Function and Regulation of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) / CRISPR Associated (Cas) Systems
Abstract
:1. Introduction
2. Discovery of CRISPR/Cas Systems
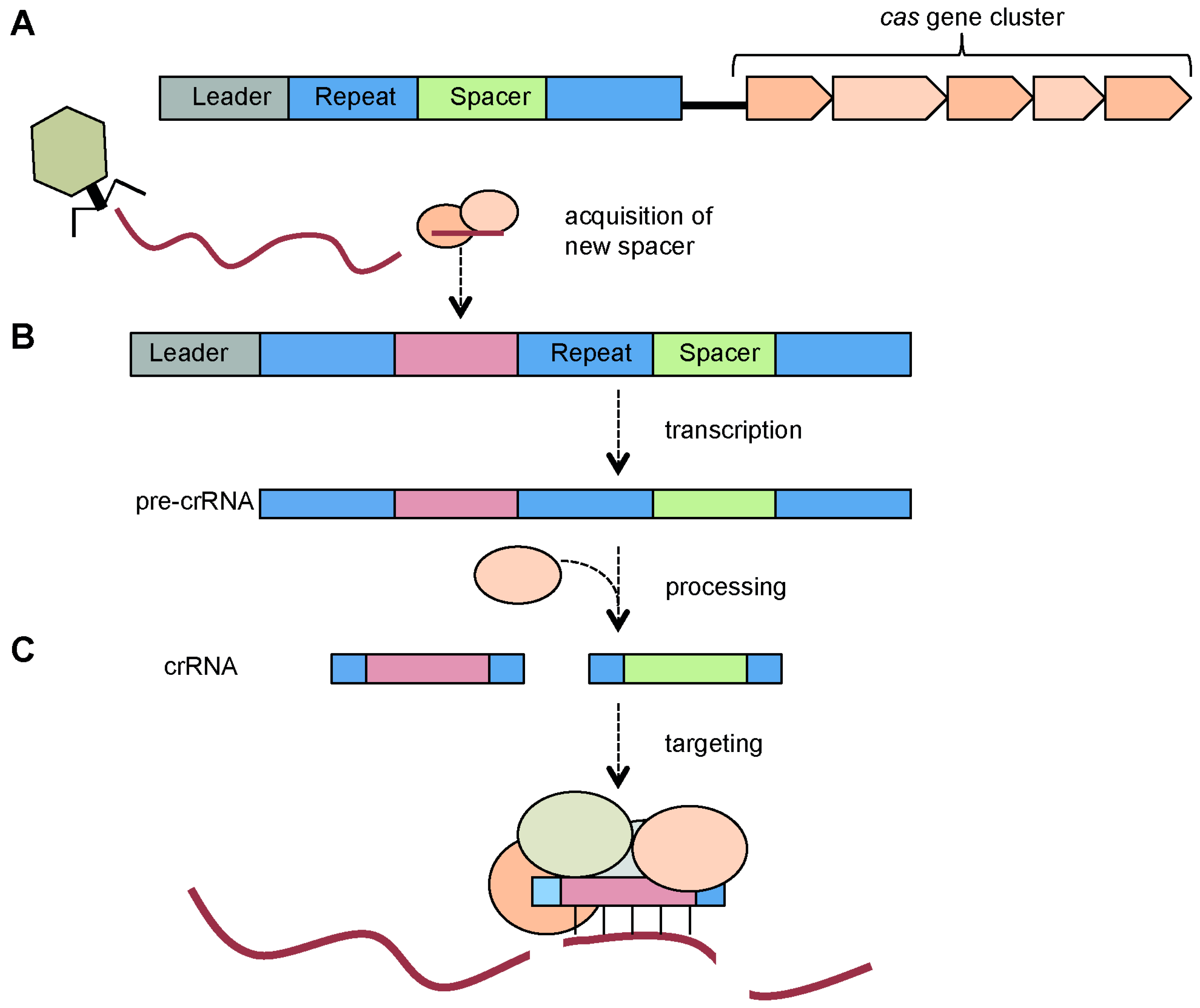
3. Components of CRISPR/Cas Systems
3.1. CRISPR Arrays: Repeats, Spacers and Leader Sequence
3.2. Cas Proteins
4. CRISPR/Cas Mechanism
4.1. Adaptation via Spacer Acquisition
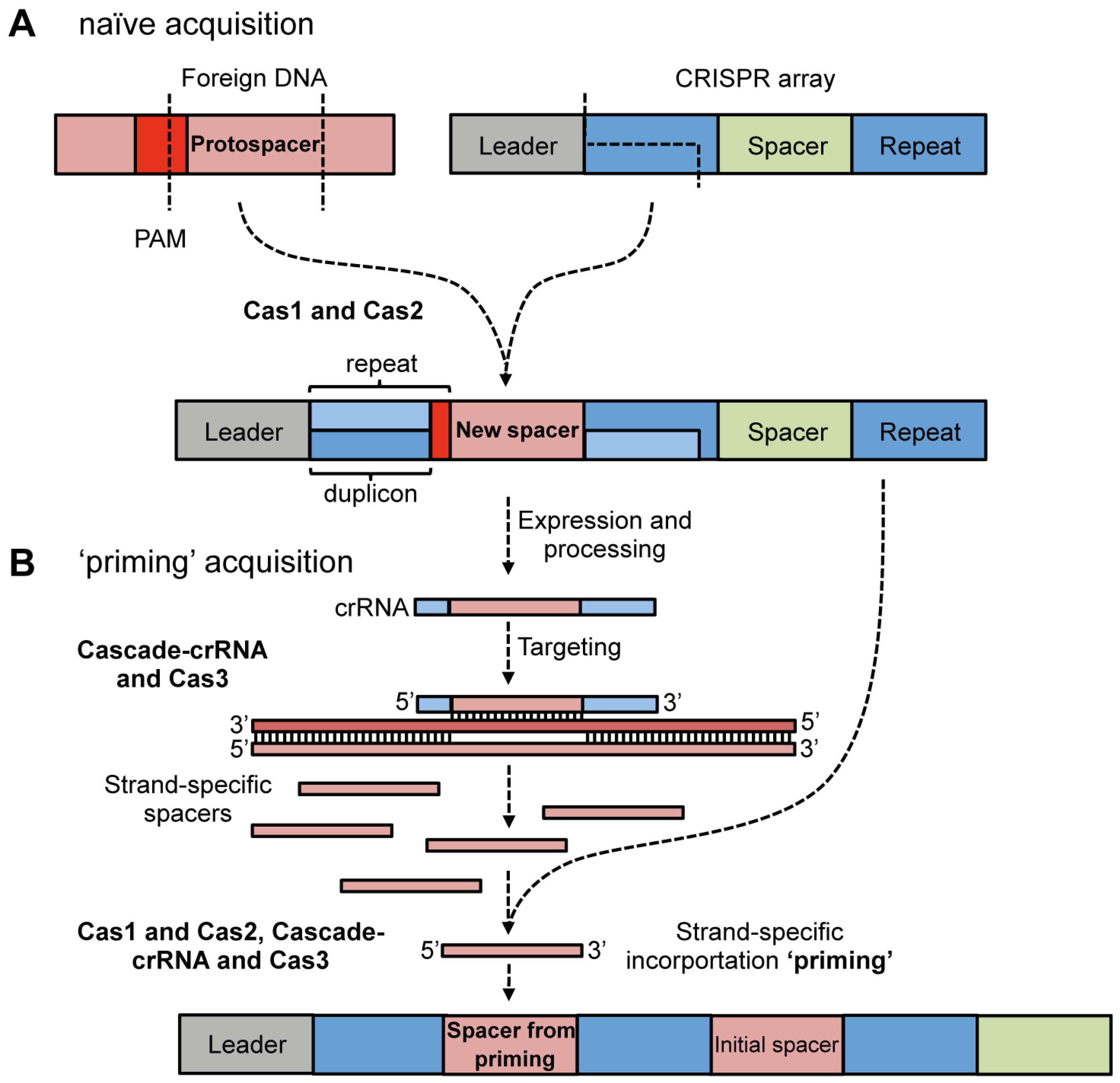
4.1.1. Cas Proteins Required for Adaptation
4.1.2. Protospacer Selection and Incorporation into CRISPR Arrays
4.1.3. Priming Acquisition
4.2. Expression, crRNA Generation and Interference
4.2.1. Type I Systems
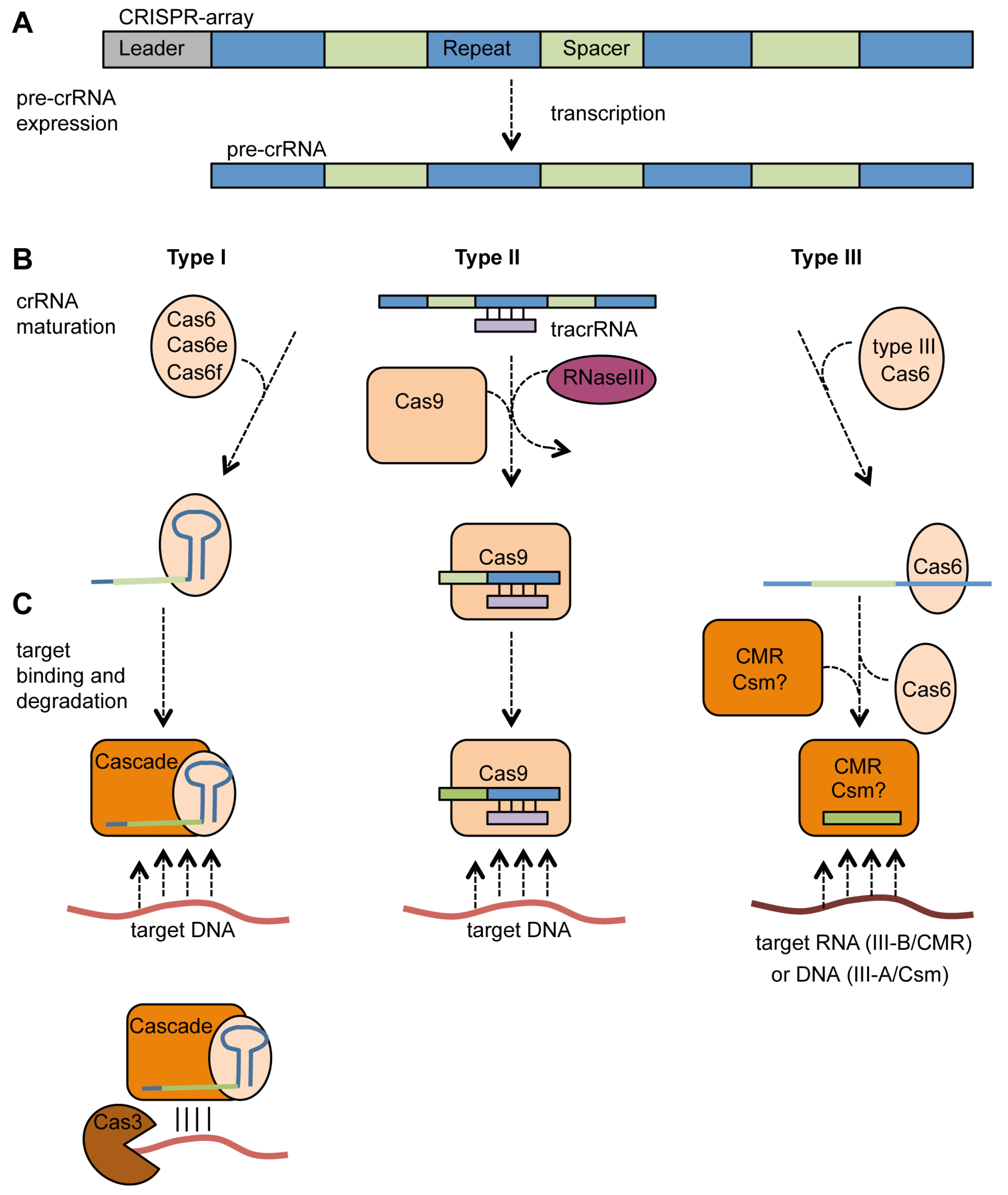
4.2.2. Type II Systems
4.2.3. Type III Systems
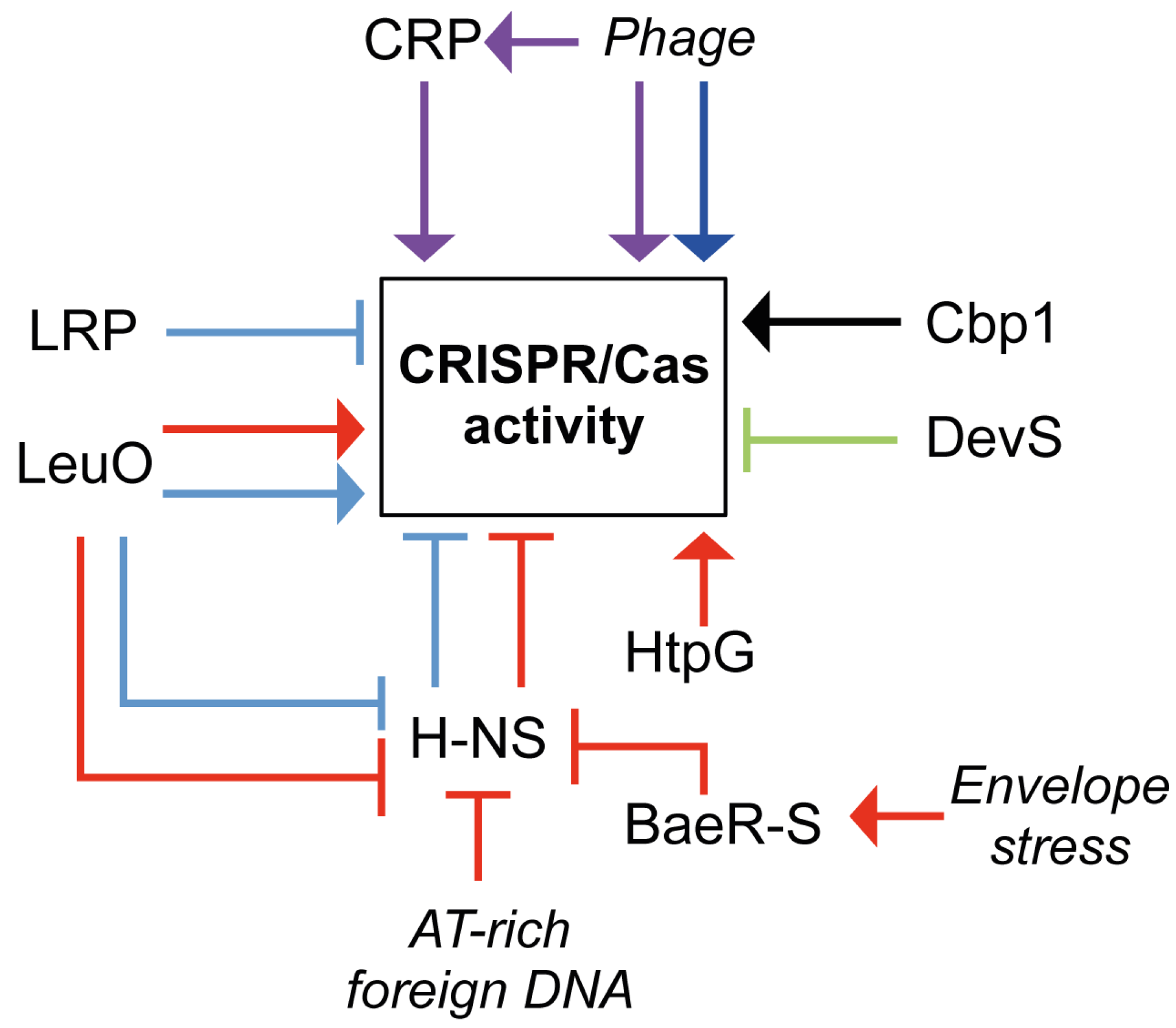
5. Regulation of CRISPR/Cas Systems
6. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Frost, L.S.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef]
- Koonin, E.V.; Wolf, Y.I. Genomics of bacteria and archaea: The emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008, 36, 6688–6719. [Google Scholar] [CrossRef]
- Fuhrman, J.A. Marine viruses and their biogeochemical and ecological effects. Nature 1999, 399, 541–548. [Google Scholar] [CrossRef]
- Hendrix, R.W. Bacteriophage genomics. Curr. Opin. Microbiol. 2003, 6, 506–511. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Petty, N.K.; Evans, T.J.; Fineran, P.C.; Salmond, G.P. Biotechnological exploitation of bacteriophage research. Trends Biotechnol. 2007, 25, 7–15. [Google Scholar] [CrossRef]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010, 11, 181–190. [Google Scholar]
- Terns, M.P.; Terns, R.M. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011, 14, 321–327. [Google Scholar]
- Wiedenheft, B.; Sternberg, S.H.; Doudna, J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012, 482, 331–338. [Google Scholar] [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar]
- Groenen, P.M.; Bunschoten, A.E.; van Soolingen, D.; van Embden, J.D. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 1993, 10, 1057–1065. [Google Scholar] [CrossRef]
