Stem Cell-Derived Models of Viral Infections in the Gastrointestinal Tract
Abstract
:1. Introduction
2. Defining Gastrointestinal Models
3. Development of Mini-Intestinal Models
3.1. Enteroid Development
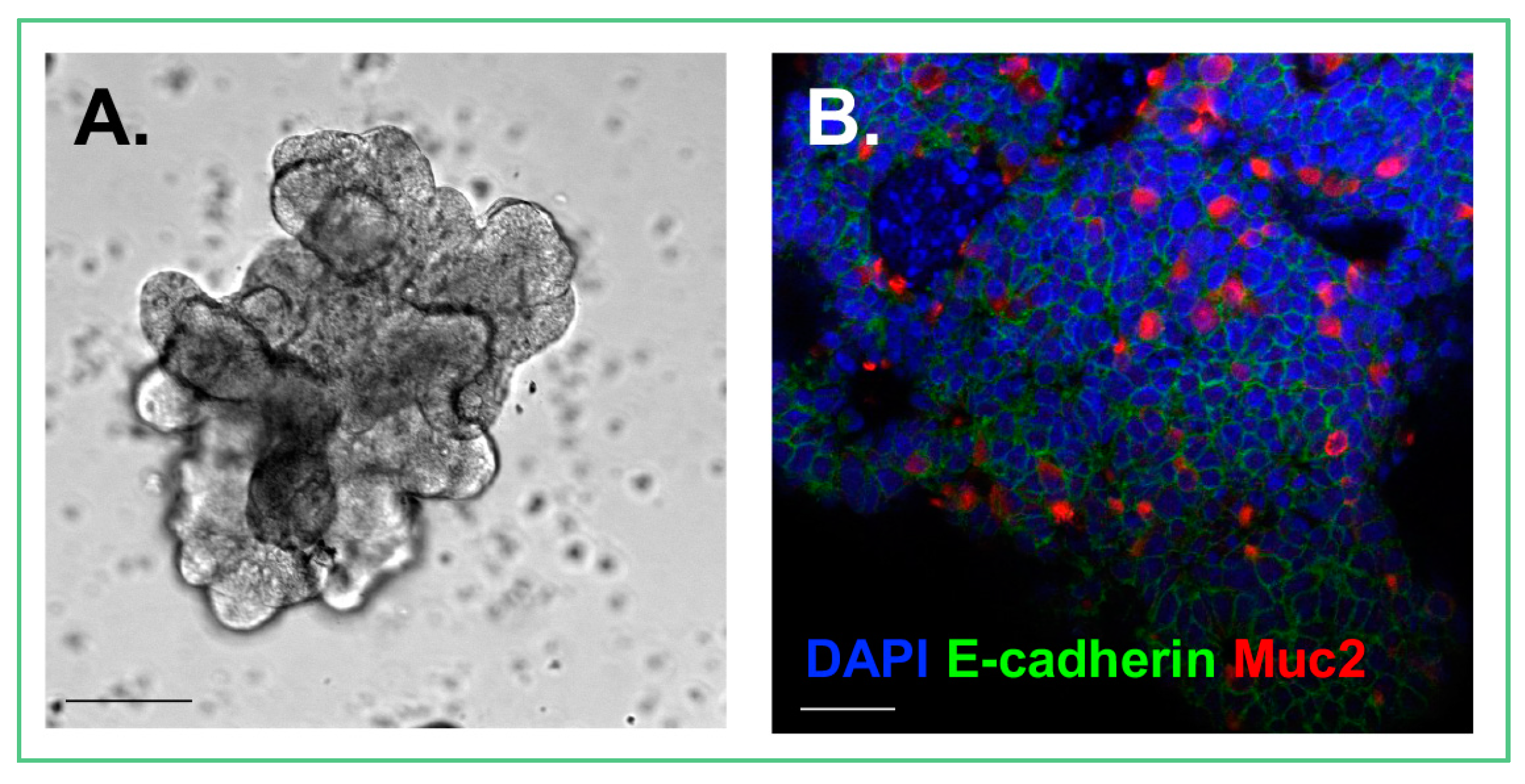
3.2. Organoid Development
4. Mini-Intestinal Differentiation and Morphology
4.1. Region-Specific Differentiation
4.2. Crypt-Villus Development
4.3. Cell Types within Crypts
5. In Vitro Models to Study Viral Infections
5.1. Human Intestinal Organoids to Model Rotaviral Infections
5.2. Human Intestinal Enteroids to Model Rotaviral Infection
5.3. Cultivating Norovirus in Human Intestinal Enteroids
5.4. Enteroids as a Model of Enterovirus Infection
5.5. Enteroid Model of Adenoviral Infection
6. Conclusions and Future Directions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Sinagoga, K.L.; Wells, J.M. Generating human intestinal tissues from pluripotent stem cells to study development and disease. EMBO J. 2015, 34, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Buczacki, S.J.; Zecchini, H.I.; Nicholson, A.M.; Russell, R.; Vermeulen, L.; Kemp, R.; Winton, D.J. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 2013, 495, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Kiyono, H. Epithelial barrier: An interface for the cross-communication between gut flora and immune system. Immunol. Rev. 2012, 245, 147–163. [Google Scholar] [CrossRef] [PubMed]
- Bevins, C.L.; Salzman, N.H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 2011, 9, 356–368. [Google Scholar] [CrossRef] [PubMed]
- O’Ryan, M.; Prado, V.; Pickering, L.K. A millennium update on pediatric diarrheal illness in the developing world. Semin. Pediatr. Infect. Dis. 2005, 16, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef] [PubMed]
- McCracken, K.W.; Howell, J.C.; Wells, J.M.; Spence, J.R. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat. Protoc. 2011, 6, 1920–1928. [Google Scholar] [CrossRef] [PubMed]
- Stelzner, M.; Helmrath, M.; Dunn, J.C.; Henning, S.J.; Houchen, C.W.; Kuo, C.; Lynch, J.; Li, L.; Magness, S.T.; Martin, M.G.; et al. A nomenclature for intestinal in vitro cultures. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G1359–G1363. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S.R.; Hill, D.R.; Altheim, C.H.; Dedhia, P.H.; Taylor, M.J.; Tsai, Y.H.; Chin, A.M.; Mahe, M.M.; Watson, C.L.; Freeman, J.J.; et al. Transcriptome-wide Analysis Reveals Hallmarks of Human Intestine Development and Maturation In Vitro and In Vivo. Stem Cell Rep. 2015, 4, 1140–1155. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.R.; Spence, J.R. Gastrointestinal Organoids: Understanding the Molecular Basis of the Host-Microbe Interface. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Dye, B.R.; Hill, D.R.; Ferguson, M.A.; Tsai, Y.H.; Nagy, M.S.; Dyal, R.; Wells, J.M.; Mayhew, C.N.; Nattiv, R.; Klein, O.D.; et al. In vitro generation of human pluripotent stem cell derived lung organoids. Elife 2015, 4, e05098. [Google Scholar] [CrossRef] [PubMed]
- Watson, C.L.; Mahe, M.M.; Munera, J.; Howell, J.C.; Sundaram, N.; Poling, H.M.; Schweitzer, J.I.; Vallance, J.E.; Mayhew, C.N.; Sun, Y.; et al. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 2014, 20, 1310–1314. [Google Scholar] [CrossRef] [PubMed]
