Environmentally Sensitive Fluorescent Sensors Based on Synthetic Peptides
Abstract
:1. Introduction
2. Synthetic Peptide Sensors: Peptides as Recognition Elements
2.1. Applications of Peptide Biosensors
2.2. Construction of Peptide Biosensors
2.2.1. Strategies for the incorporation of fluorescent dye in the peptide
2.2.2. Position of the fluorophore in the amino acid sequence
3. Synthetic Peptide Sensors: Recognition Element Grafted on a Peptide Scaffold
4. Affinity Characterization of the Interaction between Biosensor and Analyte
5. Limit of Detection
6. Perspectives for Peptide Biosensors
Acknowledgments
References
- Hellinga, H.W.; Marvin, J.S. Protein engineering and the development of generic biosensors. Trends. Biotechnol 1998, 16, 183–189. [Google Scholar]
- Loving, G.S.; Sainlos, M.; Imperiali, B. Monitoring protein interactions and dynamics with solvatochromic fluorophores. Trends Biotechnol 2010, 28, 73–83. [Google Scholar]
- Wang, H.; Nakata, E.; Hamachi, I. Recent progress in strategies for the creation of protein-based fluorescent biosensors. Chembiochem 2009, 10, 2560–2577. [Google Scholar]
- Mims, M.P.; Sturgis, C.B.; Sparrow, J.T.; Morrisett, J.D. Acrylodan can label amino as well as sulfhydryl groups: results with low-density lipoprotein, lipoprotein[a], and lipid-free proteins. Biochemistry 1993, 32, 9215–9120. [Google Scholar]
- Yem, A.W.; Epps, D.E.; Mathews, W.R.; Guido, D.M.; Richard, K.A.; Staite, N.D.; Deibel, M.R. Sitespecific chemical modification of interleukin-1-beta by acrylodan at cysteine-8 and lysine-103. J. Biol. Chem 1992, 267, 3122–3128. [Google Scholar]
- Cohen, B.E.; McAnaney, T.B.; Park, E.S.; Jan, Y.N.; Boxer, S.G.; Jan, L.Y. Probing protein electrostatics with a synthetic fluorescent amino acid. Science 2002, 296, 1700–1703. [Google Scholar]
- Bock, P.E. Active-site-selective labeling of blood coagulation proteinases with fluorescence probes by the use of thioester peptide chloromethyl ketones. J. Biol. Chem 1992, 267, 14974–14981. [Google Scholar]
- Verhamme, I.M.; Olson, S.T.; Tollefsen, D.M.; Bock, P.E. Binding of exosite ligands to human thrombin. Re-evaluation of allosteric linkage between thrombin exosites I and II. J. Biol. Chem 2002, 277, 6788–6798. [Google Scholar]
- Pollack, S.J.; Nakayama, G.R.; Schultz, P.G. Introduction of nucleophiles and spectroscopic probes into antibody combining sites. Science 1988, 242, 1038–1040. [Google Scholar]
- Hamachi, I.; Nagase, T.; Shinkai, S. A general semisynthetic method for fluorescent saccharide-biosensors based on a lectin. J. Am. Chem. Soc 2000, 122, 12065–12066. [Google Scholar]
- Nakata, E.; Nagase, T.; Shinkai, S.; Hamachi, I. Coupling a natural receptor protein with an artificial receptor to afford a semisynthetic fluorescent biosensor. J. Am. Chem. Soc 2004, 126, 490–495. [Google Scholar]
- Nakata, E.; Wang, H.; Hamachi, I. Ratiometric fluorescent biosensor for real-time and label-free monitoring of fine saccharide metabolic pathways. ChemBioChem 2008, 9, 25–28. [Google Scholar]
- Ojida, A.; Tsutsumi, H.; Kasagi, N.; Hamachi, I. Suzuki coupling for protein modification. Tetrahedron Lett 2005, 46, 3301–3305. [Google Scholar]
- Becker, C.F.W.; Hunter, C.L.; Seidel, R.P.; Kent, S.B.H.; Goody, R.S.; Engelhard, M. A sensitive fluorescence monitor for the detection of activated Ras: total chemical synthesis of site-speci¢cally labeled Ras binding domain of c-Raf1 immobilized on a surface. Chem. Biol 2001, 8, 243–252. [Google Scholar]
