Circadian and Light Regulated Expression of CBFs and their Upstream Signalling Genes in Barley
Abstract
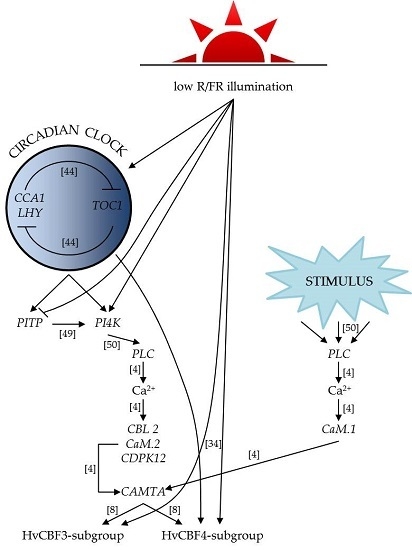
:1. Introduction
2. Results
2.1. Expression Analysis of the Core Circadian Clock Genes
2.2. Expression Analysis of the Phospholipid Signalling Pathway Genes
2.3. Gene Expression Patterns of the Calcium Signalling Elements
2.4. Expression Analysis of the HvCBF Genes
3. Discussion
3.1. Clock Genes
3.2. Ca2+ Signalling
3.3. HvCBF Genes
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Light Treatment
4.3. Sampling for Gene Expression Studies
4.4. Gene Expression Studies
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ranty, B.; Aldon, D.; Cotelle, V.; Galaud, J.-P.; Thuleau, P.; Mazars, C. Calcium Sensors as Key Hubs in Plant Responses to Biotic and Abiotic Stresses. Front. Plant Sci. 2016, 7, 327. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.-W.; Chen, X.; Mei, Y. Function and regulation of phospholipid signalling in plants. Biochem. J. 2009, 421, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Ruelland, E.; Kravets, V.; Derevyanchuk, M.; Martinec, J.; Zachowski, A.; Pokotylo, I. Role of phospholipid signalling in plant environmental responses. Environ. Exp. Bot. 2015, 114, 129–143. [Google Scholar] [CrossRef]
- Yáñez, M.; Gil-Longo, J.; Campos-Toimil, M. Calcium Binding Proteins. Adv. Exp. Med. Biol. 2012, 740, 461–482. [Google Scholar] [CrossRef] [PubMed]
- Day, I.S.; Reddy, V.S.; Shad Ali, G.; Reddy, A. Analysis of EF-hand-containing proteins in arabidopsis. Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- DeFalco, T.A.; Bender, K.W.; Snedden, W.A. Breaking the code: Ca2+ sensors in plant signalling. Biochem. J. 2010, 425, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Ikura, M.; Osawa, M.; Ames, J.B. The role of calcium-binding proteins in the control of transcription: Structure to function. BioEssays 2002, 24, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Doherty, C.J.; van Buskirk, H.A.; Myers, S.J.; Thomashow, M.F. Roles for Arabidopsis CAMTA Transcription Factors in Cold-Regulated Gene Expression and Freezing Tolerance. Plant Cell 2009, 21, 972–984. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Jaglo, K.R.; Kleff, S.; Amundsen, K.L.; Zhang, X.; Haake, V.; Zhang, J.Z.; Deits, T.; Thomashow, M.F. Components of the Arabidopsis C-Repeat/Dehydration- Responsive Element Binding Factor Cold-Response Pathway Are Conserved in Brassica napus and Other Plant Species. Plant Physiol. 2001, 127, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.S.; Wilhelm, K.S.; Thomashow, M.F. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 1994, 24, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.W.; Zhu, B.; Close, T.J. The barley (Hordeum vulgare L.) dehydrin multigene family: Sequences, allele types, chromosome assignments, and expression characteristics of 11 Dhn genes of cv Dicktoo. Theor. Appl. Genet. 1999, 98, 1234–1247. [Google Scholar] [CrossRef]
- Thomashow, M.F. PLANT COLD ACCLIMATION: Freezing Tolerance Genes and Regulatory Mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Tello, A.; Ouellet, F.; Sarhan, F. Low temperature-stimulated phosphorylation regulates the binding of nuclear factors to the promoter of Wcs120, a cold-specific gene in wheat. Mol. Gen. Genet. 1998, 257, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Fowler, S.G.; Thomashow, M.F. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004, 54, 767–781. [Google Scholar] [CrossRef] [PubMed]
- Haake, V.; Cook, D.; Riechmann, J.L.; Pineda, O.; Thomashow, M.F.; Zhang, J.Z. Transcription Factor CBF4 Is a Regulator of Drought Adaptation in Arabidopsis. Plant Physiol. 2002, 130, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Skinner, J.S.; von Zitzewitz, J.; Szűcs, P.; Marquez-Cedillo, L.; Filichkin, T.; Amundsen, K.; Stockinger, E.J.; Thomashow, M.F.; Chen, T.H.H.; Hayes, P.M. Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol. Biol. 2005, 59, 533–551. [Google Scholar] [CrossRef] [PubMed]