- Mojica, F.J.; Ferrer, C.; Juez, G.; Rodríguez-Valera, F. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol. Microbiol. 1995, 17, 85–93. [Google Scholar] [CrossRef]
- Klenk, H.P.; Clayton, R.A.; Tomb, J.F.; White, O.; Nelson, K.E.; Ketchum, K.A.; Dodson, R.J.; Gwinn, M.; Hickey, E.K.; Peterson, J.D.; et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 1997, 390, 364–370. [Google Scholar] [CrossRef]
- Jansen, R.; van Embden, J.D.; Gaastra, W.; Schouls, L.M. Identification of a novel family of sequence repeats among prokaryotes. OMICS 2002, 6, 23–33. [Google Scholar] [CrossRef]
- Mojica, F.J.; Díez-Villaseñor, C.; Soria, E.; Juez, G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 2000, 36, 244–246. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 2007, 8, 172. [Google Scholar] [CrossRef]
- van Belkum, A.; Scherer, S.; van Alphen, L.; Verbrugh, H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 1998, 62, 275–293. [Google Scholar]
- Jansen, R.; Embden, J.D.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef]
- Makarova, K.S.; Aravind, L.; Grishin, N.V.; Rogozin, I.B.; Koonin, E.V. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002, 30, 482–496. [Google Scholar] [CrossRef]
- Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered regularly interspaced short palindromerepeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef]
- Pourcel, C.; Salvignol, G.; Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005, 151, 653–663. [Google Scholar] [CrossRef]
- Makarova, K.S.; Grishin, N.V.; Shabalina, S.A.; Wolf, Y.I.; Koonin, E.V. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 2006, 1. [Google Scholar] [CrossRef]
- Haft, D.H.; Selengut, J.; Mongodin, E.F.; Nelson, K.E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005, 1, e60. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science 2008, 322, 1843–1845. [Google Scholar] [CrossRef]