- Viljanen, J.; Larsson, J.; Larsson, A.; Broo, K.S. A multipurpose receptor composed of promiscuous proteins. Analyte detection through pattern recognition. Bioconjugate Chem 2007, 18, 1935–1945. [Google Scholar]
- de Lorimier, R.M.; Tian, Y.; Hellinga, H.W. Binding and signaling of surface-immobilized reagentless fluorescent biosensors derived from periplasmic binding proteins. Protein Sci 2006, 15, 1936–1944. [Google Scholar]
- Wada, A; Mie, M.; Aizawa, M.; Lahoud, P.; Cass, A.E.G.; Kobatake, E. Design and construction of glutamine binding proteins with a self-adhering capability to unmodified hydrophobic surfaces as reagentless fluorescence sensing devices. J. Am. Chem. Soc 2003, 125, 16228–16234. [Google Scholar]
- Gilardi, G.; Zhou, L.Q.; Hibbert, L.; Cass, A.E.G. Engineering the maltose binding protein for reagentless fluorescence sensing. Anal. Chem 1994, 66, 3840–3847. [Google Scholar]
- Schmid, D.; Baici, A.; Gehring, H.; Christen, P. Kinetics of molecular chaperone action. Science 1994, 263, 971–973. [Google Scholar]
- Fekkes, P; den Blaauwen, T.; Driessen, A.J. Diffusion-limited interaction between unfolded polypeptides and the Escherichia coli chaperone SecB. Biochemistry 1995, 34, 10078–10085. [Google Scholar]
- Harikumar, K.G.; Pinon, D.I.; Wessels, W.S.; Prendergast, F.G.; Miller, L.J. Environment and mobility of a series of fluorescent reporters at the amino terminus of structurally related peptide agonists and antagonists bound to the cholecystokinin receptor. J. Biol. Chem 2002, 277, 18552–18560. [Google Scholar]
- Harikumar, K.G.; Clain, J.; Pinon, D.I.; Dong, M.; Miller, L.J. Distinct molecular mechanisms for agonist peptide binding to types A and B cholecystokinin receptors demonstrated using fluorescence spectroscopy. J. Biol. Chem 2005, 280, 1044–1050. [Google Scholar]
- Harikumar, K.G.; Hosohata, K.; Pinon, D.I.; Miller, L.J. Use of probes with fluorescence indicator distributed throughout the pharmacophore to examine the peptide agonist-binding environment of the family B G protein-coupled secretin receptor. J Biol Chem 2006, 281, 2543–2550. [Google Scholar]
- Takahashi, M.; Nokihara, K.; Mihara, H. Construction of a protein-detection system using a loop peptide library with a fluorescence label. Chem Biol 2003, 10, 53–60. [Google Scholar]
- Usui, K.; Ojima, T.; Takahashi, M.; Nokihara, K.; Mihara, H. Peptide arrays with designed secondary structures for protein characterization using fluorescent fingerprint patterns. Biopolymers 2004, 76, 129–39. [Google Scholar]
- Tomizaki, K.; Mihara, H. A novel fluorescence sensing system using a photochromism-based assay (P-CHROBA) technique for the detection of target proteins. J. Mater. Chem 2005, 15, 2732–2740. [Google Scholar]
- Usui, K.; Tomizaki, K.Y.; Ohyama, T.; Nokihara, K.; Mihara, H. A novel peptide microarray for protein detection and analysis utilizing a dry peptide array system. Mol. Biosyst 2006, 2, 113–121. [Google Scholar]
- Wearsch, P.A.; Voglino, L.; Nicchitta, C.V. Structural transitions accompanying the activation of peptide binding to the endoplasmic reticulum Hsp90 chaperone GRP94. Biochemistry 1998, 37, 5709–5719. [Google Scholar]
- Enander, K.; Choulier, L.; Olsson, A.L.; Yushchenko, D.A.; Kanmert, D.; Klymchenko, A.S.; Demchenko, A.P.; Mely, Y.; Altschuh, D. A peptide-based, ratiometric biosensor construct for direct fluorescence detection of a protein analyte. Bioconjugate Chem 2008, 19, 1864–1870. [Google Scholar]
- Choulier, L.; Shvadchak, V.V.; Naidoo, A.; Klymchenko, A.S.; Mély, Y.; Altschuh, D. A peptide-based fluorescent ratiometric sensor for quantitative detection of proteins. Anal. Biochem. (in press)..
