1. Background
Tremendous amounts of data are currently publicly available, and more is being produced at an unprecedented rate in all domains of life sciences worldwide, promising to generate solutions to diverse problems in health and medicine. Hence, the hallmark of the “big data” era in life sciences involves searching for and integrating biological, medical, and pharmaceutical data from the multitude of independent databases online. However, most of our meta-data is still source bound, in a sense that critical parameters or the conditions the data is generated in are often not disclosed, being disparately stored and in different, unstructured formats. Although some of the databases include links to other resources, these often lack the information required to understand the intent or limitations of the relationship. Hence, the ability of a life scientist to search and retrieve relevant data is hindered by non-standard search interfaces backed by an enormous diversity of data formats that are poorly linked. These problems not only lead to difficulties in navigating through available databases presenting a tremendous barrier to their reuse but also contribute to the non-reproducibility of scientific findings, hindering scientific progress.
Recently, the Linked Data (LD) paradigm has emerged as a mechanism for employing the Web as a means for data and knowledge integration wherein both documents and data are linked [
1]. Importantly, the structure and semantics of the underlying data are kept intact, making this the Semantic Web (SW). LD essentially comprises a set of best practices for publishing and linking structure data on the Web, which allows for publishing and exchanging information in an interoperable and reusable fashion. Ontologies not only help in integrating data from multiple sources in an interoperable way, but also assist in querying and retrieving the specific required information easily. With the widespread adoption of Linked Data, there are several use cases, which are possible due to the interlinking of disparate data into one uniform and global information space [
2]. Linked Data, in these cases, not only assists in creating mashups but also empowers the uncovering of meaningful and impactful relationships and discoveries. These discoveries have enabled scientists to explore the existing data and uncover meaningful outcomes that could not have been uncovered previously.
In this review, we propose avenues for the use of Linked Data in the Life Sciences by identifying three tiers of research practice that are data intensive, time consuming, and lack support from the data science perspective, which would potentially benefit from a paradigm shift. These three tiers are (1) performing a systematic review of the existing body of knowledge, (2) performing a meta-analysis from the literature retrieved as a result of the systematic review, and (3) knowledge discovery of novel links across different evidence streams and databases. Conventionally, each of the tasks associated with these tiers are difficult to perform since combining information from different data sets (which are also in different formats) is not only time consuming but also non-reproducible and results in accidental omission of important information.
The central pillars of evidence-based medicine are constituted of systematic reviews and meta-analyses (SR-MAs) [
3,
4]. They are indispensable in answering specific research (for example, clinical) questions by summarizing the evidence available. They are also an important component of clinical practice guidelines, thus providing the connection between clinical research and practice [
5]. However, currently the reporting of SR-MAs suffers from problems such as incomplete, unclear, and non-transparent reports. As a result, researchers cannot judge their reliability and usefulness, and this adds a significant delay to the meta-analysis process, which is already time consuming. As a result, the translation of clinical research findings to clinical practice is delayed. There are various reasons for this. For example, occasionally access to a key article is behind a paywall, which invokes the paradox that we would not know whether it is worth it to pay for the access to an article unless we were to pay and get access to it. Additionally, retrieving, accessing, and analyzing relevant data is difficult because the data is in different locations and data formats, thus making it non-interoperable. When the researchers gain access to the relevant data it requires significant effort to clean, normalize, and integrate this data, which usually requires assistance from a computer scientist or a bioinformatician. Therefore, there is an urgent need to empower the life scientist in the age of big data with solutions that are freely available, interlinked, and user friendly.
We examine the state of the art followed by demonstrating, through mock examples, the utility of LD using the case study starting with a research question: “Can obesity be a potential cause for breast cancer in later life?” In particular, we start off with (1) analyzing the systematical review process from the existing body of knowledge from published articles, including all the mechanisms described, linking obesity to breast cancer via different mechanisms. Then we select the most likely mechanism—the Insulin-Like Growth Factor 1 Receptor (IGF1R)—for our purposes of demonstration, to focus on to investigate whether this mechanism can explain the association between obesity and breast cancer. Then, we show the ease of access to interoperable data to (2) perform meta-analysis to test this hypothesis and, finally, we show how LD enables (3) knowledge discovery of novel links across different evidence streams, such as drugs and other compounds, which could be causing/promoting/preventing this effect. We argue that utilizing LD in the life sciences will enable data sets to be better Findable, Accessible, Interoperable, and Reusable. Furthermore, integration of LD workflows together will enable a significant improvement in efficiency and reproducibility.