- Bikard, D.; Hatoum-Aslan, A.; Mucida, D.; Marraffini, L.A. CRISPR Interference Can Prevent Natural Transformation and Virulence Acquisition during In Vivo Bacterial Infection. Cell Host Microbe 2012, 12, 177–186. [Google Scholar] [CrossRef]
- Lillestøl, R.K.; Redder, P.; Garrett, R.A.; Brugger, K. A putative viral defence mechanism in archaeal cells. Archaea 2006, 2, 59–72. [Google Scholar] [CrossRef]
- Nakata, A.; Amemura, M.; Makino, K. Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J. Bacteriol. 1989, 171, 3553–3556. [Google Scholar]
- She, Q.; Singh, R.K.; Confalonieri, F.; Zivanovic, Y.; Allard, G.; Awayez, M.J.; Chan-Weiher, C.C.; Clausen, I.G.; Curtis, B.A.; De Moors, A.; et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 2001, 98, 7835–7840. [Google Scholar]
- van Embden, J.D.; van Gorkom, T.; Kremer, K.; Jansen, R.; van der Zeijst, B.A.; Schouls, L.M. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 2000, 182, 2393–2401. [Google Scholar]
- Horvath, P.; Romero, D.A.; Côuté-Monvoisin, A.C.; Richards, M.; Deveau, H.; Moineau, S.; Boyaval, P.; Fremaux, C.; Barrangou, R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008, 190, 1401–1412. [Google Scholar] [CrossRef]
- Kunin, V.; Sorek, R.; Hugenholtz, P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007, 8. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef]
- Brodt, A.; Lurie-Weinberger, M.N.; Gophna, U. CRISPR loci reveal networks of gene exchange in archaea. Biol. Direct 2011, 6. [Google Scholar] [CrossRef]
- Deveau, H.; Barrangou, R.; Garneau, J.E.; Labonté, J.; Fremaux, C.; Boyaval, P.; Romero, D.A.; Horvath, P.; Moineau, S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008, 190, 1390–1400. [Google Scholar]
- Stern, A.; Keren, L.; Wurtzel, O.; Amitai, G.; Sorek, R. Self-targeting by CRISPR: Gene regulation or autoimmunity? Trends Genet. 2010, 26, 335–340. [Google Scholar] [CrossRef]
- Agari, Y.; Sakamoto, K.; Tamakoshi, M.; Oshima, T.; Kuramitsu, S.; Shinkai, A. Transcription profileof Thermus thermophilus CRISPR systems after phage infection. J. Mol. Biol. 2009, 395, 270–281. [Google Scholar]