- Thompson, M. Synthesis, photophysical effects, and DNA targeting properties of oxazole yellow-peptide bioconjugates. Bioconjugate Chem 2006, 17, 507–513. [Google Scholar]
- Chen, H.; Chung, N.N.; Lemieux, C.; Zelent, B.; Vanderkooi, J.M.; Gryczynski, I.; Wilkes, B.C.; Schiller, P.W. [Aladan3]TIPP: A fluorescent δ-opioid antagonist with high δ-receptor binding affinity and δ selectivity. Biopolymers 2005, 80, 325–331. [Google Scholar]
- Vázquez, M.E.; Nitz, M.; Stehn, J.; Yaffe, M.B.; Barbara Imperiali, B. Fluorescent caged phosphoserine peptides as probes to investigate phosphorylation-dependent protein associations. J. Am. Chem. Soc 2003, 125, 10150–10151. [Google Scholar]
- Harikumar, K.G.; Pinon, D.I.; Miller, L.J. Fluorescent indicators distributed throughout the pharmacophore of cholecystokinin provide insights into distinct modes of binding and activation of type A and B cholecystokinin receptors. J. Biol. Chem 2006, 281, 27072–27080. [Google Scholar]
- Vázquez, M.E.; Rothman, D.M.; Imperiali, B. A new environment-sensitive fluorescent amino acid for Fmoc-based solid phase peptide synthesis. Org. Biomol.Chem 2004, 2, 1965–1966. [Google Scholar]
- Sainlos, M.; Iskenderian, W.S.; Imperiali, B. A general screening strategy for peptide-based fluorogenic ligands: probes for dynamic studies of PDZ domain-mediated interactions. J. Am. Chem. Soc 2009, 131, 6680–6682. [Google Scholar]
- Vázquez, M.E.; Blanco, J.B.; Imperiali, B. Photophysics and biological applications of the environment-sensitive fluorophore 6-N,N-dimethylamino-2,3-naphthalimide. J. Am. Chem. Soc 2005, 127, 1300–1306. [Google Scholar]
- Venkatraman, P.; Nguyen, T.T.; Sainlos, M.; Bilsel, O.; Chitta, S.; Imperiali, B.; Stern, L.J. Fluorogenic probes for monitoring peptide binding to class II MHC proteins in living cells. Nat. Chem. Biol 2007, 3, 222–228. [Google Scholar]
- Wang, Q.; Cahill, S.M.; Blumenstein, M.; Lawrence, D.S. Self-reporting fluorescent substrates of protein tyrosine kinases. J. Am. Chem. Soc 2006, 128, 1808–1809. [Google Scholar]
- Wang, Q.; Dai, Z.; Cahill, S.M.; Blumenstein, M.; Lawrence, D.S. Light-regulated sampling of protein tyrosine kinase activity. J. Am. Chem. Soc 2006, 128, 14016–14017. [Google Scholar]
- Wang, Q.; Lawrence, D.S. Phosphorylation-driven protein–protein interactions: a protein kinase sensing system. J. Am. Chem. Soc 2005, 127, 7684–7685. [Google Scholar]
- Sharma, V.; Agnes, R.S.; Lawrence, D.S. Deep quench: an expanded dynamic range for protein kinase sensors. J. Am. Chem. Soc 2007, 129, 2742–2743. [Google Scholar]