This paper is structured as follows: In
Section 2, we specifically describe the available concepts of the Semantic Web technologies as well as data sets, which are openly available. Then, we propose an LD-based approach that uses these Semantic Web technologies and data sets to enable all three tiers of the research process namely, (1) systematic reviews, (2) meta-analysis, and (3) knowledge discovery. In
Section 3, we describe the proposed framework, followed by
Section 4 demonstrating each of the use cases in detail by describing the state of the art followed by our approach. In
Section 5, we discuss the advantages and challenges of our proposed approach, and we conclude in
Section 6.
2. Preliminaries
2.1. Semantic Web Technologies
Linked Data: The World Wide Web greatly facilitated the share of information around the world. This Web infrastructure supports a vast distributed network of web pages that can refer to each another with global links called URLs (Uniform Resource Locator). However, the idea of the Semantic Web is to not only support a distributed Web at the level of the data but also at the level of the representation. The main idea is that instead of having one web page refer to another, individual data items can indicate another using global references called URI (Uniform Resource Identifiers). This has given rise to Linked Data. Linked Data (w3.org/DesignIssues/LinkedData.html) [
1] involves the formation of typed links between data from different sources on the Web. It has the properties of being machine-readable, having a meaning that is explicitly defined, being linked to other data sets external to itself, and being able to be linked to from external data sets [
1]. The Linked Data principles define the use of Web technologies to establish data-level links among diverse data sources. Linked Data is very useful in cases where exchanges of heterogeneous data are required between distributed systems [
6].
RDF: Resource Description Framework (RDF) (w3.org/TR/rdf-concepts/) is the data model used by the Semantic Web infrastructure to represent this distributed web of data. RDF is an XML-based language for describing information contained in a Web resource. This Web resource can be anything; for example, a Web page or a site. RDF leverages the infrastructure of the Web, using many of its familiar and successful features, while extending them to provide a foundation for a distributed network of data. The properties of RDF are:
Language recommended by W3C [
7], which serves in managing the distributed data.
Used to describe any fact, independent of any domain.
Provides a basis for coding and reusing structured data as well as metadata.
It is structured, i.e., machine-readable as well as human-readable. Machines can do useful operations with the knowledge expressed in RDF.
Enables interoperability among applications by exchanging machine-understandable information on the Web.
RDF Triples: In the Semantic Web, we refer to the things in the world that are described by an RDF expression as resources or entities. A resource is identified by a Uniform Resource Identifier (URI). The use of URIs ensures that the name of a resource is globally unique. A property is a resource that has a name and can also be used to describe some specific relation of the given resource. The property has an object value, which can either be a literal or another resource referenced by a URI. The resource, property, and object value together form a triple (denoting the three components).
RDF triples are written as URIs in angular brackets ending with a period at the end in the form <subject> <predicate> <objective>. Two examples are shown in
Listing 1.
Listing 1. Two examples of RDF triples are shown with the first denoting as Amsterdam as the capital of Netherlands, and the second denoting Amsterdam as the resource of type City (denoting a class in an ontology).
This first triple states that the resource ‘Netherlands’ has the capital ‘Amsterdam’, which is also a resource. The second triple states that the resource ‘Amsterdam’ is of type ‘City’, which is a class in the DBpedia ontology (discussed further in this section). This way, we can link individual facts together, either in the same data set or even across data sets (when the object value resource can be from a different data set), thus calling it Linked Data.
Triple store and SPARQL: RDF data is stored in triple stores, which are software programs that are equipped to store and index RDF data efficiently, in order to enable querying this data easily and effectively. A triple store for RDF data is like DBMS for relational databases. Virtuoso [
8], Sesame [
9], and BigOWLIM [
10] are typical examples of triple stores for desktop and server computers. To access the triples (data) stored in triple stores, there is a query language called SPARQL. “The SPARQL Protocol and RDF Query Language (SPARQL) is the W3C standard query language and protocol for RDF (w3.org/TR/rdf-sparql-query/)” [
11]. SPARQL facilitates the user to write queries that consist of triple patterns along with conjunctions (logical “and”), disjunctions (logical “or”) and/or a set of optional patterns [
12]. The SPARQL query specifies the pattern(s) that the resulting data should satisfy. The results of SPARQL queries can be result sets or RDF graphs.
