Transcriptome Profiling of the Lungs Reveals Molecular Clock Genes Expression Changes after Chronic Exposure to Ambient Air Particles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Exposure Protocol
2.3. Lung Morphometric Analysis
2.4. Blood and Tissue Biomarker Assays
2.5. Second-Generation RNA Sequencing for Gene Expression Analysis
2.6. Real-Time PCR for Verification of Selected Genes
2.7. Statistical Analysis
3. Results
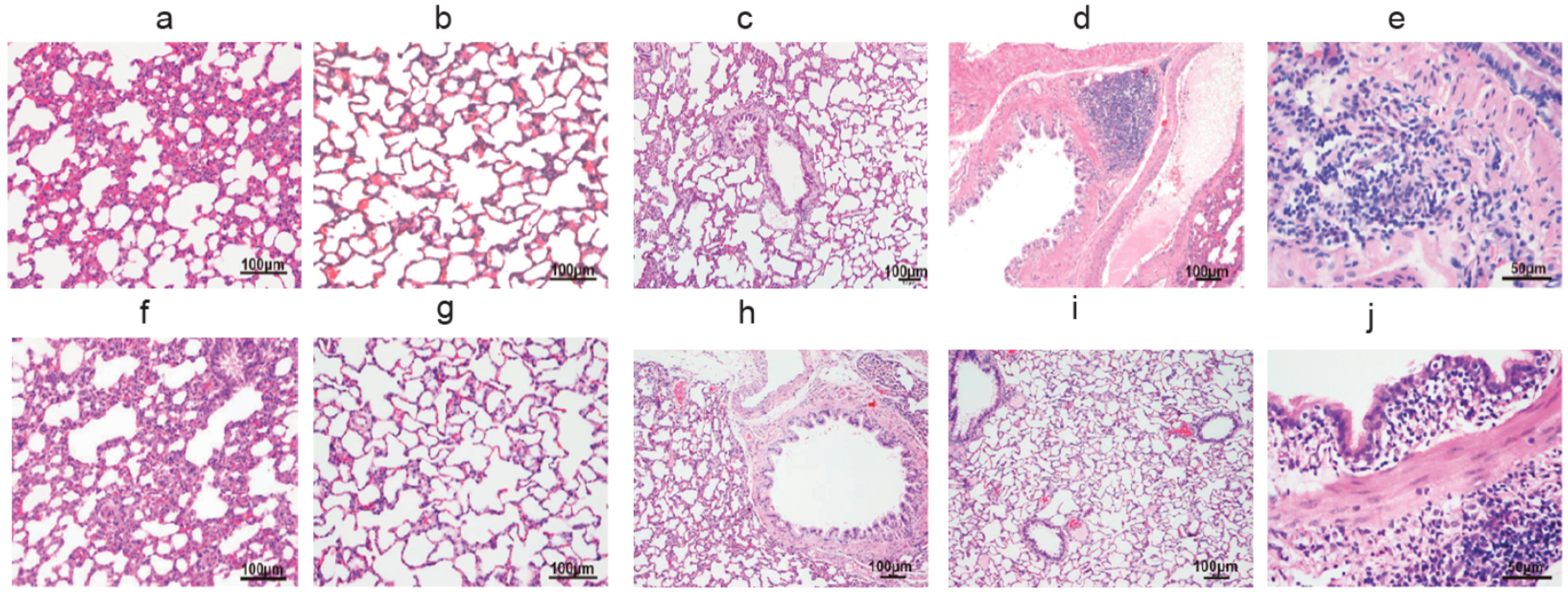
3.1. The Pulmonary Inflammation of Lung Histology
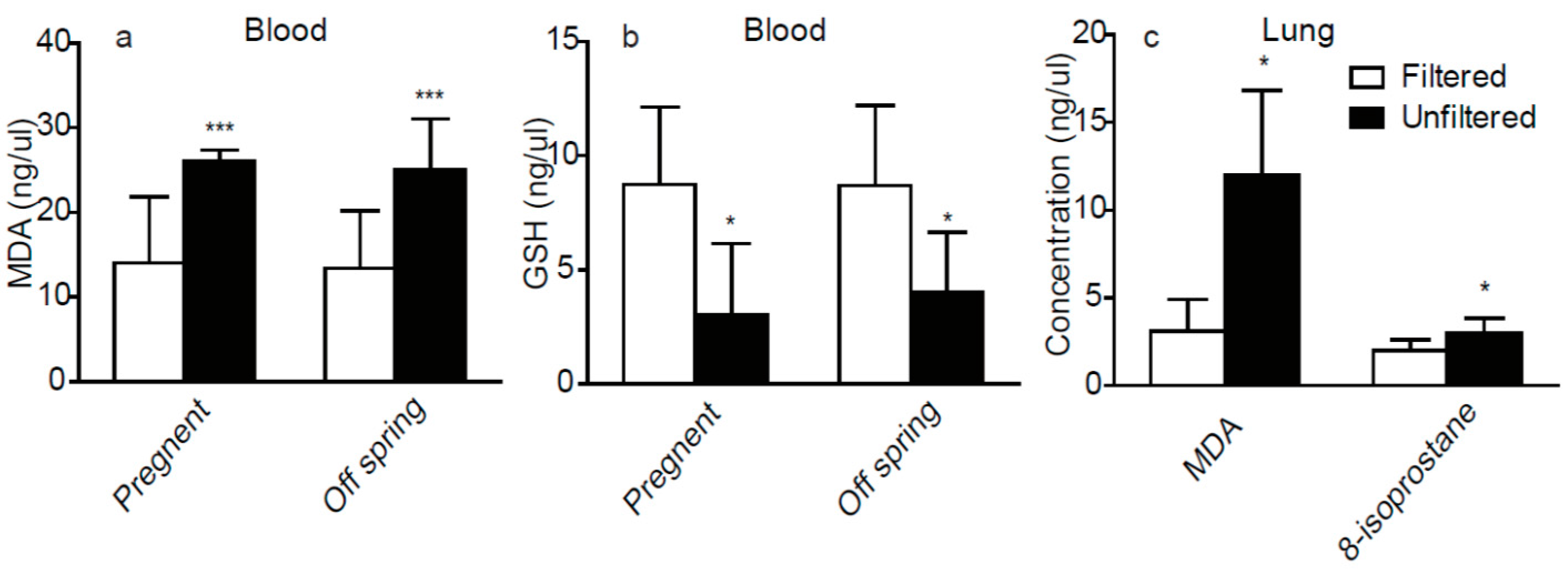
3.2. The Oxidative Stress Factors in Plasma and Lung
3.3. The Circadian Rhythm Genes in Lung
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| COPD | Chronic Obstructive Pulmonary Disease |
| GSH | Glutathione |
| GAPDH | Glyceraldehyde 3-phostaphate Dehydrogenase |
| HE stained | Hematoxylin-Eosin Stained |
| HEPA | High-Efficiency Particulate Air |
| MDA | Malondialdehyde |
| NF-κB | Nuclear Factor-κB |
| PM2.5 | Particulate Matter with an aerodynamic diameter less than 2.5 μm |
| qPCR | Real-Time PCR |
| ROS | Reactive Oxygen Species |
| RNA-Seq | Second Generation RNA Sequencing |
| SCN | Suprachiasmatic Nucleus |
| TTFL | Cell-Autonomous Transcription-Translation Feedback Loops |
References
- Albrecht, U. Timing to perfection: The biology of central and peripheral circadian clocks. Neuron 2012, 74, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Hirota, T.; Fukada, Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zool. Sci. 2004, 21, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Sun, K.K.; Wang, K.; Sun, Z.S.; Zhao, M.; Wang, J. Sex-related difference in food-anticipatory activity of mice. Horm. Behav. 2015, 70, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Bando, H.; Nishio, T.; van der Horst, G.T.; Masubuchi, S.; Hisa, Y.; Okamura, H. Vagal regulation of respiratory clocks in mice. J. Neurosci. 2007, 27, 4359–4365. [Google Scholar] [CrossRef] [PubMed]
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012, 35, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Dibner, C.; Schibler, U.; Albrecht, U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010, 72, 517–549. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Lazar, M.A. Clocks, metabolism, and the epigenome. Mol. Cell. 2012, 47, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Brum, M.C.; Filho, F.F.; Schnorr, C.C.; Bottega, G.B.; Rodrigues, T.C. Shift work and its association with metabolic disorders. Diabetol. Metab. Syndr. 2015, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Rahman, I. Current concepts on oxidative/carbonyl stress, inflammation and epigenetics in pathogenesis of chronic obstructive pulmonary disease. Toxicol. Appl. Pharmacol. 