- Aurora, M.; Spence, J.R. hPSC-derived lung and intestinal organoids as models of human fetal tissue. Dev. Biol. 2016, 420, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S.R.; Freeman, J.J.; Wieck, M.M.; El-Nachef, W.; Altheim, C.H.; Tsai, Y.H.; Huang, S.; Dyal, R.; White, E.S.; Grikscheit, T.C.; et al. Generation of tissue-engineered small intestine using embryonic stem cell-derived human intestinal organoids. Biol. Open 2015, 4, 1462–1472. [Google Scholar] [CrossRef] [PubMed]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Shimokawa, M.; Date, S.; Takano, A.; Matano, M.; Nanki, K.; Ohta, Y.; Toshimitsu, K.; Nakazato, Y.; Kawasaki, K.; et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell 2016, 18, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.K.; Stange, D.E.; Sato, T.; Karthaus, W.; Farin, H.F.; Huch, M.; van Es, J.H.; Clevers, H. Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods 2011, 9, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Saxena, K.; Blutt, S.E.; Ettayebi, K.; Zeng, X.L.; Broughman, J.R.; Crawford, S.E.; Karandikar, U.C.; Sastri, N.P.; Conner, M.E.; Opekun, A.R.; et al. Human Intestinal Enteroids: A New Model to Study Human Rotavirus Infection, Host Restriction, and Pathophysiology. J. Virol. 2015, 90, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Ootani, A.; Li, X.; Sangiorgi, E.; Ho, Q.T.; Ueno, H.; Toda, S.; Sugihara, H.; Fujimoto, K.; Weissman, I.L.; Capecchi, M.R.; et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 2009, 15, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Gregorieff, A.; Begthel, H.; Clevers, H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003, 17, 1709–1713. [Google Scholar] [CrossRef] [PubMed]
- Kuhnert, F.; Davis, C.R.; Wang, H.T.; Chu, P.; Lee, M.; Yuan, J.; Nusse, R.; Kuo, C.J. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc. Natl. Acad. Sci. USA 2004, 101, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Carmon, K.S.; Gong, X.; Lin, Q.; Thomas, A.; Liu, Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 11452–11457. [Google Scholar] [CrossRef] [PubMed]
- Dignass, A.U.; Sturm, A. Peptide growth factors in the intestine. Eur. J. Gastroenterol. Hepatol. 2001, 13, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Haramis, A.P.; Begthel, H.; van den Born, M.; van Es, J.; Jonkheer, S.; Offerhaus, G.J.; Clevers, H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 2004, 303, 1684–1686. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Li, Y.; Zhao, B.; Xu, C.; Liu, Y.; Li, H.; Zhang, B.; Wang, X.; Yang, X.; Xie, W.; et al. Bmp restricts stemness of intestinal lgr5(+) stem cells by directly suppressing their signature genes. Nat. Commun. 2017, 8, 13824. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Giltay, R.; Talts, U.; Timpl, R.; Talts, J.F. Expression and distribution of laminin α1 and α2 chains in embryonic and adult mouse tissues: An immunochemical approach. Exp. Cell Res. 2002, 275, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Fuller, M.K.; Faulk, D.M.; Sundaram, N.; Mahe, M.M.; Stout, K.M.; von Furstenberg, R.J.; Smith, B.J.; McNaughton, K.K.; Shroyer, N.F.; Helmrath, M.A.; et al. Intestinal stem cells remain viable after prolonged tissue storage. Cell Tissue Res. 2013, 354, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Clevers, H. Primary mouse small intestinal epithelial cell cultures. In Epithelial Cell Culture Protocols; Humana Press: Totowa, NJ, USA, 2013; pp. 319–328. [Google Scholar]