- Shults, M.D.; Imperiali, B. Versatile fluorescence probes of protein kinase activity. J. Am. Chem. Soc 2003, 125, 14248–14249. [Google Scholar]
- Shults, M.D.; Carrico-Moniz, D.; Imperiali, B. Optimal Sox-based fluorescent chemosensor design for serine/threonine protein kinases. Anal. Biochem 2006, 352, 198–207. [Google Scholar]
- Wu, J.; Zheng, Y.G. Fluorescent reporters of the histone acetyltransferase. Anal. Biochem 2008, 380, 106–110. [Google Scholar]
- Feng, Y.; Xie, N.; Wu, J.; Yang, C.; Zheng, Y.G. Inhibitory study of protein arginine methyltransferase 1 using a fluorescent approach. Biochem. Biophys. Res. Commun 2009, 379, 567–572. [Google Scholar]
- de Lorimier, R.M.; Smith, J.J.; Dwyer, M.A.; Looger, L.L.; Sali, K.M.; Paavola, C.D.; Rizk, S.S.; Sadigov, S.; Conrad, D.W.; Loew, L.; Hellinga, H.W. Construction of a fluorescent biosensor family. Protein Sci 2002, 11, 2655–2675. [Google Scholar]
- Medintz, I.L.; Mauro, J.M. Use of a cyanine dye as a reporter probe in reagentless maltose sensors based on E. coli maltose binding protein. Anal. Lett 2004, 37, 191–202. [Google Scholar]
- Sherman, D.B.; Pitner, J.B.; Ambroise, A.; Thomas, K.J. Synthesis of Thiol-derivative, long-wavelength fluorescent phenoxazine derivatives for biosensor applications. Bioconjugate Chem 2006, 17, 387–392. [Google Scholar]
- Tian, Y.; Cuneo, M.J.; Changela, A.; Höcker, B.; Beese, L.S.; Hellinga, H.W. Structure-based design of robust glucose biosensors using a Thermotoga maritima periplasmic glucose-binding protein. Protein Sci 2007, 16, 2240–2250. [Google Scholar]
- Morii, T.; Sugimoto, K.; Makino, K.; Otsuka, M.; Imoto, K.; Mori, Y. A new fluorescent biosensor for inositol trisphosphate. J. Am. Chem. Soc 2002, 124, 2002,. 1138–1139. [Google Scholar]
- Renard, M.; Belkadi, L.; Hugo, N.; England, P.; Altschuh, D.; Bedouelle, H. Knowledge-based design of reagentless fluorescent biosensors from recombinant antibodies. J. Mol. Biol 2002, 318, 429–442. [Google Scholar]
- Renard, M.; Belkadi, L.; Bedouelle, H. Deriving topological constraints from functional data for the design of reagentless fluorescent immunosensors. J. Mol. Biol 2003, 326, 167–175. [Google Scholar]
- Marvin, J.S.; Corcoran, E.E.; Hattangadi, N.A.; Zhang, J.V.; Gere, S.A.; Hellinga, H.W. The rational design of allosteric interactions in a monomeric protein and its applications to the construction of biosensors. Proc. Natl. Acad. Sci. USA 1997, 94, 4366–4371. [Google Scholar]
- Choi, E.J.; Mao, J.; Mayo, S.L. Computational design and biochemical characterization of maize nonspecific lipid transfer protein variants for biosensor applications. Protein Sci 2007, 16, 582–588. [Google Scholar]