Ontology: In order to semantically represent data, W3C introduces the concept of an ontology, which is defined as “the terms used to describe and represent an area of knowledge” [
13]. An ontology is specific to one domain area of knowledge. The ontology contains concepts/terms, also called classes, and relationships among those terms. The relationships between these classes can be expressed by using a hierarchy, i.e., superclasses represent higher-level concepts and subclasses represent finer, more specific, concepts. The finer classes inherit all the features and attributes that the higher classes have. Additionally, properties express another level of relationship. In other words, an ontology defines a set of classes (e.g., “Writer”, “Book”, “Person”), and their hierarchy, i.e., which class is a subclass of another one (e.g., “Writer” is a subclass of “Person”). The ontology also defines how these classes interact with each other, i.e., how different classes are connected to each other via properties (e.g., a “Book” has an author of type “Writer”).
FAIR principles: Another important concept that has been recently introduced is making all this well-structured data as well as metadata on the Web FAIR [
14]—that is, making it Findable, Accessible, Interoperable, and Reusable. The FAIR Principles focus on enhancing the ability of machines to automatically find and use the data, in addition to supporting its reuse by individuals.
smartAPI: Lastly, it is important to provide Application Programming Interfaces (APIs) to retrieve the data. APIs are a set of functions and procedures that allow the creation of applications, which access the features or data of an operating system, application, or other service. Our previous work introduced smartAPI [
15] (smart-api.info), with the aim to make APIs FAIR: Findable with the API metadata and the registry; Accessible with the detailed API operations metadata; Interoperable with the responseDataType metadata (profiler); and Reusable with the access to existing APIs stored in an open repository. smartAPI leverages the use of semantic technologies such as ontologies and Linked Data for the annotation, discovery, and reuse of APIs.
2.2. Data Sets
Life sciences is rapidly becoming the most data intensive branch of sciences that is generating data from the molecular level to the population level. As a part of our case study we have chosen representative data sources, which are described in this section as part of our user scenario. It is important to mention, however, that the database selection in the case study is not exhaustive and inclusion of other databases is not only possible but also intended within the LD framework.
MEDLINE: MEDLINE is the U.S. National Library of Medicine bibliographic database which contains more than 24 million references to journal articles in life sciences with a concentration on biomedicine. MEDLINE is continuously indexing articles from 5400 of the world’s leading life sciences journals. A distinctive feature of MEDLINE is that the records are indexed with NLM Medical Subject Headings (MeSH). PubMed (
https://www.ncbi.nlm.nih.gov/pubmed) provides free access to MEDLINE and links to full text articles when possible.
Medical Subject Headings (MeSH): MeSH is curated and maintained by the National Library of Medicine and consists of sets of terms naming descriptors in a hierarchical structure that permits searching at various levels of specificity. At the most general level of the hierarchical structure are very broad headings. More specific headings are found at more narrow levels of the thirteen-level hierarchy. In the 2016 version of MeSH are defined 27,883 descriptors associated with over 87,000 entry terms to assist the users in finding the most appropriate MeSH Heading to their topic of interest.
Gene Expression Omnibus (GEO): GEO (
https://www.ncbi.nlm.nih.gov/geo/) is a publicly available, functional genomics data repository supporting MIAME-compliant (Minimum Information about a Microarray Experiment) data submissions. Data that is array- and sequence-based is accepted. There are internal tools available to help users to query and download experiments from curated database profiles. There is also an online graphical user interface for data export and basic analysis [
16].
ArrayExpress: ArrayExpress (
https://www.ebi.ac.uk/arrayexpress/) is the major repository for archiving functional genomics data from microarray and sequencing platforms to support reproducible research. ArrayExpress also facilitates submissions in compliance with Minimum Information about a Microarray Experiment (MIAME) and Minimum Information about a Sequencing Experiment (MINSEQE) guidelines [
17].
STRING-db: STRING (
https://string-db.org/) is a database of known and predicted protein–protein interactions. The interactions include direct (physical) and indirect (functional) associations; they stem from computational prediction, from knowledge transfer between organisms, and from interactions aggregated from other databases [
18]. As of 31 October 2017, STRING database covers 9,643,763 proteins from 2031 organisms.
DrugBank: The DrugBank database (
https://www.drugbank.ca/) is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e., chemical, pharmacological, and pharmaceutical) data with comprehensive drug target information. The database contains 9591 drug entries including 2037 FDA-approved small molecule drugs, 241 FDA-approved biotech (protein/peptide) drugs, 96 nutraceuticals, and over 6000 experimental drugs. Additionally, 4661 non-redundant protein sequences are linked to these drug entries. Each entry (DrugCard) contains more than 200 data fields with half of the information being devoted to drug/chemical data and the other half devoted to drug target or protein data [
19].