- Yu, H.; Hasan, N.M.; In, J.G.; Estes, M.K.; Kovbasnjuk, O.; Zachos, N.C.; Donowitz, M. The Contributions of Human Mini-Intestines to the Study of Intestinal Physiology and Pathophysiology. Annu. Rev. Physiol. 2017, 79, 291–312. [Google Scholar] [CrossRef] [PubMed]
- VanDussen, K.L.; Marinshaw, J.M.; Shaikh, N.; Miyoshi, H.; Moon, C.; Tarr, P.I.; Ciorba, M.A.; Stappenbeck, T.S. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 2015, 64, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Ameri, J.; Stahlberg, A.; Pedersen, J.; Johansson, J.K.; Johannesson, M.M.; Artner, I.; Semb, H. FGF2 specifies hESC-derived definitive endoderm into foregut/midgut cell lineages in a concentration-dependent manner. Stem Cells 2010, 28, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Gibson, J.D.; Miyamoto, S.; Sail, V.; Verma, R.; Rosenberg, D.W.; Nelson, C.E.; Giardina, C. Intestinal lineage commitment of embryonic stem cells. Differentiation 2011, 81, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.M.; Spence, J.R. How to make an intestine. Development 2014, 141, 752–760. [Google Scholar] [CrossRef] [PubMed]
- McKie, A.T.; Barrow, D.; Latunde-Dada, G.O.; Rolfs, A.; Sager, G.; Mudaly, E.; Mudaly, M.; Richardson, C.; Barlow, D.; Bomford, A.; et al. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science 2001, 291, 1755–1759. [Google Scholar] [CrossRef] [PubMed]
- Krasinski, S.D.; Upchurch, B.H.; Irons, S.J.; June, R.M.; Mishra, K.; Grand, R.J.; Verhave, M. Rat lactase-phlorizin hydrolase/human growth hormone transgene is expressed on small intestinal villi in transgenic mice. Gastroenterology 1997, 113, 844–855. [Google Scholar] [CrossRef]
- Colombo, V.; Lorenz-Meyer, H.; Semenza, G. Small intestinal phlorizin hydrolase: The “beta-glycosidase complex”. Biochim. Biophys. Acta 1973, 327, 412–424. [Google Scholar] [CrossRef]
- Dawson, P.A.; Lan, T.; Rao, A. Bile acid transporters. J. Lipid Res. 2009, 50, 2340–2357. [Google Scholar] [CrossRef] [PubMed]
- Middendorp, S.; Schneeberger, K.; Wiegerinck, C.L.; Mokry, M.; Akkerman, R.D.; van Wijngaarden, S.; Clevers, H.; Nieuwenhuis, E.E. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells 2014, 32, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Nijenhuis, T.; Hoenderop, J.G.; van der Kemp, A.W.; Bindels, R.J. Localization and regulation of the epithelial Ca2+ channel TRPV6 in the kidney. J. Am. Soc. Nephrol. 2003, 14, 2731–2740. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yamamoto, Y.; Wilson, L.H.; Zhang, T.; Howitt, B.E.; Farrow, M.A.; Kern, F.; Ning, G.; Hong, Y.; Khor, C.C.; et al. Cloning and variation of ground state intestinal stem cells. Nature 2015, 522, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Shaffiey, S.A.; Jia, H.; Keane, T.; Costello, C.; Wasserman, D.; Quidgley, M.; Dziki, J.; Badylak, S.; Sodhi, C.P.; March, J.C.; et al. Intestinal stem cell growth and differentiation on a tubular scaffold with evaluation in small and large animals. Regen. Med. 2016, 11, 45–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gunasekara, D.B.; Reed, M.I.; DiSalvo, M.; Bultman, S.J.; Sims, C.E.; Magness, S.T.; Allbritton, N.L. