1. Introduction
Tidal salt marshes are among the most complex and productive ecosystems in the world [
1,
2]. Occurring at the interface between terrestrial and aquatic ecosystems and often between saline and non-saline water bodies, they provide valuable ecological services such as nutrient cycling, pollutant filtration, floodwater absorption, storm surge dissipation, and shoreline stabilization [
2,
3]. Salt marshes have some of the highest levels of net primary production, with aboveground biomass production from marsh grasses of up to 4000 g∙m
−2∙yr
−1 and below-ground production estimated at over 6000 g∙m
−2∙yr
−1 in inland areas [
4]. These high rates of primary production, in turn, support a diverse meso- and macrofauna including large populations of migratory waterfowl and wading birds and economically important species of fish and shellfish [
2]. In addition to the tangible economic benefits from commercial fisheries, salt marshes provide a variety of other ecosystem services such as birdwatching, fishing, and other forms of recreation and tourism [
3].
Despite their enormous value, salt marshes worldwide have been degraded or destroyed by a variety of anthropogenic threats including eutrophication due to agricultural and industrial runoff, drainage and reclamation for residential and commercial development, modification of hydrological flow regimes, relative sea level rise, increased air and water surface temperatures, and higher acidification associated with elevated atmospheric CO
2 [
1,
5,
6]. In the northern Gulf of Mexico, coastal marshes are threatened by accelerating submergence due to the loss of upstream sediments combined with rising sea levels [
4]. In addition to rapid shoreline retreat from long-term coastal erosion, subsidence has led to the loss of thousands of square kilometers of salt marsh in the northern Gulf of Mexico through conversion to open water [
1]. Even more troubling, estimates for global sea level rise based on the most recently available data suggest that an increase of 1 m or more by the year 2100 is a strong possibility given the accelerating loss of the polar ice sheets [
7,
8,
9,
10]. If realized, these projected rates of sea level rise in excess of 10 mm∙yr
−1 would cause substantial losses of coastal marsh areas in the northern Gulf of Mexico [
11].
To monitor the status of salt marshes on global to regional scales, it is vital to develop accurate quantitative relationships between metrics derived from remotely sensed data and indicators of salt marsh productivity such as aboveground biomass. Previous studies established strong correlations between live biomass of the salt marsh grass
Spartina alterniflora (smooth cordgrass) and the canopy radiance corresponding to the red and near-infrared (NIR) spectral bands captured by Landsat Thematic Mapper (TM) sensors, as well as vegetation indices derived from those spectral bands [
12]. Zhang
et al. [
13] reported statistically significant relationships between spectral vegetation indices derived from Landsat TM imagery and various measures of salt marsh biomass collected in San Pablo Bay, California. Among the models considered, the simple vegetation index (
i.e., the near-infrared band divided by the red band) explained the most variation in green fresh biomass, with an R
2 value of 0.58 [
13]. Jensen
et al. [
3] used sub-meter resolution color-infrared (CIR) aerial photography in conjunction with field samples of biomass collected from salt marsh areas in coastal South Carolina to develop linear regression models relating the CIR spectral data to aboveground biomass of
S. alterniflora. They found the strongest relationship between biomass and the NIR band, with an R
2 value of 0.70 for the model containing this variable.
One of the major limitations of using satellite or CIR imagery to estimate aboveground biomass is the so-called “saturation problem,” wherein optical/infrared sensors are unable to distinguish variations in green biomass in areas of dense vegetation canopies. This saturation occurs when the canopy closure and leaf area density reach a threshold (varies by vegetation type) beyond which the optical/infrared sensor cannot detect increasing levels of green biomass response [
14,
15]. To address this problem, it has been proposed that biomass estimation could be improved by combining imagery from optical/infrared sensors with data acquired from active remote sensors, such as radar and airborne laser scanning (ALS, commonly referred to as “lidar” (light detection and ranging)) [
14,
16]. These active sensors provide additional information about topography and/or vegetation height that could potentially improve the quantitative models developed to predict aboveground biomass and other vegetation characteristics. Waring
et al. [
14] argued that Landsat imagery could be combined with synthetic aperture radar (SAR) data to improve biomass estimation in areas of dense vegetation, and Moghaddam
et al. [
17] reported significant improvements in regression models relating foliage biomass to Landsat TM data when combined with airborne SAR data collected over old-growth coniferous forests in the Pacific Northwest. In their study of woodland, shrub, grass, and mixed vegetation ecosystems in the Canadian arctic, Chen
et al. [
18] found that, in all cases, regression models combining Landsat TM and JERS-1 SAR data explained more of the variation in aboveground biomass than models containing either data source alone. Another type of active remote sensing technology, interferometric synthetic aperture radar (InSAR or IfSAR), requires that two SAR images be acquired over the same geographic area from a pair of close-range antennas. Because IfSAR can correct for terrain effects on radar backscatter intensity, it can potentially improve biomass estimation [
19].