- Pougach, K.; Semenova, E.; Bogdanova, E.; Datsenko, K.A.; Djordjevic, M.; Wanner, B.L.; Severinov, K. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol. 2010, 77, 1367–1379. [Google Scholar] [CrossRef]
- Pul, Ü.; Wurm, R.; Arslan, Z.; Geissen, R.; Hofmann, N.; Wagner, R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 2010, 75, 1495–1512. [Google Scholar] [CrossRef]
- Przybilski, R.; Richter, C.; Gristwood, T.; Clulow, J.S.; Vercoe, R.B.; Fineran, P.C. Csy4 is responsible for CRISPR RNA processing in Pectobacterium atrosepticum. RNA Biol. 2011, 8, 517–528. [Google Scholar] [CrossRef]
- Yosef, I.; Goren, M.G.; Qimron, U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012, 40, 5569–5576. [Google Scholar] [CrossRef]
- Godde, J.S.; Bickerton, A. The repetitive DNA elements called CRISPRs and their associated genes: Evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 2006, 62, 718–729. [Google Scholar] [CrossRef]
- Brouns, S.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.; Snijders, A.P.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; van der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar]
- Jore, M.M.; Lundgren, M.; van Duijn, E.; Bultema, J.B.; Westra, E.R.; Waghmare, S.P.; Wiedenheft, B.; Pul, Ü.; Wurm, R.; Wagner, R.; et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011, 18, 529–536. [Google Scholar] [CrossRef]
- Wiedenheft, B.; Lander, G.C.; Zhou, K.; Jore, M.M.; Brouns, S.J.; van der Oost, J.; Doudna, J.A.; Nogales, E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 2011, 477, 486–489. [Google Scholar] [CrossRef]
- Wiedenheft, B.; van Duijn, E.; Bultema, J.B.; Waghmare, S.P.; Zhou, K.; Barendregt, A.; Westphal, W.; Heck, A.J.; Boekema, E.J.; Dickman, M.J.; et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. USA 2011, 108, 10092–10097. [Google Scholar]
- Nam, K.H.; Haitjema, C.; Liu, X.; Ding, F.; Wang, H.; DeLisa, M.P.; Ke, A. Cas5d Protein Processes Pre-crRNA and Assembles into a Cascade-like Interference Complex in Subtype I-C/Dvulg CRISPR-Cas System. Structure 2012, 20, 1574–1584. [Google Scholar] [CrossRef]