2011, 254, 72–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Zhang, J.J.; Li, Z.; Gow, A.; Chung, K.F.; Hu, M.; Sun, Z.; Zeng, L.; Zhu, T.; Jia, G.; et al. Chronic exposure to air pollution particles increases the risk of obesity and metabolic syndrome: Findings from a natural experiment in Beijing. FASEB J. 2016, 30, 2115–2122. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.J.; Savenkova, M.I.; Karatsoreos, I.N. Environmental disruption of the circadian clock leads to altered sleep and immune responses in mouse. Brain Behav. Immun. 2015, 47, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Burioka, N.; Fukuoka, Y.; Takata, M.; Endo, M.; Miyata, M.; Chikumi, H.; Tomita, K.; Kodani, M.; Touge, H.; Takeda, K.; et al. Circadian rhythms in the CNS and peripheral clock disorders: Function of clock genes: Influence of medication for bronchial asthma on circadian gene. J. Pharmacol. Sci. 2007, 103, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Ferraz, E.; Borges, M.C.; Terra-Filho, J.; Martinez, J.A.; Vianna, E.O. Comparison of 4 AM and 4 PM bronchial responsiveness to hypertonic saline in asthma. Lung 2006, 184, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.J.; Banks-Schlegel, S. Chronobiology of asthma. Am. J. Respir. Crit. Care Med. 1998, 158, 1002–1007. [Google Scholar] [CrossRef] [PubMed]
- Panzer, S.E.; Dodge, A.M.; Kelly, E.A.; Jarjour, N.N. Circadian variation of sputum inflammatory cells in mild asthma. J. Allergy Clin. Immunol. 2003, 111, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Petty, T.L. Circadian variations in chronic asthma and chronic obstructive pulmonary disease. Am. J. Med. 1988, 85, 21–23. [Google Scholar] [CrossRef]
- Tsai, C.L.; Brenner, B.E.; Camargo, C.A., Jr. Circadian-rhythm differences among emergency department patients with chronic obstructive pulmonary disease exacerbation. Chronobiol. Int. 2007, 24, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Agusti, A.; Hedner, J.; Marin, J.M.; Barbe, F.; Cazzola, M.; Rennard, S. Night-time symptoms: A forgotten dimension of COPD. Eur. Respir. Rev. 2011, 20, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Karakatsani, A.; Analitis, A.; Perifanou, D.; Ayres, J.G.; Harrison, R.M.; Kotronarou, A.; Kavouras, I.G.; Pekkanen, J.; Hameri, K.; Kos, G.P.; et al. Particulate matter air pollution and respiratory symptoms in individuals having either asthma or chronic obstructive pulmonary disease: A European multicentre panel study. Environ. Health 2012, 11, 75. [Google Scholar] [CrossRef] [PubMed]