- Skinner, J.S.; Szűcs, P.; von Zitzewitz, J.; Marquez-Cedillo, L.; Filichkin, T.; Stockinger, E.J.; Thomashow, M.F.; Chen, T.H.H.; Hayes, P.M. Mapping of barley homologs to genes that regulate low temperature tolerance in Arabidopsis. Theor. Appl. Genet. 2006, 112, 832–842. [Google Scholar] [CrossRef] [PubMed]
- Galiba, G.; Quarrie, S.A.; Sutka, J.; Morgounov, A.; Snape, J.W. RFLP mapping of the vernalization (Vrn1) and frost resistance (Fr1) genes on chromosome 5A of wheat. Theor. Appl. Genet. 1995, 90, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Galiba, G.; Vágújfalvi, A.; Li, C.; Soltész, A.; Dubcovsky, J. Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Sci. 2009, 176, 12–19. [Google Scholar] [CrossRef]
- Vágújfalvi, A.; Galiba, G.; Cattivelli, L.; Dubcovsky, J. The cold-regulated transcriptional activator Cbf3 is linked to the frost-tolerance locus Fr-A2 on wheat chromosome 5A. Mol. Genet. Genom. 2003, 269, 60–67. [Google Scholar] [CrossRef]
- Vágújfalvi, A.; Aprile, A.; Miller, A.; Dubcovsky, J.; Delugu, G.; Galiba, G.; Cattivelli, L. The expression of several Cbf genes at the Fr-A2 locus is linked to frost resistance in wheat. Mol. Genet. Genom. 2005, 274, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Francia, E.; Rizza, F.; Cattivelli, L.; Stanca, A.M.; Galiba, G.; Tóth, B.; Hayes, P.M.; Skinner, J.S.; Pecchioni, N. Two loci on chromosome 5H determine low-temperature tolerance in a “Nure” (winter) × “Tremois” (spring) barley map. Theor. Appl. Genet. 2004, 108, 670–680. [Google Scholar] [CrossRef] [PubMed]
- Francia, E.; Barabaschi, D.; Tondelli, A.; Laidó, G.; Rizza, F.; Stanca, A.M.; Busconi, M.; Fogher, C.; Stockinger, E.J.; Pecchioni, N. Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor. Appl. Genet. 2007, 115, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.K.; Galiba, G.; Dubcovsky, J. A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-Am2 in Triticum monococcum. Mol. Genet. Genom. 2006, 275, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Tondelli, A.; Francia, E.; Barabaschi, D.; Aprile, A.; Skinner, J.S.; Stockinger, E.J.; Stanca, A.M.; Pecchioni, N. Mapping regulatory genes as candidates for cold and drought stress tolerance in barley. Theor. Appl. Genet. 2006, 112, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Tondelli, A.; Pagani, D.; Ghafoori, I.N.; Rahimi, M.; Ataei, R.; Rizza, F.; Flavell, A.J.; Cattivelli, L. Allelic variation at Fr-H1/Vrn-H1 and Fr-H2 loci is the main determinant of frost tolerance in spring barley. Environ. Exp. Bot. 2014, 106, 148–155. [Google Scholar] [CrossRef]
- Stockinger, E.J.; Skinner, J.S.; Gardner, K.G.; Francia, E.; Pecchioni, N. Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J. 2007, 51, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Von Zitzewitz, J.; Cuesta-Marcos, A.; Condon, F.; Castro, A.J.; Chao, S.; Corey, A.; Filichkin, T.; Fisk, S.P.; Gutierrez, L.; Haggard, K.; et al. The Genetics of Winterhardiness in Barley: Perspectives from Genome-Wide Association Mapping. Plant Genome J. 2011, 4, 76–91. [Google Scholar] [CrossRef]
- Maibam, P.; Nawkar, G.M.; Park, J.H.; Sahi, V.P.; Lee, S.Y.; Kang, C.H. The influence of light quality, circadian rhythm, and photoperiod on the CBF-mediated freezing tolerance. Int. J. Mol. Sci. 2013, 14, 11527–11543. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Kim, Y.-K.; Park, J.-Y.; Kim, J. Light signalling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J. 2002, 29, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-M.; Thomashow, M.F. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 15054–15059. [Google Scholar] [CrossRef] [PubMed]
