Bioenerg. 2017, 1858, 231–238. [Google Scholar] [CrossRef]
- Melin, F.; Xie, H.; Meyer, T.; Ahn, Y.O.; Gennis, R.B.; Michel, H.; Hellwig, P. The unusual redox properties of C-type oxidases. Biochim. Biophys. Acta 2016, 1857, 1892–1899. [Google Scholar] [CrossRef]
- Peng, H.; Zhang, P.; Bilal, M.; Wang, W.; Hu, H.; Zhang, X. Enhanced biosynthesis of phenazine-1-carboxamide by engineered Pseudomonas chlororaphis HT66. Microb. Cell Fact. 2018, 17, 117. [Google Scholar] [CrossRef]
- Moreno-Avitia, F.; Utrilla, J.; Bolívar, F.; Nogales, J.; Escalante, A. Metabolic reconstruction of Pseudomonas chlororaphis ATCC 9446 to understand its metabolic potential as a phenazine-1-carboxamide-producing strain. Appl. Microbiol. Biotechnol. 2020, 104, 10119–10132. [Google Scholar] [CrossRef]
- Cotter, P.D.; Gahan, C.G.; Hill, C. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 2001, 40, 465–475. [Google Scholar] [CrossRef]
- Jiang, J.; Sun, Y.F.; Tang, X.; He, C.N.; Shao, Y.L.; Tang, Y.J.; Zhou, W.W. Alkaline pH shock enhanced production of validamycin A in fermentation of Streptomyces hygroscopicus. Bioresour. Technol. 2018, 249, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, S.Y.; Park, S.; Middelberg, A.P. Control of fed-batch fermentations. Biotechnol. Adv. 1999, 17, 29–48. [Google Scholar] [CrossRef]
- Qu, L.; Ren, L.J.; Sun, G.N.; Ji, X.J.; Nie, Z.K.; Huang, H. Batch, fed-batch and repeated fed-batch fermentation processes of the marine thraustochytrid Schizochytrium sp. for producing docosahexaenoic acid. Bioprocess. Biosyst. Eng. 2013, 36, 1905–1912. [Google Scholar] [CrossRef] [PubMed]






| Run | X1 Glycerol (g/L) | X2 Soy Peptone (g/L) | X3 Soybean Cake (g/L) | X4 MgSO4 (g/L) | X5 K2HPO4 (g/L) | PCN Production (g/L) |
|---|---|---|---|---|---|---|
| 1 | 20 | 20 | 20 | 0 | 0.6 | 4.24 |
| 2 | 40 | 10 | 20 | 0 | 0 | 4.04 |
| 3 | 20 | 10 | 10 | 0 | 0 | 2.88 |
| 4 | 20 | 20 | 10 | 0 | 0 | 3.85 |
| 5 | 20 | 10 | 20 | 0.3 | 0.6 | 3.17 |
| 6 | 40 | 10 | 10 | 0 | 0.6 | 2.15 |
| 7 | 20 | 20 | 20 | 0.3 | 0 | 4.66 |
| 8 | 40 | 20 | 10 | 0.3 | 0.6 | 3.52 |
| 9 | 40 | 20 | 20 | 0 | 0.6 | 4.34 |
| 10 | 40 | 20 | 10 | 0.3 | 0 | 4.20 |
| 11 | 40 | 10 | 20 | 0.3 | 0 | 3.97 |
| 12 | 20 | 10 | 10 | 0.3 | 0.6 | 2.16 |
| Parameter | Effect | Coefficient | Standard Error | T Value | p Value |
|---|---|---|---|---|---|
| Constant | 3.5894 | 0.0395 | 91.203 | 0.00000 | |
| X1-Glycerol | 0.2085 | 0.1043 | 0.0395 | 2.642 | 0.03842 |
| X2-Soy peptone | 1.0758 | 0.5379 | 0.0395 | 13.633 | 0.00001 |
| X3-Soybean cake | 0.9406 | 0.4703 | 0.0395 | 11.920 | 0.00002 |
| X4-MgSO4 | 0.0310 | 0.0155 | 0.0395 | 0.393 | 0.70767 |
| X5-K2HPO4 | −0.6710 | −0.3355 | 0.0395 | −8.503 | 0.00015 |
| Run | X1 Glycerol (g/L) | X2 Soy Peptone (g/L) | X3 Soybean Cake (g/L) | PCN Production (g/L) |
|---|---|---|---|---|
| 1 | 30.0 | 15.0 | 15.0 | 4.28 |
| 2 | 32.5 | 21.5 | 20.6 | 4.45 |
| 3 | 35.0 | 27.9 | 26.3 | 4.30 |