- Andersson, A.F.; Banfield, J.F. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 2008, 320, 1047–1050. [Google Scholar] [CrossRef]
- Tyson, G.W.; Banfield, J.F. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ. Microbiol. 2008, 10, 200–207. [Google Scholar]
- Pride, D.T.; Salzman, J.; Haynes, M.; Rohwer, F.; Davis-Long, C.; White, R.A., 3rd.; Loomer, P.; Armitage, G.C.; Relman, D.A. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012, 6, 915–926. [Google Scholar] [CrossRef]
- Garneau, J.E.; Dupuis, M.E.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadan, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef]
- Swarts, D.C.; Mosterd, C.; van Passel, M.W.; Brouns, S.J. CRISPR interference directs strand specific spacer acquisition. PLoS One 2012, 7, e35888. [Google Scholar]
- Datsenko, K.A.; Pougach, K.; Tikhonov, A.; Wanner, B.L.; Severinov, K.; Semenova, E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012, 3. [Google Scholar] [CrossRef]
- Cady, K.C.; Bondy-Denomy, J.; Heussler, G.E.; Davidson, A.R.; O'Toole, G.A. The CRISPR/Cas Adaptive Immune System of Pseudomonas aeruginosa Mediates Resistance to Naturally Occurring and Engineered Phages. J. Bacteriol. 2012, 194, 5728–5738. [Google Scholar] [CrossRef]
- Lopez-Sanchez, M.J.; Sauvage, E.; Da Cunha, V.; Clermont, D.; Ratsima Hariniaina, E.; Gonzalez-Zorn, B.; Poyart, C.; Rosinski-Chupin, I.; Glaser, P. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol. Microbiol. 2012, 85, 1057–1071. [Google Scholar] [CrossRef]
- Erdmann, S.; Garrett, R.A. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 2012, 85, 1044–1056. [Google Scholar] [CrossRef]
- Sapranauskas, R.; Gasiunas, G.; Fremaux, C.; Barrangou, R.; Horvath, P.; Siksnys, V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011, 39, 9275–9282. [Google Scholar] [CrossRef]
- Hatoum-Aslan, A.; Maniv, I.; Marraffini, L.A. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc. Natl. Acad. Sci. USA 2011, 108, 21218–21222. [Google Scholar] [CrossRef]