- Brook, R.D.; Rajagopalan, S.; Pope, C.A., 3rd; Brook, J.R.; Bhatnagar, A.; Diez-Roux, A.V.; Holguin, F.; Hong, Y.; Luepker, R.V.; Mittleman, M.A.; et al. Particulate matter air pollution and cardiovascular disease: An update to the scientific statement from the American Heart Association. Circulation 2010, 121, 2331–2378. [Google Scholar] [CrossRef] [PubMed]
- Finkel, T. Oxidant signals and oxidative stress. Curr. Opin. Cell Biol. 2003, 15, 247–254. [Google Scholar] [CrossRef]
- McKnight, S.L.; Rutter, J.; Reick, M.; Wu, L.C. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 2001, 293, 510–514. [Google Scholar]
- Rutter, J.; Reick, M.; McKnight, S.L. Metabolism and the control of circadian rhythms. Annu. Rev. Biochem. 2002, 71, 307–331. [Google Scholar] [CrossRef] [PubMed]
- Giebultowicz, J.M.; Krishnan, N.; Kretzschmar, D.; Rakshit, K.; Chow, E. The circadian clock gene period extends healthspan in aging Drosophila melanogaster. Aging 2009, 1, 937–948. [Google Scholar]
- Takahashi, J.S. Molecular components of the circadian clock in mammals. Diabetes Obes. Metab. 2015, 17 (Suppl. 1), 6–11. [Google Scholar] [CrossRef] [PubMed]
- Duez, H.; Staels, B. Rev-erb-alpha: An integrator of circadian rhythms and metabolism. J. Appl. Physiol. (1985) 2009, 107, 1972–1980. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.C.; Guarente, L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 2013, 153, 1448–1460. [Google Scholar] [CrossRef] [PubMed]
- Lowrey, P.L.; Takahashi, J.S. Genetics of the mammalian circadian system: Photic entrainment, circadian pacemaker mechanisms, and posttranslational regulation. Annu. Rev. Genet. 2000, 34, 533–562. [Google Scholar] [CrossRef] [PubMed]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Sato, T.K.; Panda, S.; Miraglia, L.J.; Reyes, T.M.; Rudic, R.D.; McNamara, P.; Naik, K.A.; FitzGerald, G.A.; Kay, S.A.; Hogenesch, J.B. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 2004, 43, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Eide, E.J.; Vielhaber, E.L.; Hinz, W.A.; Virshup, D.M. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. J. Biol. Chem. 2002, 277, 17248–17254. [Google Scholar] [CrossRef] [PubMed]
- Burioka, N.; Fukuoka, Y.; Koyanagi, S.; Miyata, M.; Takata, M.; Chikumi, H.; Takane, H.; Watanabe, M.; Endo, M.; Sako, T.; et al. Asthma: Chronopharmacotherapy and the molecular clock. Adv. Drug Deliv. Rev. 2010, 62, 946–955. [Google Scholar] [CrossRef] [PubMed]
- Burioka, N.; Koyanagi, S.; Fukuoka, Y.; Okazaki, F.; Fujioka, T.; Kusunose, N.; Endo, M.; Suyama, H.; Chikumi, H.; Ohdo, S.; et al. Influence of intermittent hypoxia on the signal transduction pathways to inflammatory response and circadian clock regulation. Life Sci. 2009, 85, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Okada, K.; Doki, Y.; Azama, T.; Iwanaga, H.; Miki, H.; Nakayama, M.; Miyata, H.; Takiguchi, S.; Fujiwara, Y.; et al. Injection of LPS causes transient suppression of biological clock genes in rats. J. Surg. Res. 2008, 145, 5–12. [Google Scholar]