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 2017, 128, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, W.; Roh, T.; Estes, M.K.; Kaplan, D.L. In vitro enteroid-derived three-dimensional tissue model of human small intestinal epithelium with innate immune responses. PLoS ONE 2017, 12, e0187880. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Lee, J.; Choi, J.H.; Bahinski, A.; Ingber, D.E. Co-culture of Living Microbiome with Microengineered Human Intestinal Villi in a Gut-on-a-Chip Microfluidic Device. J. Vis. Exp. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A.; et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 2871. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Leblond, C.P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I. Columnar cell. Am. J. Anat. 1974, 141, 461–479. [Google Scholar] [CrossRef] [PubMed]
- Bjerknes, M.; Cheng, H. Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology 1999, 116, 7–14. [Google Scholar] [CrossRef]
- Bjerknes, M.; Cheng, H. Intestinal epithelial stem cells and progenitors. Methods Enzymol. 2006, 419, 337–383. [Google Scholar] [PubMed]
- Tetteh, P.W.; Basak, O.; Farin, H.F.; Wiebrands, K.; Kretzschmar, K.; Begthel, H.; van den Born, M.; Korving, J.; de Sauvage, F.; van Es, J.H.; et al. Replacement of lost lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell 2016, 18, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Van Es, J.H.; Sato, T.; van de Wetering, M.; Lyubimova, A.; Yee Nee, A.N.; Gregorieff, A.; Sasaki, N.; Zeinstra, L.; van den Born, M.; Korving, J.; et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 2012, 14, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, U.; Saxena, M.; O’Neill, N.K.; Saadatpour, A.; Yuan, G.C.; Herbert, Z.; Murata, K.; Shivdasani, R.A. Dynamic reorganization of chromatin accessibility signatures during dedifferentiation of secretory precursors into lgr5+ intestinal stem cells. Cell Stem Cell 2017, 21, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Metcalfe, C.; Kljavin, N.M.; Ybarra, R.; de Sauvage, F.J. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell 2014, 14, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for LGR5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, H.B.; Estes, M.K. Rotaviruses: From pathogenesis to vaccination. Gastroenterology 2009, 136, 1939–1951. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Steele, A.D.; Duque, J.; Parashar, U.D. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 136–141. [Google Scholar] [CrossRef]
- Arnold, M.; Patton, J.T.; McDonald, S.M. Culturing, storage, and quantification of rotavirus. Curr. Protoc. Microbiol. 2009. [Google Scholar]
- Sheth, R.; Anderson, J.; Sato, T.; Oh, B.; Hempson, S.J.; Rollo, E.; Mackow, E.R.; Shaw, R.D. Rotavirus stimulates IL-8 secretion from cultured epithelial cells. Virology 1996, 221, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Pedron, T.; Thibault, C.; Sansonetti, P.J. The invasive phenotype of Shigella flexneri directs a distinct gene expression pattern in the human intestinal epithelial cell line Caco-2. J. Biol. Chem. 2003, 278, 33878–33886. [Google Scholar] [CrossRef] [PubMed]