Bio2RDF: Bio2RDF [
20] is a biological database that uses Semantic Web technologies to provide interlinked life science data. Bio2RDF is built from RDFizer programs written in JSP, and uses the Sesame open source triple store and an OWL ontology. Documents from 35 public bioinformatics databases such as KEGG, PDB, MGI, HGNC, and several of NCBI’s databases are made available in RDF and follow a unique URL pattern in the form of
http://bio2rdf.org/namespace:id. Bio2RDF is based on a three-step approach to build mashups of bioinformatics data.
PubChem: PubChem (
https://pubchem.ncbi.nlm.nih.gov/) is the largest database of chemical molecules and their activities against biological assays. It is open access and many compound structures as well as data sets can be downloaded for free. It also contains descriptions of substances and small molecules with fewer than 1000 atoms and 1000 bonds [
21].
ChEBI: Chemical Entities of Biological Interest (ChEBI) (
http://www.ebi.ac.uk/chebi/) is a database and ontology of molecular entities focused on ‘small’ chemical compounds. The molecular entities in ChEBI are either products of nature or synthetic products that have potential bioactivity. Molecules directly encoded by the genome, such as nucleic acids, proteins, and peptides derived from proteins by proteolytic cleavage, are not as a rule included in ChEBI [
22].
OMIM: Online Mendelian Inheritance in Man (OMIM) (
https://www.omim.org/) is a continuously updated catalog of human genes and genetic disorders and traits, with a particular focus on the gene–phenotype relationship. As of 31 October 2017, approximately 8488 of the over 24,000 entries in OMIM represented phenotypes; the rest represented genes, many of which were related to known phenotypes [
23].
HPA: The Human Protein Atlas (HPA) has been established with the aim of mapping all the human proteins in cells, tissues, and organs using integration of various-omics technologies, including antibody-based imaging, mass-spectrometry-based proteomics, transcriptomics, and systems biology [
24].
3. Proposed Framework
We propose an approach to couple the above-mentioned databases to facilitate the three tiers of the research process in a continuous fashion.
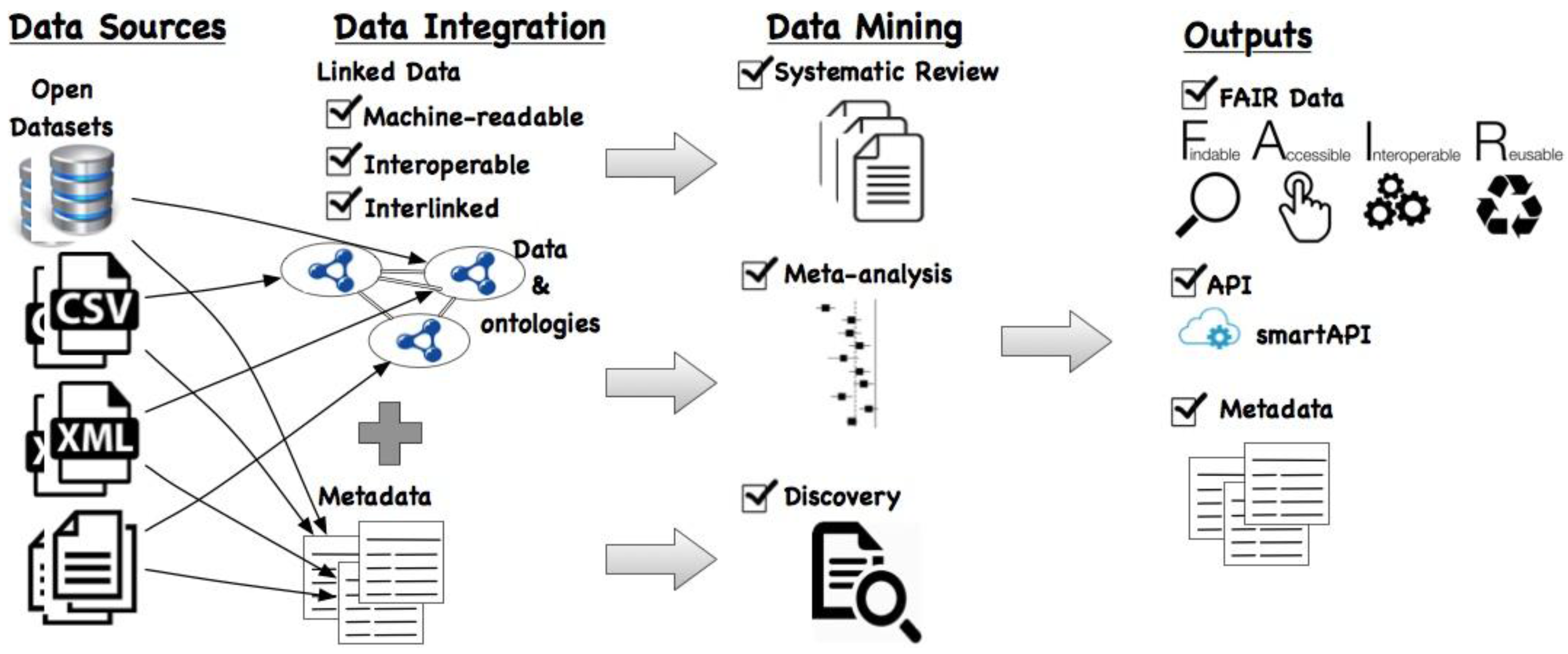
Figure 1 depicts a general overview of our proposed framework. As the input, (open) data from sources—which can be in any of proprietary formats such as database, CSV, XML, or documents—are considered. We propose to convert this data into Linked Data using existing tools such as the RDF Mapping Language [
25] (RML) (
http://semweb.mmlab.be/rml/spec.html), Sparqlify (
https://github.com/AKSW/Sparqlify), OpenRefine’s RDF extension (
http://refine.deri.ie/), etc. RML is a language for specifying customized mappings from heterogeneous data structures (including databases, XML, CSV) to the RDF data model. An example of mapping a CSV file to RDF is shown using the RML mapping language (
http://rml.io/spec.html#example-CSV). Essentially, the table name is the resource, the column names map to properties, and the values in each cell correspond to the object value in the triple. Additionally, each resource is typed by associating it to a class in an (existing) ontology.
After conversion of data sets into the RDF format, they are not only machine-readable but also more interoperable since they are represented using a standardized vocabulary. To interlink the different data sets, there are several tools available such as LIMES (
http://aksw.org/Projects/LIMES.html), SILK (