| 4 | 37.5 | 34.3 | 31.9 | 4.71 |
| 5 | 40.0 | 40.8 | 37.5 | 5.27 |
| 6 | 42.5 | 47.2 | 43.2 | 4.23 |
| 7 | 45.0 | 53.7 | 48.8 | 3.27 |
| Run | X1 Glycerol (g/L) | X2 Soy Peptone (g/L) | X3 Soybean Cake (g/L) | PCN Production (g/L) | |
|---|---|---|---|---|---|
| Actual | Predicted | ||||
| 1 | 37 | 32.8 | 29.5 | 5.37 | 5.37 |
| 2 | 43 | 32.8 | 29.5 | 5.22 | 5.27 |
| 3 | 37 | 48.8 | 29.5 | 4.65 | 4.45 |
| 4 | 43 | 48.8 | 29.5 | 3.98 | 3.98 |
| 5 | 37 | 32.8 | 45.5 | 5.56 | 5.60 |
| 6 | 43 | 32.8 | 45.5 | 5.46 | 5.51 |
| 7 | 37 | 48.8 | 45.5 | 4.64 | 4.69 |
| 8 | 43 | 48.8 | 45.5 | 4.30 | 4.21 |
| 9 | 35 | 40.8 | 37.5 | 5.20 | 5.24 |
| 10 | 45 | 40.8 | 37.5 | 4.78 | 4.76 |
| 11 | 40 | 27.3 | 37.5 | 5.31 | 5.20 |
| 12 | 40 | 54.3 | 37.5 | 3.22 | 3.34 |
| 13 | 40 | 40.8 | 24 | 5.20 | 5.27 |
| 14 | 40 | 40.8 | 51 | 5.70 | 5.66 |
| 15 | 40 | 40.8 | 37.5 | 5.38 | 5.37 |
| 16 | 40 | 40.8 | 37.5 | 5.34 | 5.37 |
| 17 | 40 | 40.8 | 37.5 | 5.42 | 5.37 |
| 18 | 40 | 40.8 | 37.5 | 5.40 | 5.37 |
| 19 | 40 | 40.8 | 37.5 | 5.41 | 5.37 |
| 20 | 40 | 40.8 | 37.5 | 5.30 | 5.37 |
| Source | Degrees of Freedom | Sum of Squares | Mean Square | F Value | p Value |
|---|---|---|---|---|---|
| Model | 9 | 7.1257 | 0.79175 | 96.49 | <0.0001 |
| X1-Glycerol | 1 | 0.27817 | 0.27817 | 33.90 | 0.0002 |
| X2-Soy peptone | 1 | 4.17977 | 4.17977 | 509.41 | <0.0001 |
| X3-Soybean cake | 1 | 0.18501 | 0.18501 | 22.55 | 0.0008 |
| X1X1 | 1 | 0.14219 | 0.24596 | 29.98 | 0.0003 |
| X2X2 | 1 | 2.23439 | 2.17653 | 265.27 | <0.0001 |
| X3X3 | 1 | 0.01483 | 0.01483 | 1.81 | 0.2085 |
| X1X2 | 1 | 0.07086 | 0.07086 | 8.64 | 0.0148 |
| X1X3 | 1 | 0.01891 | 0.01891 | 2.31 | 0.1599 |
| X2X3 | 1 | 0.00157 | 0.00157 | 0.19 | 0.6712 |
| Residual | 10 | 0.08205 | 0.00821 | ||
| Lack of fit | 5 | 0.07086 | 0.01417 | 6.33 | 0.032 |
| Pure error | 5 | 0.01119 | 0.00224 | ||
| Total | 19 | 7.20777 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, J.; Wang, W.; Hu, H.; Zhang, H.; Zhang, X. Enhanced Phenazine-1-Carboxamide Production in Pseudomonas chlororaphis H5△fleQ△relA through Fermentation Optimization. Fermentation 2022, 8, 188. https://doi.org/10.3390/fermentation8040188
Cui J, Wang W, Hu H, Zhang H, Zhang X. Enhanced Phenazine-1-Carboxamide Production in Pseudomonas chlororaphis H5△fleQ△relA through Fermentation Optimization. Fermentation. 2022; 8(4):188. https://doi.org/10.3390/fermentation8040188
Chicago/Turabian StyleCui, Jiajia, Wei Wang, Hongbo Hu, Hongyan Zhang, and Xuehong Zhang. 2022. "Enhanced Phenazine-1-Carboxamide Production in Pseudomonas chlororaphis H5△fleQ△relA through Fermentation Optimization" Fermentation 8, no. 4: 188. https://doi.org/10.3390/fermentation8040188
APA StyleCui, J., Wang, W., Hu, H., Zhang, H., & Zhang, X. (2022). Enhanced Phenazine-1-Carboxamide Production in Pseudomonas chlororaphis H5△fleQ△relA through Fermentation Optimization. Fermentation, 8(4), 188. https://doi.org/10.3390/fermentation8040188