- Drummond, C.G.; Bolock, A.M.; Ma, C.; Luke, C.J.; Good, M.; Coyne, C.B. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc. Natl. Acad. Sci. USA 2017, 114, 1672–1677. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.L.; Clemens, J.D.; Sack, D.A.; Knowlton, D.R.; McNeal, M.M.; Huda, N.; Ahmed, F.; Rao, M.; Schiff, G.M. Culture adaptation and characterization of group A rotaviruses causing diarrheal illnesses in Bangladesh from 1985 to 1986. J. Clin. Microbiol. 1991, 29, 1915–1923. [Google Scholar] [PubMed]
- Finkbeiner, S.R.; Zeng, X.L.; Utama, B.; Atmar, R.L.; Shroyer, N.F.; Estes, M.K. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio 2012, 3, e00159-12. [Google Scholar] [CrossRef] [PubMed]
- Hale, A.; Mattick, K.; Lewis, D.; Estes, M.; Jiang, X.; Green, J.; Eglin, R.; Brown, D. Distinct epidemiological patterns of Norwalk-like virus infection. J. Med. Virol. 2000, 62, 99–103. [Google Scholar] [CrossRef]
- Jones, M.K.; Watanabe, M.; Zhu, S.; Graves, C.L.; Keyes, L.R.; Grau, K.R.; Gonzalez-Hernandez, M.B.; Iovine, N.M.; Wobus, C.E.; Vinje, J.; et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014, 346, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Ramani, S.; Atmar, R.L.; Estes, M.K. Epidemiology of human noroviruses and updates on vaccine development. Curr. Opin. Gastroenterol. 2014, 30, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.R.; Neill, F.H.; Blutt, S.E.; Zeng, X.L.; Qu, L.; et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Hutson, A.M.; Estes, M.K.; Prasad, B.V. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. USA 2008, 105, 9175–9180. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.S.; Bromme, B.A.; Holly, M.K.; Wiens, M.E.; Gounder, A.P.; Sul, Y.; Smith, J.G. Alpha-defensin-dependent enhancement of enteric viral infection. PLoS Pathog. 2017, 13, e1006446. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.G.; Silvestry, M.; Lindert, S.; Lu, W.; Nemerow, G.R.; Stewart, P.L. Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLoS Pathog. 2010, 6, e1000959. [Google Scholar] [CrossRef] [PubMed]
- Bohm, R.; Fleming, F.E.; Maggioni, A.; Dang, V.T.; Holloway, G.; Coulson, B.S.; von Itzstein, M.; Haselhorst, T. Revisiting the role of histo-blood group antigens in rotavirus host-cell invasion. Nat. Commun. 2015, 6, 5907. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lanik, W.E.; Mara, M.A.; Mihi, B.; Coyne, C.B.; Good, M. Stem Cell-Derived Models of Viral Infections in the Gastrointestinal Tract. Viruses 2018, 10, 124. https://doi.org/10.3390/v10030124
Lanik WE, Mara MA, Mihi B, Coyne CB, Good M. Stem Cell-Derived Models of Viral Infections in the Gastrointestinal Tract. Viruses. 2018; 10(3):124. https://doi.org/10.3390/v10030124
Chicago/Turabian StyleLanik, Wyatt E., Madison A. Mara, Belgacem Mihi, Carolyn B. Coyne, and Misty Good. 2018. "Stem Cell-Derived Models of Viral Infections in the Gastrointestinal Tract" Viruses 10, no. 3: 124. https://doi.org/10.3390/v10030124