- Novák, A.; Boldizsár, Á.; Ádám, É.; Kozma-Bognár, L.; Majláth, I.; Båga, M.; Tóth, B.; Chibbar, R.; Galiba, G. Light-quality and temperature-dependent CBF14 gene expression modulates freezing tolerance in cereals. J. Exp. Bot. 2016, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Toledo-Ortiz, G.; Pyott, D.E.; Halliday, K.J. Interaction of light and temperature signalling. J. Exp. Bot. 2014, 65, 2859–2871. [Google Scholar] [CrossRef] [PubMed]
- Casal, J.J. Phytochromes, cryptochromes, phototropin: Photoreceptor interactions in plants. Photochem. Photobiol. 2000, 71, 1–11. [Google Scholar] [CrossRef]
- Kim, W.-Y.; Fujiwara, S.; Suh, S.-S.; Kim, J.; Kim, Y.; Han, L.; David, K.; Putterill, J.; Nam, H.G.; Somers, D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 2007, 449, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Sharrock, R.A. The phytochrome red/far-red photoreceptor superfamily. Genome Biol. 2008, 9, 230. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Su, Y.-S.; Lagarias, J.C. Phytochrome Structure and Signaling Mechanisms. Annu. Rev. Plant Biol. 2006, 57, 837–858. [Google Scholar] [CrossRef] [PubMed]
- Blom, T.J.; Tsujita, M.J.; Roberts, G.L. Far-red at End of Day and Reduced Irradiance Affect Plant Height of Easter and Asiatic Hybrid Lilies. HortScience 1995, 30, 1009–1012. [Google Scholar]
- Franklin, K.A.; Whitelam, G.C. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet. 2007, 39, 1410–1413. [Google Scholar] [CrossRef] [PubMed]
- Grundy, J.; Stoker, C.; Carré, I.A. Circadian regulation of abiotic stress tolerance in plants. Front. Plant Sci. 2015, 6, 648. [Google Scholar] [CrossRef] [PubMed]
- Nohales, M.A.; Kay, S.A. Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Biol. 2016, 23, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Mas, P. STRESSing the role of the plant circadian clock. Trends Plant Sci. 2015, 20, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Webb, A.A.R. The physiology of circadian rhythms in plants. New Phytol. 2003, 160, 281–303. [Google Scholar] [CrossRef]
- Jung, J.-Y.; Kim, Y.-W.; Kwak, J.M.; Hwang, J.-U.; Young, J.; Schroeder, J.I.; Hwang, I.; Lee, Y. Phosphatidylinositol 3- and 4-Phosphate Are Required for Normal Stomatal Movements. Plant Cell 2002, 14, 2399–2412. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, G.; De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, 165–183. [Google Scholar] [CrossRef]
- Munnik, T.; Testerink, C. Plant phospholipid signaling: “In a nutshell”. J. Lipid Res. 2009, 50, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Novák, A.; Boldizsár, Á.; Gierczik, K.; Vágújfalvi, A.; Ádám, É.; Kozma-Bognár, L.; Galiba, G. Light and Temperature Signalling at the Level of CBF14 Gene Expression in Wheat and Barley. Plant Mol. Biol. Rep. 2017, 35, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Nusinow, D.A. Into the Evening: Complex Interactions in the Arabidopsis circadian clock. Trends Genet. 2016, 32, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Clausen, J.; Boden, S.; Oliver, S.N.; Casao, M.C.; Ford, B.; Anderssen, R.S.; Trevaskis, B. Dawn and dusk set states of the circadian oscillator in sprouting barley (Hordeum vulgare) seedlings. PLoS ONE 2015, 10, e0129781. [Google Scholar] [CrossRef] [PubMed]
- Ford, B.; Deng, W.; Clausen, J.; Oliver, S.; Boden, S.; Hemming, M.; Trevaskis, B. Barley (Hordeum vulgare) circadian clock genes can respond rapidly to temperature in an EARLY FLOWERING 3-dependent manner. J. Exp. Bot. 2016, 67, 5517–5528. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Shimizu, H.; Nohales, M.A.; Araki, T.; Kay, S.A. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 2014, 515, 419–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.H.; Knight, M.R.; Kondo, T.; Masson, P.; Sedbrook, J.; Haley, A.; Trewavas, A. Circadian Oscillations of Cytosolic and Chloroplastic Free Calcium in Plants. Science 1995, 269, 1863–1865. [Google Scholar] [CrossRef] [PubMed]