- Murphy, B.A.; Vick, M.M.; Sessions, D.R.; Cook, R.F.; Fitzgerald, B.P. Acute systemic inflammation transiently synchronizes clock gene expression in equine peripheral blood. Brain Behav. Immun. 2007, 21, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Shibata, S.; Takahashi, S.; Yokota, S.I.; Hara, R.; Kobayashi, T.; Akiyama, M.; Moriya, T. Physical and inflammatory stressors elevate circadian clock gene mPer1 mRNA levels in the paraventricular nucleus of the mouse. Endocrinology 2001, 142, 4910–4917. [Google Scholar]
- Hwang, J.W.; Sundar, I.K.; Yao, H.; Sellix, M.T.; Rahman, I. Circadian clock function is disrupted by environmental tobacco/cigarette smoke, leading to lung inflammation and injury via a SIRT1-BMAL1 pathway. FASEB J. 2014, 28, 176–194. [Google Scholar] [CrossRef] [PubMed]
- Musiek, E.S.; Lim, M.M.; Yang, G.; Bauer, A.Q.; Qi, L.; Lee, Y.; Roh, J.H.; Ortiz-Gonzalez, X.; Dearborn, J.T.; Culver, J.P.; et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J. Clin. Investig. 2013, 123, 5389–5400. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J.E.; Blaikley, J.; Beesley, S.; Matthews, L.; Simpson, K.D.; Boyce, S.H.; Farrow, S.N.; Else, K.J.; Singh, D.; Ray, D.W.; et al. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl. Acad. Sci. USA 2012, 109, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Migita, H.; Morser, J.; Kawai, K. Rev-erbalpha upregulates NF-kappaB-responsive genes in vascular smooth muscle cells. FEBS Lett. 2004, 561, 69–74. [Google Scholar] [CrossRef]

© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, P.; Li, Z.; Li, X.; Yang, L.; Zhang, L.; Li, N.; Guo, C.; Lu, S.; Wei, Y. Transcriptome Profiling of the Lungs Reveals Molecular Clock Genes Expression Changes after Chronic Exposure to Ambient Air Particles. Int. J. Environ. Res. Public Health 2017, 14, 90. https://doi.org/10.3390/ijerph14010090
Song P, Li Z, Li X, Yang L, Zhang L, Li N, Guo C, Lu S, Wei Y. Transcriptome Profiling of the Lungs Reveals Molecular Clock Genes Expression Changes after Chronic Exposure to Ambient Air Particles. International Journal of Environmental Research and Public Health. 2017; 14(1):90. https://doi.org/10.3390/ijerph14010090
Chicago/Turabian StyleSong, Pengcheng, Zhigang Li, Xiaoqian Li, Lixin Yang, Lulu Zhang, Nannan Li, Chen Guo, Shuyu Lu, and Yongjie Wei. 2017. "Transcriptome Profiling of the Lungs Reveals Molecular Clock Genes Expression Changes after Chronic Exposure to Ambient Air Particles" International Journal of Environmental Research and Public Health 14, no. 1: 90. https://doi.org/10.3390/ijerph14010090