- Wiedenheft, B.; Zhou, K.; Jinek, M.; Coyle, S.M.; Ma, W.; Doudna, J.A. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure 2009, 17, 904–912. [Google Scholar] [CrossRef]
- Han, D.; Lehmann, K.; Krauss, G. SSO1450-A CAS1 protein from Sulfolobus solfataricus P2 with high affinity for RNA and DNA. FEBS Lett. 2009, 583, 1928–1932. [Google Scholar] [CrossRef]
- Babu, M.; Beloglazova, N.; Flick, R.; Graham, C.; Skarina, T.; Nocek, B.; Gagarinova, A.; Pogoutse, O.; Brown, G.; Binkowski, A.; et al. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 2011, 79, 484–502. [Google Scholar] [CrossRef]
- Beloglazova, N.; Brown, G.; Zimmerman, M.D.; Proudfoot, M.; Makarova, K.S.; Kudritska, M.; Kochinyan, S.; Wang, S.; Chruszcz, M.; Minor, W.; et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem. 2008, 283, 20361–20371. [Google Scholar]
- Samai, P.; Smith, P.; Shuman, S. Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 2010, 66, 1552–1556. [Google Scholar] [CrossRef]
- Nam, K.H.; Ding, F.; Haitjema, C.; Huang, Q.; Delisa, M.P.; Ke, A. Double-stranded Endonuclease Activity in B. halodurans Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated Cas2 Protein. J. Biol. Chem. 2012. Epub ahead of print. [Google Scholar]
- Plagens, A.; Tjaden, B.; Hagemann, A.; Randau, L.; Hensel, R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J. Bacteriol. 2012, 194, 2491–2500. [Google Scholar] [CrossRef]
- Goren, M.G.; Yosef, I.; Auster, O.; Qimron, U. Experimental Definition of a Clustered Regularly Interspaced Short Palindromic Duplicon in Escherichia coli. J. Mol. Biol. 2012, 423, 14–16. [Google Scholar] [CrossRef]
- Westra, E.R.; Brouns, S.J. The rise and fall of CRISPRs-Dynamics of spacer acquisition and loss. Mol. Microbiol. 2012, 85, 1021–1025. [Google Scholar] [CrossRef]
- Mojica, F.J.; Diez-Villasenor, C.; Garcia-Martinez, J.; Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009, 155, 733–740. [Google Scholar] [CrossRef]
- Richter, H.; Zoephel, J.; Schermuly, J.; Maticzka, D.; Backofen, R.; Randau, L. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 2012. Epub ahead of print.