- Love, J.; Dodd, A.N.; Webb, A.A.R. Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell 2004, 16, 956–966. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-C.; Jinn, T.-L. Oscillation regulation of Ca2+/calmodulin and heat-stress related genes in response to heat stress in rice (Oryza sativa L.). Plant Signal. Behav. 2012, 7, 1056–1057. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Trewavas, A.J.; Knight, M.R. Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 1996, 8, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Marozsán-Tóth, Z.; Vashegyi, I.; Galiba, G.; Tóth, B. The cold response of CBF genes in barley is regulated by distinct signaling mechanisms. J. Plant Physiol. 2015, 181, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Badawi, M.; Danyluk, J.; Boucho, B.; Houde, M.; Sarhan, F. The CBF gene family in hexaploid wheat and its relationship to the phylogenetic complexity of cereal CBFs. Mol. Genet. Genom. 2007, 277, 533–554. [Google Scholar] [CrossRef] [PubMed]
- Knox, A.K.; Dhillon, T.; Cheng, H.; Tondelli, A.; Pecchioni, N.; Stockinger, E.J. CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor. Appl. Genet. 2010, 121, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Francia, E.; Morcia, C.; Pasquariello, M.; Mazzamurro, V.; Milc, J.A.; Rizza, F.; Terzi, V.; Pecchioni, N. Copy number variation at the HvCBF4–HvCBF2 genomic segment is a major component of frost resistance in barley. Plant Mol. Biol. 2016, 92, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wu, D.; Yang, Q.; Zeng, J.; Jin, G.; Chen, Z.-H.; Zhang, G.; Dai, F. Identification of Mild Freezing Shock Response Pathways in Barley Based on Transcriptome Profiling. Front. Plant Sci. 2016, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.B. Cold acclimation threshold induction temperatures in cereals. Crop Sci. 2008, 48, 1147–1154. [Google Scholar] [CrossRef]
- Dong, M.A.; Farre, E.M.; Thomashow, M.F. CIRCADIAN CLOCK-ASSOCIATED 1 and LATE ELONGATED HYPOCOTYL regulate expression of the C-REPEAT BINDING FACTOR (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7241–7246. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Park, M.-J.; Lim, M.-H.; Kim, S.-G.; Lee, M.; Baldwin, I.T.; Park, C.-M. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 2012, 24, 2427–2442. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Burton, R.A.; Shirley, N.J.; King, B.J.; Harvey, A.J.; Fincher, G.B. The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol. 2004, 134, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Morran, S.; Eini, O.; Pyvovarenko, T.; Parent, B.; Singh, R.; Ismagul, A.; Eliby, S.; Shirley, N.; Langridge, P.; Lopato, S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 2011, 9, 230–249. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, T.M.S. Molecular and Genetic Analyses of Freezing Tolerance in the Triticeae Cereals. Ph.D. thesis, The Ohio State University, Columbus, OH, USA, 2012. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]






© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gierczik, K.; Novák, A.; Ahres, M.; Székely, A.; Soltész, A.; Boldizsár, Á.; Gulyás, Z.; Kalapos, B.; Monostori, I.; Kozma-Bognár, L.; et al. Circadian and Light Regulated Expression of CBFs and their Upstream Signalling Genes in Barley. Int. J. Mol. Sci. 2017, 18, 1828. https://doi.org/10.3390/ijms18081828
Gierczik K, Novák A, Ahres M, Székely A, Soltész A, Boldizsár Á, Gulyás Z, Kalapos B, Monostori I, Kozma-Bognár L, et al. Circadian and Light Regulated Expression of CBFs and their Upstream Signalling Genes in Barley. International Journal of Molecular Sciences. 2017; 18(8):1828. https://doi.org/10.3390/ijms18081828
Chicago/Turabian StyleGierczik, Krisztián, Aliz Novák, Mohamed Ahres, András Székely, Alexandra Soltész, Ákos Boldizsár, Zsolt Gulyás, Balázs Kalapos, István Monostori, László Kozma-Bognár, and et al. 2017. "Circadian and Light Regulated Expression of CBFs and their Upstream Signalling Genes in Barley" International Journal of Molecular Sciences 18, no. 8: 1828. https://doi.org/10.3390/ijms18081828