In recent years, lidar data have been used to estimate aboveground biomass and other biophysical parameters for a variety of forest, shrub, and grassland ecosystems [
15,
20]. More commonly, they have been used to develop digital terrain models for a variety of applications including coastal modeling [
21]. However, it has been shown that the accuracy of lidar DEMs is reduced by vegetation, especially the tall, dense grasses typical of coastal salt marsh such as
S. alterniflora and
Juncus roemerianus (black rush) [
22]. The term “lidar error” used herein is defined as the difference between the lidar DEM elevation and the true elevation at a specific location. This error has a significant impact on coastal modeling results in areas where dense vegetation degrades the accuracy of lidar-derived DEMs. In these areas the DEM estimates the elevation of the marsh platform (also known as the marsh table) on the order of 0.20 to 0.80 m above its true bare earth elevation. In terms of coastal modeling, this can prevent flooding of the marsh surface in a simulation under both normal tide and storm surge conditions. This has ramifications in engineering [
23], ecological [
24], and integrated models assessing major processes such as sea level rise and climate change [
25,
26,
27].
In this study, a combination of field sampling and remote sensing was used to develop linear regression models relating the aboveground biomass density of salt marsh vegetation to variables derived from the remotely sensed data. This study differs from similar work in the past [
22] because it is species independent. This paper demonstrates that a combination of optical satellite imagery and data obtained from active remote sensing sources, such as SAR and/or lidar sensors, will explain more of the variation in aboveground biomass than would either data source alone. These species-independent biomass estimates can then be used to classify the aboveground biomass density and develop corresponding lidar DEM correction values.
5. Results and Discussion
Using this newly developed regression model, spatially variable biomass densities were estimated for the study area (
Figure 6).
Figure 6.
Biomass density classifications for study area. (A) Three-class scheme: High, Medium, and Low; (B) Two-class scheme: High and Low.
Figure 6.
Biomass density classifications for study area. (A) Three-class scheme: High, Medium, and Low; (B) Two-class scheme: High and Low.
The results of the first comparison overwhelmingly showed that the mean errors for all adjustment scenarios, including the control, were significantly less than the lidar DEM (therefore, we do not show those results here in detail). The errors between the DEM for each scenario and the GPS-RTK measured elevation at the 229 testing points, including the
p value for the second z-test, are shown in
Table 7.
Table 7.
Errors in the lidar and biomass-adjusted DEMs. The p value represents the probability that the mean error of the control (single-class median) scenario is greater than the mean error for a particular adjustment scenario.
Table 7.
Errors in the lidar and biomass-adjusted DEMs. The p value represents the probability that the mean error of the control (single-class median) scenario is greater than the mean error for a particular adjustment scenario.
| Measure | Lidar DEM (m) | 3-Class, Quartile Adjustment (m) | 3-Class, Median Adjustment (m) | 2-Class, Quartile Adjustment (m) | 2-Class, Median Adjustment (m) | 1-Class, Median Adjustment (m) |
|---|
| RMSE | 0.65 | 0.42 | 0.42 | 0.40 | 0.42 | 0.44 |
| Mean Error | 0.61 | 0.34 | 0.34 | 0.32 | 0.35 | 0.37 |
| Std. Dev. Error | 0.24 | 0.25 | 0.24 | 0.24 | 0.24 | 0.24 |
| p value | 1.000 | 0.050 | 0.060 | 0.001 | 0.134 | n/a |
As shown, the biomass-adjusted DEMs resulted in an average reduction in the Root Mean Square (RMS) error from 0.65 m to 0.42 m, representing a 35% decrease. All adjustment scenarios performed better than simply using the overall median adjustment across the entire DEM; however, only the quartile-based adjustments were statistically significant. Also, the two-class scheme performed slightly better than the three-class scheme, indicating that detecting natural breaks in the biomass density sample population was a better approach than arbitrarily splitting it into three classes with an approximately equal number of samples. The best performing adjustment scenario was the two-class quartile scenario. This was likely due to the better performance of the two-class scheme and the better capture of the non-linear nature of the lidar error/biomass density relationship. For comparison, the lidar DEM and the two-class, quartile-adjusted DEM are shown in
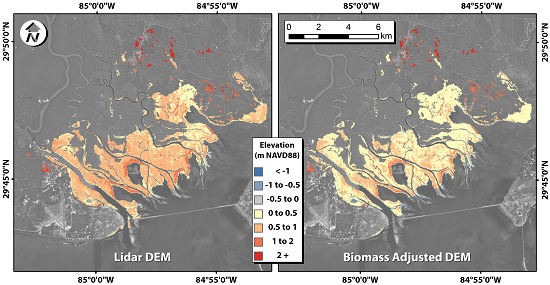
Figure 7.