- Sashital, D.G.; Jinek, M.; Doudna, J.A. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat. Struct. Mol. Biol. 2011, 18, 680–687. [Google Scholar] [CrossRef]
- Gesner, E.M.; Schellenberg, M.J.; Garside, E.L.; George, M.M.; MacMillan, A.M. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat. Struct. Mol. Biol. 2011, 18, 688–692. [Google Scholar] [CrossRef]
- Haurwitz, R.E.; Jinek, M.; Wiedenheft, B.; Zhou, K.; Doudna, J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 2010, 329, 1355–1358. [Google Scholar]
- Sternberg, S.H.; Haurwitz, R.E.; Doudna, J.A. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA 2012, 18, 661–672. [Google Scholar] [CrossRef]
- Haurwitz, R.E.; Sternberg, S.H.; Doudna, J.A. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J. 2012, 31, 2824–2832. [Google Scholar] [CrossRef]
- Lintner, N.G.; Kerou, M.; Brumfield, S.K.; Graham, S.; Liu, H.; Naismith, J.H.; Sdano, M.; Peng, N.; She, Q.; Copie, V.; et al. Structural and Functional Characterization of an Archaeal Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-associated Complex for Antiviral Defense (CASCADE). J. Biol. Chem. 2011, 286, 21643–21656. [Google Scholar]
- Sashital, D.G.; Wiedenheft, B.; Doudna, J.A. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol. Cell. 2012, 46, 606–615. [Google Scholar] [CrossRef]
- Semenova, E.; Jore, M.M.; Datsenko, K.A.; Semenova, A.; Westra, E.R.; Wanner, B.; van der Oost, J.; Brouns, S.J.; Severinov, K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. USA 2011, 108, 10098–10103. [Google Scholar]
- Westra, E.R.; van Erp, P.B.; Kunne, T.; Wong, S.P.; Staals, R.H.; Seegers, C.L.; Bollen, S.; Jore, M.M.; Semenova, E.; Severinov, K.; et al. CRISPR Immunity Relies on the Consecutive Binding and Degradation of Negatively Supercoiled Invader DNA by Cascade and Cas3. Mol. Cell 2012, 46, 595–605. [Google Scholar] [CrossRef]
- Howard, J.A.; Delmas, S.; Ivančić-Baće, I.; Bolt, E.L. Helicase dissociation and annealing of RNA-DNA hybrids by Escherichia coli Cas3 protein. Biochem. J. 2011, 439, 85–95. [Google Scholar] [CrossRef]
- Sinkunas, T.; Gasiunas, G.; Fremaux, C.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011, 30, 1335–1342. [Google Scholar] [CrossRef]
- Beloglazova, N.; Petit, P.; Flick, R.; Brown, G.; Savchenko, A.; Yakunin, A.F. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011, 30, 4616–4627. [Google Scholar] [CrossRef]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Carte, J.; Wang, R.; Li, H.; Terns, R.M.; Terns, M.P. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008, 22, 3489–3496. [Google Scholar] [CrossRef]
- Carte, J.; Pfister, N.T.; Compton, M.M.; Terns, R.M.; Terns, M.P. Binding and cleavage of CRISPR RNA by Cas6. RNA 2010, 16, 2181–2188. [Google Scholar] [CrossRef]
- Wang, R.; Preamplume, G.; Terns, M.P.; Terns, R.M.; Li, H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: Recognition and cleavage. Structure 2011, 19, 257–264. [Google Scholar] [CrossRef]
- Hale, C.R.; Majumdar, S.; Elmore, J.; Pfister, N.; Compton, M.; Olson, S.; Resch, A.M.; Glover, C.V., 3rd.; Graveley, B.R.; Terns, R.M.; et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell 2012, 45, 292–302. [Google Scholar]
- Hale, C.R.; Zhao, P.; Olson, S.; Duff, M.O.; Graveley, B.R.; Wells, L.; Terns, R.M.; Terns, M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 2009, 139, 945–956. [Google Scholar] [CrossRef]
- Hale, C.; Kleppe, K.; Terns, R.M.; Terns, M.P. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA 2008, 14, 2572–2579. [Google Scholar] [CrossRef]
- Zhang, J.; Rouillon, C.; Kerou, M.; Reeks, J.; Brugger, K.; Graham, S.; Reimann, J.; Cannone, G.; Liu, H.; Albers, S.V.; et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell 2012, 45, 303–313. [Google Scholar] [CrossRef]
- Marraffini, L.A.; Sontheimer, E.J. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 2010, 463, 568–571. [Google Scholar] [CrossRef]
- Young, J.C.; Dill, B.D.; Pan, C.; Hettich, R.L.; Banfield, J.F.; Shah, M.; Fremaux, C.; Horvath, P.; Barrangou, R.; VerBerkmoes, N.C. Phage-induced expression of CRISPR-associated proteins is revealed by shotgun proteomics in Streptococcus thermophilus. PLoS One 2012, 7, e38077. [Google Scholar]
- Dorman, C.J. H-NS, the genome sentinel. Nat. Rev. Microbiol. 2007, 5, 157–161. [Google Scholar] [CrossRef]