- Rizk, S.S.; Cuneo, M.J.; Hellinga, H.W. Identification of cognate ligands for the Escherichia coli phnD protein product and engineering of a reagentless fluorescent biosensor for phosphonates. Protein Sci 2006, 15, 1745–1751. [Google Scholar]
- Thomas, J.; Sherman, D.B.; Amiss, T.J.; Andaluz, S.A.; Pitner, J.B. Synthesis and biosensor performance of a near-IR thiol-derivative fluorophore based on benzothiazolium squaraine. Bioconjugate Chem 2007, 18, 1841–1846. [Google Scholar]
- Dattelbaum, J.D.; Looger, L.L.; Benson, D.E.; Sali, K.M.; Thompson, R.B.; Hellinga, H.W. Analysis of allosteric signal transduction mechanisms in an engineered fluorescent maltose biosensor. Protein Sci 2005, 14, 284–291. [Google Scholar]
- Matsumura, S.; Sakamoto, S.; Ueno, A.; Mihara, H. Construction of α-Helix peptides with β-cyclodextrin and dansyl units and their conformational and molecular sensing properties. Chem. Eur. J 2000, 6, 1781–1788. [Google Scholar]
- Hossain, M.A.; Hamasaki, K.; Mihara, H.; Ueno, A. Association and guest-induced dissociation of a novel α-helix peptide bearing pyrene and γ-cyclodextrin in the side chains. Chem. Lett 2000, 29, 252–253. [Google Scholar]
- Hossain, M.A.; Matsumura, S.; Kanai, T.; Hamasaki, K.; Mihara, H.; Ueno, A. Association of α-helix peptides that have γ-cyclodextrin and pyrene units in their side chain, and induction of dissociation of the association dimer by external stimulant molecules. J. Chem. Soc. Perkin Trans 2000, 2, 1527–1533. [Google Scholar]
- Toyoda, T.; Matsumura, S.; Mihara, H.; Ueno, A. Guest-responsive excimer emission in an α-helix peptide bearing γ-cyclodextrin and two naphthalene units. Macromol. Rapid Commun 2000, 21, 485–488. [Google Scholar]
- Hossain, M.A.; Hamasaki, K.; Takahashi, K.; Mihara, H.; Ueno, A. Guest-induced diminishment in fluorescence quenching and molecule sensing ability of a novel cyclodextrin-peptide conjugate. J. Am. Chem. Soc 2001, 123, 7435–7436. [Google Scholar]
- Yana, D.; Shimizu, T.; Hamasaki, K.; Mihara, H.; Ueno, A. Double naphthalene-tagged cyclodextrin-peptide capable of exhibiting guest-induced naphthalene excimer fluorescence. Macromol. Rapid. Commun 2002, 23, 11–15. [Google Scholar]
- Toyoda, T.; Mihara, H.; Ueno, A. Fluorescent cyclodextrin/peptide hybrids with a novel guest-responsive chemosensor in the peptide side chain. Macromol. Rapid. Commun 2002, 23, 905–908. [Google Scholar]
- Hossain, M.A.; Takahashi, K.; Mihara, H.; Ueno, A. Molecule-responsive fluorescent sensors of α-helix peptides bearing α-cyclodextrin, pyrene and nitrobenzene units in their side chains. J. Inclusion Phenom. Macrocyclic Chem 2002, 43, 271–277. [Google Scholar]