http://silkframework.org/), etc., which perform lexical matches over the object values to (semi-)automatically find matches between data sets which are in RDF format.
As a result, with the data sets including the textual data annotated using classes from standard ontologies, retrieving similar studies to perform (1) systematic reviews and (2) meta-analysis becomes easier. Moreover, with the different data sets interlinked with one another, the (3) discovery of novel links between these data sets is now possible by querying this integrated data space.
In the proposed framework, it is then mandated that the outputs of these three analyses should be made available using the FAIR principles such that not only the data but also the metadata associated with it be maximally findable, accessible, interoperable, and reusable. This will greatly impact the reusability of data, metadata, and results to be applied in different use cases. Moreover, it is possible to make the data available via an API, specifically using the smartAPI specification. This will facilitate finding and exploring data sets relevant to the particular use case by querying the smartAPI registry. In the following sections, we describe each of the three use cases in detail that follow the proposed approach.
4. Use Cases
Using the Semantic Web technologies and Linked Data (as described in
Section 2.1), in this section we demonstrate their utility in three specific use cases: (1) systematic reviews, (2) meta-analysis, and (3) knowledge discovery. We describe the current state of the art for each, followed by proposals for solutions using SW technologies and LD.
4.1. Systematic Reviews
Today, conventional search for scientific papers in order to perform systematic reviews includes searching for articles annotated with specific keywords. The authors choose these keywords when they submit their paper. MEDLINE uses a hierarchical structure called MeSH subject terms to annotate each article and continuously manually curates every new article published in the journals that are included in MEDLINE. This manual curation is quite tedious and time consuming, and the searches often exclude the article from MEDLINE journals that have not yet been fully indexed (new articles), as well as other PubMed citations that are not indexed for MEDLINE. In this manuscript, we demonstrate the premise of LD with a real-world example that we have recently published on the systematic analysis of the body of evidence linking obesity to breast cancer. The detailed methodology can be found in the original paper [
26]. In summary, the conventional identification of relevant mechanisms linking obesity to breast cancer includes:
Identifying MeSH terms that are sufficient for describing the exposure (obesity) and the outcome (breast cancer) independently.
Identifying candidate mechanistic MeSH terms that could be the link between the exposure and the outcome.
Performing two independent searches: one with the exposure and one with the outcome to retrieve all the articles with all their metadata and associated MeSH terms.
Performing an enrichment analysis on the candidate mechanistic MeSH terms with the results of these two searches to rank the candidate MeSH terms for their association with the exposure and/or the outcome according to their preferential association in both exposure and outcome.
The top candidate mechanistic link and all associated studies are then downloaded to be further studied for assessing the overall body of evidence linking obesity to the mechanistic link (first subreview) and the mechanistic link to the breast cancer (second subreview).