- Westra, E.R.; Pul, Ü.; Heidrich, N.; Jore, M.M.; Lundgren, M.; Stratmann, T.; Wurm, R.; Raine, A.; Mescher, M.; van Heereveld, L.; et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. In Mol. Microbiol.; 2010; Volume 77, pp. 1380–1393. [Google Scholar]
- Lucchini, S.; Rowley, G.; Goldberg, M.D.; Hurd, D.; Harrison, M.; Hinton, J.C. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006, 2, e81. [Google Scholar] [CrossRef]
- Navarre, W.W.; Porwollik, S.; Wang, Y.; McClelland, M.; Rosen, H.; Libby, S.J.; Fang, F.C. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 2006, 313, 236–238. [Google Scholar] [CrossRef]
- Mojica, F.J.; Díez-Villaseñor, C. The on-off switch of CRISPR immunity against phages in Escherichia coli. Mol. Microbiol. 2010, 77, 1341–1345. [Google Scholar] [CrossRef]
- Medina-Aparicio, L.; Rebollar-Flores, J.E.; Gallego-Hernández, A.L.; Vázquez, A.; Olvera, L.; Gutiérrez-Ríos, R.M.; Calva, E.; Hernández-Lucas, I. The CRISPR/Cas Immune System Is an Operon Regulated by LeuO, H-NS, and Leucine-Responsive Regulatory Protein in Salmonella enterica Serovar Typhi. J. Bacteriol. 2011, 193, 2396–2407. [Google Scholar] [CrossRef]
- Hernández-Lucas, I.; Gallego-Hernández, A.L.; Encarnación, S.; Fernández-Mora, M.; Martínez-Batallar, A.G.; Salgado, H.; Oropeza, R.; Calva, E. The LysR-type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar Typhi. J. Bacteriol. 2008, 190, 1658–1670. [Google Scholar] [CrossRef]
- Majumder, A.; Fang, M.; Tsai, K.J.; Ueguchi, C.; Mizuno, T.; Wu, H.Y. LeuO expression in response to starvation for branched-chain amino acids. J. Biol. Chem. 2001, 276, 19046–19051. [Google Scholar]
- Drahos, D.J.; Hendrix, R.W. Effect of bacteriophage lambda infection on synthesis of groE protein and other Escherichia coli proteins. J. Bacteriol. 1982, 149, 1050–1063. [Google Scholar]
- MacRitchie, D.M.; Buelow, D.R.; Price, N.L.; Raivio, T.L. Two-component signaling and gram negative envelope stress response systems. Adv. Exp. Med. Biol. 2008, 631, 80–110. [Google Scholar] [CrossRef]
- Perez-Rodriguez, R.; Haitjema, C.; Huang, Q.; Nam, K.H.; Bernardis, S.; Ke, A.; DeLisa, M.P. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol. Microbiol. 2011, 79, 584–599. [Google Scholar] [CrossRef]
- Baranova, N.; Nikaido, H. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 2002, 184, 4168–4176. [Google Scholar] [CrossRef]
- Yosef, I.; Goren, M.G.; Kiro, R.; Edgar, R.; Qimron, U. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proc. Natl. Acad. Sci. USA 2011, 108, 20136–20141. [Google Scholar] [CrossRef]
- Chen, C.C.; Ghole, M.; Majumder, A.; Wang, Z.; Chandana, S.; Wu, H.Y. LeuO-mediated transcriptional derepression. J. Biol. Chem. 2003, 278, 38094–38103. [Google Scholar]
- Viswanathan, P.; Murphy, K.; Julien, B.; Garza, A.G.; Kroos, L. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J. Bacteriol. 2007, 189, 3738–3750. [Google Scholar]
- Shinkai, A.; Kira, S.; Nakagawa, N.; Kashihara, A.; Kuramitsu, S.; Yokoyama, S. Transcription activation mediated by a cyclic AMP receptor protein from Thermus thermophilus HB8. J. Bacteriol. 2007, 189, 3891–3901. [Google Scholar] [CrossRef]
- Deng, L.; Kenchappa, C.S.; Peng, X.; She, Q.; Garrett, R.A. Modulation of CRISPR locus transcription by the repeat-binding protein Cbp1 in Sulfolobus. Nucleic Acids Res. 2012, 40, 2470–2480. [Google Scholar]
- Peng, X.; Brugger, K.; Shen, B.; Chen, L.; She, Q.; Garrett, R.A. Genus-specific protein binding to the large clusters of DNA repeats (short regularly spaced repeats) present in Sulfolobus genomes. J. Bacteriol. 2003, 185, 2410–2417. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Richter, C.; Chang, J.T.; Fineran, P.C. Function and Regulation of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) / CRISPR Associated (Cas) Systems. Viruses 2012, 4, 2291-2311. https://doi.org/10.3390/v4102291
Richter C, Chang JT, Fineran PC. Function and Regulation of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) / CRISPR Associated (Cas) Systems. Viruses. 2012; 4(10):2291-2311. https://doi.org/10.3390/v4102291
Chicago/Turabian StyleRichter, Corinna, James T. Chang, and Peter C. Fineran. 2012. "Function and Regulation of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) / CRISPR Associated (Cas) Systems" Viruses 4, no. 10: 2291-2311. https://doi.org/10.3390/v4102291