- Furukawa, S.; Mihara, H.; Ueno, A. Sensing behavior of fluorescent cyclodextrin/peptide hybrids bearing a macrocyclic metal complex. Macromol. Rapid Commun 2003, 24, 202–206. [Google Scholar]
- Enander, K.; Dolphin, G.T.; Andersson, L.K.; Liedberg, B.; Lundstrom, I.; Baltzer, L. Designed, folded polypeptide scaffolds that combine key biosensing events of recognition and reporting. J. Org. Chem 2002, 67, 3120–3123. [Google Scholar]
- Enander, K.; Dolphin, G.T.; Liedberg, B.; Lundström, I.; Baltzer, L. A versatile polypeptide platform for integrated recognition and reporting: affinity arrays for protein-ligand interaction analysis. Chem. Eur. J 2004, 10, 2375–2385. [Google Scholar]
- Enander, K.; Dolphin, G.T.; Baltzer, L. Designed, functionalized helix-loop-helix motifs that bind human carbonic anhydrase II: A new class of synthetic receptor molecules. J. Am. Chem. Soc 2004, 126, 4464–4465. [Google Scholar]
- Andersson, T.; Lundquist, M.; Dolphin, G.T.; Enander, K.; Jonsson, B-H.; Nilsson, J.W.; Baltzer, B. The binding of human carbonic anhydrase II by functionalized folded polypeptide receptors. Chem. Biol 2005, 12, 1245–1252. [Google Scholar]
- Salins, L.L.E.; Ware, R.A.; Ensor, C.M.; Daunert, S. A novel reagentless sensing system for measuring glucose based on the galactose/glucose-binding protein. Anal. Biochem 2001, 294, 19–26. [Google Scholar]
- Lam, H.; Kostov, Y.; Rao, G.; Tolosa, L. Low-cost optical lifetime assisted ratiometric glutamine sensor based on glutamine binding protein. Anal. Biochem 2008, 383, 61–67. [Google Scholar]
- Chan, P.H.; Liu, H.B.; Chen, Y.W.; Chan, K.C.; Tsang, C.W.; Leung, Y.C.; Wong, K.Y. Rational design of a novel fluorescent biosensor for β-lactam antibiotics from a class A β-lactamase. J. Am. Chem. Soc 2004, 126, 4074–4075. [Google Scholar]



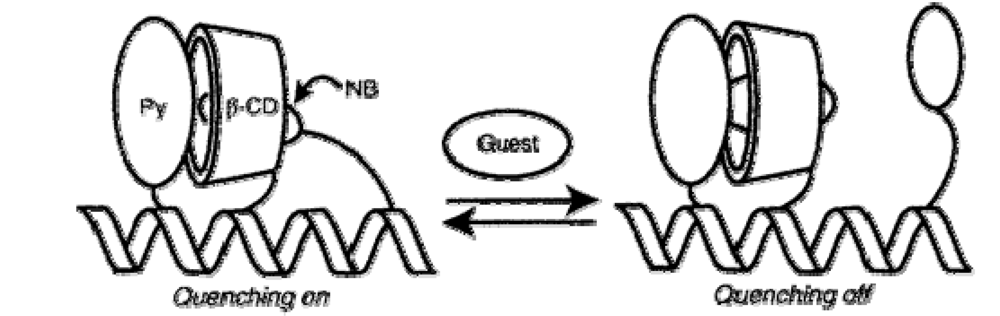
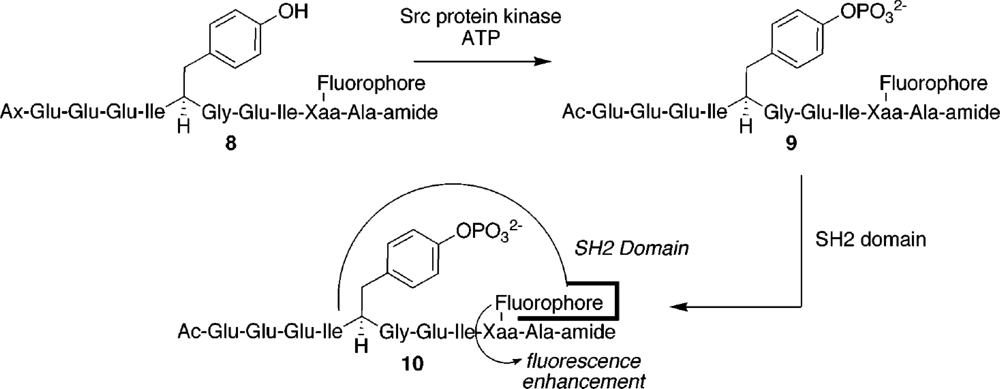
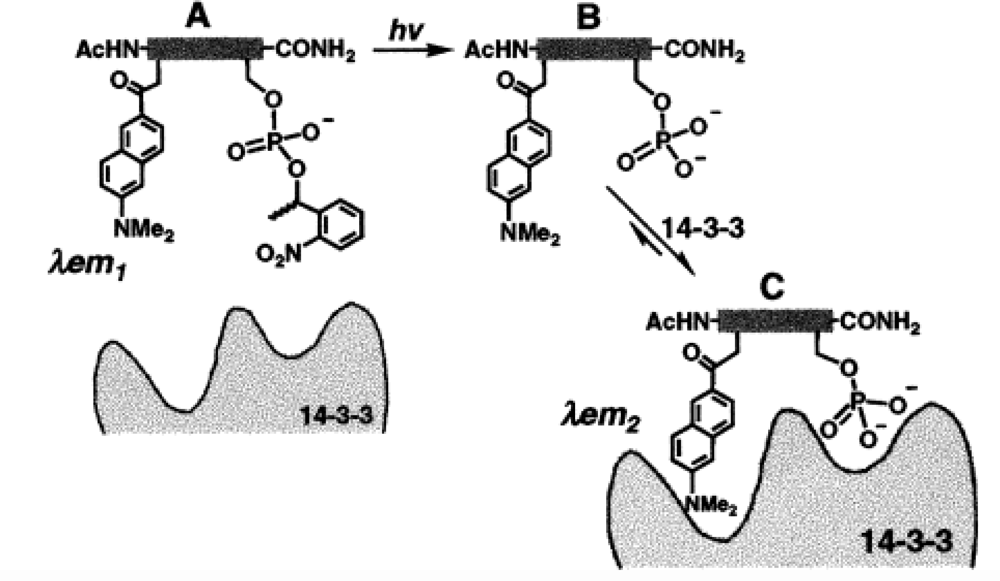
| System characteristics | Sensor affinity and performance | References | |||
|---|---|---|---|---|---|
| Analytes | Receptors | Fluorophores | Maximum fluorescent signal change upon analyte-receptor interaction | KD of the corresponding analyte-receptor complex | |
| DNaK chaperone | Targeting sequence of the precursor of mitochondrial aspartate aminotransferase | Acrylodan | 4-fold | 1.4 μM | [19] |
| SecB chaperone | Bovine pancreatic trypsin inhibitor | Acrylodan | 3.4 -fold | 5.4 nM | [20] |
| Cholecystokinin (CCK) receptor | Peptides agonist and antagonist of the CCK receptor | Alexa, NBDa, Acrylodan | NR | NDd | [21] |
| Cholecystokinin (CCK) receptor | Peptides agonist of the CCK receptor | Alexa488 | NR | NDd | [22] |