Next steps in the systematic review framework entail extracting and coding the information (human studies/animal models/cell models) from the papers manually by independent researchers, and comparing the resulting documents with a scheme such as the Grading of Recommendations Assessment, Development, and Evaluation (GRADE) assessment [
27] to evaluate the quality of the body of evidence per evidence stream (i.e., human and animal studies) within each subreview.
This systematic review process aims to integrate the body of knowledge spread over hundreds of manuscripts in various databases (findability), some behind paywalls (accessibility), and in different formats (interoperability), to test the merits of a specific scientific hypothesis. However, the sheer volume of literature in combination with the heterogeneity of the types of studies (human/animal/cell) and methods being used in those studies hinders its reproducibility [
26,
28].
Our proposal is to use ontologies as a method for creating reporting standards for research articles. Ontologies provide structured vocabularies, terminologies, and reasoning capabilities that enable standardized and semantically interconnected machine-readable sections in a research article. Annotating individual facts in articles using an ontology can enable and speed up the search for and retrieval of relevant articles, as well as ensure that all related articles are included. Additionally, this facilitates the integration of the sparse data that is currently been stored in different locations and formats in a central repository. Then, by storing all the articles annotated using ontologies in a central repository/triple store, this will address the problem of finding and retrieving relevant articles by performing a single query. Another problem is dealing with the sheer volume of the data. That is, in our use case, when we query repositories using MeSH terms, we tend to retrieve a huge number of articles linked more to the parent class (e.g., neoplasms vs breast neoplasms). Using SPARQL queries, we can then specify particular MeSH terms from any level in the hierarchy in one query to retrieve only and all of the relevant articles.
In particular, the queries one can ask are: “Give me all the studies from MEDLINE which have the MeSH term “Obesity” between 1990 and today.” “Give me all the studies from MEDLINE which have the MeSH term “Breast Neoplasms” between years 1990 and today.” “Find the MeSH terms that are linking those two sets of studies together.”
Listing 2 shows an example SPARQL query to retrieve all studies that are linked using two specific keywords. In our case study, detailed in our previous paper, performing the above-mentioned steps resulted in a list of mechanistic processes potentially linking obesity to breast cancer. After analyzing the papers and based on careful consideration, the Insulin-like Growth Factor Type 1 Receptor (IGF1R) was chosen as a potential candidate to follow up [
26]. Additionally,
Figure 2 shows detailed steps of the systematic review process along with the advantages of using Semantic Web/Linked Data.
Listing 2. An example SPARQL query to query the repository to retrieve articles with two specific MeSH terms (concepts) from the year 1990 onwards. Part of the query (without the triple and filter for year) can be performed on
https://id.nlm.nih.gov/mesh/query1.
PREFIX meshv: <http://id.nlm.nih.gov/mesh/vocab#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX ex: <http://example.com/>
SELECT ?article ?label ?year
WHERE
{
?article ex:meshConcept ?meshdescriptor .
?article ex:year ?year .
?meshdescriptor a meshv:Descriptor .
?meshdescriptor meshv:concept ?concept .
?concept rdfs:label ?cName .
FILTER(REGEX(?concept ,’breast neoplasm ’,’i’) || REGEX(?concept ,’obesity ’,’i’)) FILTER (?year > 1990)
} 4.2. Meta-Analysis
After the systematic review process and identifying candidate mechanistic links in the previous step, the next step is to identify the key publications with their data available in publicly accessible databases. The time-consuming step here is to go through all the publications screened at the systematic review phase to identify the type of data available from each publication, assess its relevance, extract the links to the data, and download the data from the repositories.
After downloading the data from various repositories, the problem of interoperability arises due to the non-standard collection of metadata about the studies across different data types and data repositories such as GEO and ArrayExpress. However, some of the interoperability issues can be addressed by simply searching for the information in the publication or contacting the authors of the manuscript to which the data belongs. However, this is also time consuming and error-prone. Finally, the complete data set is corrected for systematic batch effects present due to having multiple studies in the data set and normalized together. This allows for assessing the potential background noise and determining the signal/noise ratio in the data set.
In this case, we propose the use of the ontology-annotated documents stored in a central repository (triple store) as the starting point of performing the meta-analysis. That is, once we have queried the triple store and retrieved the relevant articles (as part of the systematic review step), we will then (automatically by querying the triple store) retrieve all the links in these constituent studies. These links correspond to the actual data from relevant databases. This data in these databases should also be represented in the machine-readable RDF format along with good quality metadata. This will facilitate the easy retrieval of the data by means of SPARQL queries. Additionally, by linking the data to specific classes in an ontology, it will make it easier to extract specific quantitative information and directly output to a desired format for further (meta-)analysis. This will reduce the burden of manually extracting the individual information from each database and combining it to perform the meta-analysis.