| Secretin receptor | Analogues of the hormone secretin | Alexa488 | NR | NDd | [23] |
| α-amylase# | Library of designed loop peptides | Fluorescein | > 4-fold | 1.1 μM | [24] |
| β-lactoglobulin# | Mini-library of designed β-strand peptides | Fluorescein | 2.5-fold | ND | [25] |
| PKA, α-amylase, β-galactosidase, lysozyme, hexokinase, S-100 | Peptides derived from substrates of 4 kinases (PKA, c-Src kinase, c-Abl tyrosine kinase & PKC) | Spiropyran | NR | ND | [26] |
| Calmodulin# | Mini-library of designed α-helical peptides | TAMRAb | 4-fold | 1.5 μM | [27] |
| GRP94 | VSV8, the immonudominant peptide epitope of the vesicular stomatitis virus | Acrylodan, Nile-red | NR | NDd | [28] |
| Fab 57P | Sequence 134–151 of the tobacco mosaic virus coat protein | 3-hydroxychromonec* | 1.4-folde | 2.4 nM | [29] |
| scFv 1F4 | N-terminus sequence of the E6 protein of human papillomavirus 16 | 3-hydroxychromonec* | 1.5 -folde | 1nM | [30] |
| Double-stranded DNA | Polypeptide derived from the Hin recombinase of Salmonella typhimurium | Oxazole yellow | >1.1-fold | 10 nM | [31] |
| System characteristics | Sensor affinity and performance | References | |||
|---|---|---|---|---|---|
| Analytes | Receptors | Fluorescent amino acids | Maximum fluorescent signal change upon analyte-receptor interaction | KD of the corresponding analyte-receptor complex | |
| δ-opioid receptor | δ-opioid antagonists | DANAa (Aladan) | NR | NDd | [32] |
| 14-3-3 protein | Caged phosphopeptides | DANAa (Aladan) | 4-fold | 700 nM | [33] |
| Cholecystokinin (CCK) receptor | Peptides agonist of the CCK receptor | DANAa (Aladan) | NR | NDd | [34] |
| 14-3-3 protein | Phosphopeptides | 4-DAPAb | 6-fold | 4.6 μM | [35] |
| PDZ domains | C-terminal sequence of stargazin, CRIPT, NR2a and GluR1 | 4-DAPAb | 265-fold | 0.2 μM | [36] |
| SH2 phosphotyrosine-binding domains | SH2 domains | 6-DMNAc* | 11-fold | 2.4 μM | [37] |
| Class II MHC proteins | HLA-DR-binding peptides | 4-DAPAb and 6-DMNAc* | 1100-fold | NDd | [38] |
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Choulier, L.; Enander, K. Environmentally Sensitive Fluorescent Sensors Based on Synthetic Peptides. Sensors 2010, 10, 3126-3144. https://doi.org/10.3390/s100403126
Choulier L, Enander K. Environmentally Sensitive Fluorescent Sensors Based on Synthetic Peptides. Sensors. 2010; 10(4):3126-3144. https://doi.org/10.3390/s100403126
Chicago/Turabian StyleChoulier, Laurence, and Karin Enander. 2010. "Environmentally Sensitive Fluorescent Sensors Based on Synthetic Peptides" Sensors 10, no. 4: 3126-3144. https://doi.org/10.3390/s100403126