Listing 3 shows an example SPARQL query for the following question: Give me all the data from the samples in GEO and ArrayExpress that are from human breast samples, that are associated with studies which have the MeSH term “Breast Neoplasms” and “Receptor, IGF Type 1” or “Obesity” and “Receptor, IGF Type 1”, and conducted using the platform “Affymetrix Human Transcriptome Array 2.0”.
Listing 3. An example SPARQL query to retrieve all the data from the samples in GEO and ArrayExpress that are from human breast samples that are associated with studies which have the MeSH terms: (i) Breast Neoplasms and Receptor Insulin-Like Growth Factor (IGF) Type 1 or (ii) Obesity and Receptor IGF Type 1 and conducted using the platform Affymetrix Human Transcriptome Array 2.0.
4.3. Knowledge Discovery
After performing meta-analysis of the data extracted from various studies to confirm or refute the mechanistic link as the mechanism linking obesity to breast cancer, we want to investigate existing compounds or drugs that are either designed to act on the mechanism directly or effect the mechanism indirectly, where information is available in different data sources. This allows us to discover compounds that can be tested experimentally or by clinical trials to potentially alleviate the effect of the exposure (obesity) on the outcome (breast cancer).
In searching for compounds that have an effect on IGF1R, we first access the connectivity map (
https://portals.broadinstitute.org/cmap/) [
29] or CancerDR (
http://crdd.osdd.net/raghava/cancerdr/) [
30] to analyze the existing data on more than 7000 expression profiles representing 1309 compounds in the connectivity map or 148 anticancer drugs and their effectiveness against around 1000 cancer cell lines, respectively. After identifying candidate compounds, the final step is to assess their suitability as a follow-up study, which requires the extraction of the PubChem Compound Identifier, DrugBank ID, and/or ChEBI IDs of the compounds to search in various databases for reported efficacy and adverse effects.
In our proposed framework, with all the biologically relevant data sets available as Linked Data (specifically biological data sets as part of the Bio2RDF project) we can perform this analysis by querying the data sets and traversing the interlinks between them. For example, by querying Bio2RDF, we can extract the IDs from PubChem, DrugBank, and ChEBI directly in one SPARQL query along with the associated information from each of these data sources.
Listing 4 shows an example SPARQL query for the following question: “Find all direct interactions of IGF1R protein from STRING-db and give me all the compounds that are described to have DrugBank interactions with IGF1R or any other interacting partners.”
Listing 4. An example SPARQL query to retrieve all direct interactions of the Insulin-Like Growth Factor 1 Receptor (IGF1R) protein from STRING-db and all the compounds that are described to have DrugBank interactions with IGF1R or any other interacting partners.
Additionally, by making the data as well as the metadata and the results FAIR, the methodology and the results can be reproduced for any other similar use case. Also, exposing the data using the smartAPI specification will facilitate the finding and exploring of connections pertaining to the use case, especially those that were unknown before and could not have been discovered.
5. Discussion
Deployment of Linked Data in Life Sciences requires a consistent, functioning, and overarching framework that is able to integrate various steps of the research cycle. We have identified three fundamental tiers of the research cycle in life sciences (conducting systematic reviews, meta-analysis of existing data, and knowledge discovery of novel links across evidence streams and databases) that would greatly profit from the Linked Data implementation. The systematic review and meta-analysis (SR-MA) of available data from comparable studies of interest has been proven to be a very powerful tool, especially in epidemiology (clinical trials) and medicine (effects of medical interventions) [
31], for confidently estimating the role of various exposures to different outcomes. Although there are success stories [
32,
33], the main challenges in the meta-analysis of existing data lies in the accessibility of the study data and its interoperability [
14]. Over the last decade, there have been numerous efforts to improve the quality of reporting for meta-analyses for diagnostic, randomized, and non-randomized study designs and observational biomedical research studies by establishing guidelines, checklists, and standards [
34,
35,
36,
37]. However, none of the reporting guidelines fully incorporate the full set of details required for the conduct of SR-MAs [
38,
39,
40] One example is that of the CONSORT (Consolidated Standards of Reporting Trials) statement, which is a checklist developed to tackle the problems arising from inadequate reporting of Randomized Control Trials. However, this checklist only includes 22 of the 100 items needed for successfully conducting SR-MAs [
41,
42].
The use of Semantic Web technologies and Linked Data, specifically ontologies, has been increasingly applied in several life science use cases. In our previous work [
41], we proposed a Center of Excellence in Research Reporting in Neurosurgery (CERR-N) and the creation of a clinically significant computational ontology to encode Randomized Controlled Trial (RCT) studies, specifically in neurosurgery. The study showed that standardized reporting for neurosurgery articles can be reliably achieved through the integration of a computational ontology within the context of a CERR-N. A similar approach can be applied for our particular use cases and further expanded to assist in meta-analysis and knowledge discovery. In fact, a number of studies have explored and successfully demonstrated the advantages of ontology-driven meta-analysis [
42,
43].
The process of annotating articles with ontologies can be a tedious one, if it has to be done manually. In February 2008, the FEBS editorial board designed an experiment asking authors to provide structured information on protein interactions along the lines of the minimum information requirement for reporting protein interaction experiments (MIMIx) recommendations [
44]. However, the author input was of poor quality, which, in turn, burdened the curators, as they had to invest a lot of time into cleaning the input. The conclusion was that having an automated system that would provide a list of relevant identifiers would be beneficial. This would likely help to save authors and curators a significant amount of time. To complement this study, the BioCreative (
http://www.biocreative.org/) organizers challenged text-mining researchers to reproduce a subset of the annotations provided by the Structured Digital Abstracts. This study concluded that text mining could help to select specific articles for database curators (highest accuracy = 92%). Recently, to automatically annotate articles using concepts from existing ontologies, there is the BioPortal annotator (
https://bioportal.bioontology.org/annotator) tool. The annotator provides an automated method to annotate input text with the 650+ (biologically relevant) ontologies available in BioPortal (
https://bioportal.bioontology.org/annotator). The annotator allows different filters and settings to narrow down to specific ontologies or UMLS semantic types. The annotations can be exported in the XML or JSON formats. We have performed this annotation using the BioPortal annotator, as a proof of principle, on two sections of our manuscript (abstract and use cases) and we provide the annotations as
Supplementary Materials in JSON format. However, one may argue that there may not always be (direct) matches for all concepts in an article. To address this issue, there are efforts such as the Semanticscience Integrated Ontology (SIO) [
45]. SIO is an upper level comprising essential types and relations for the rich description of arbitrary (real, hypothesized, virtual, fictional) objects, processes, and their attributes, with the goal of facilitating knowledge discovery. In spite of these tools and ontologies, we argue that the journals themselves should mandate the tagging of articles with standardized terms from existing ontologies/terminologies along with the article. This enriched metadata should be then published digitally alongside the article to enhance its FAIRness.
LD is dramatically growing, and currently more than 50 billion facts are represented (
http://lod-cloud.net/state/). LD has also enabled knowledge discovery in a number of interesting use cases by overcoming typical data management and consumption issues such as heterogeneity, integration, and exploration. Successful use cases of LD have been in healthcare research [
46,
47]; the biomedical domain, e.g., for drug discovery [
48,
49]; or for identifying patterns in particular diseases [
50]. LD assists in building mashups by interlinking heterogeneous and disparate data from multiple sources but also enables the uncovering of meaningful and impactful relationships between data (facts). These discoveries have paved the way for scientists to explore the existing data and uncover meaningful insights that would not have been possible previously.
Our proposal of a FAIR-enabled framework for life sciences goes beyond these existing studies and not only enables streamlined systematic review, meta-analysis, and knowledge discovery from disparate data sources, but also enables the results to be reproduced [
51]. Moreover, by storing all the data in a central repository (triple store) in a machine-readable structured format, the relevant data can be easily findable and accessible. Importantly, this would enable dynamic and up-to-date access of the data. Besides this, the use of RDF to structure the data facilitates the interoperability between sources which are currently disparate and in different formats.
Additionally, there should be a visual interface to allow researchers to search, explore, and visualize the available data as well as the results without necessarily knowing about the underlying data structure. Tools such as DISQOVER (
https://www.disqover.com/) and COREMINE (
https://www.coremine.com/medical/#search) already provide this functionality to explore and visualize a variety of data sets and specifically the constituent data types. This would be highly beneficial with added functionalities to directly perform the meta-analysis in the interface itself. Currently, however, these tools only have access to very high-level meta-data such as the publication records; hence, they do not have any information about the type of experiments conducted and whether the data from these experiments is publicly available.
Finally, successful implementation of Linked Data in the above-mentioned three tiers and benchmarking of the LD-assisted research cycle against the conventional research cycle in life sciences will constitute the first step in uniting and standardizing the available resources in the life sciences domain. This will further incentivize the use of the Linked Data paradigm in future studies.