Figure 7.
(A) Lidar DEM and (B) two-class, quartile-adjusted DEM.
Figure 7.
(A) Lidar DEM and (B) two-class, quartile-adjusted DEM.
The adjusted DEM contained many more pixels in the 0–0.5 m range than did the original lidar DEM (
Figure 7), which is the expected range for the marsh platform. The marsh platform typically is slightly higher than mean sea level, and the marsh is the most productive when an optimal periodic inundation occurs. In this region, mean sea level and mean high water are approximately 4.5 and 22.8 cm above NAVD88, respectively.
These findings must be tempered by the fact that even with the technique proposed here, the error is still more than we would like, especially in microtidal systems such as the northern Gulf of Mexico. In contrast, results achieved by Hladik and Alber [
22] showed a reduction in mean DEM error from 0.10 m to −0.01 m using a species and vegetation-height specific adjustment methodology. Notably, that study site (Sapelo Island, Georgia) had substantially less error between the lidar DEM and GPS-RTK elevations than did Apalachicola (0.10
vs. 0.61 m), so a direct comparison with the results presented here is difficult. However, if one is able to generate or obtain spatially distributed species classification along with vegetation height classes, the method of Hladik and Alber is likely superior. If these data are not available, we recommend the method proposed here. In any case, we would caution against blindly using this or any other method without first verifying that it is applicable in the tidal, vegetation type and density conditions of a particular study site. This can be accomplished by conducting an exploratory field data acquisition and remote sensing analysis campaign, using the method presented here as guidance.
To view these findings in their proper context, assessments of the processing sequence and potential sources of error are required. First, the lidar elevation accuracy standard set by the US Federal Emergency Management Agency is 36.6 cm (1.19 ft) at the 95% confidence level, so most lidar surveys in coastal areas are designed to meet this standard. Oftentimes the vertical elevation accuracy in open terrain is closer to 15 cm [
37]. The lidar data used in this study met that standard, but for microtidal systems, even acceptable lidar accuracy may not be enough for some coastal modeling applications (one of the primary uses of lidar DEMs).
Another potential source of error is the acquisition time of the remote sensing data, especially with respect to the seasonality of the marsh vegetation growth cycle. Depending on the time of year, the marsh grasses may be at their peak or dormant heights. For this study, the field biomass density samples and the ASTER imagery were collected in comparable years (June/September 2011 and May 2011), but there was a seasonal discrepancy. With optical-IR sensors, such as ASTER, it is more difficult to obtain cloud-free imagery in Florida during the summer growing season due to the general storminess of that time period.
The acquisition times of the elevation data (IfSAR DSM, lidar, and GPS-RTK) were substantially disparate (2004, 2007, and 2013, respectively). This is not unusual, as IfSAR and lidar data are expensive and are not collected on a regular basis in most areas. With respect to the difference between the lidar elevations and the GPS-RTK elevations, their separation in time could have been influenced by accretion or erosion in the marsh. Although it is true that there is likely to have been accretion on the marsh surface, it has been estimated to be about 0.8 mm∙yr
-1 in Apalachicola [
38]. This would have resulted in an accretion of approximately 5 mm, well within the accuracy bounds of the lidar, so is unlikely to have contributed significantly to the error in the regression model. However, the IfSAR data were much older and the month of acquisition could not be determined from the metadata. As the IfSAR provided a measure of canopy elevation, rather than a relative measure such as canopy height, this may have impacted the results significantly.
Lastly, the relatively small sample size for field-harvested biomass density (n = 16) requires further caution when applying the technique proposed herein. When using this technique in areas containing comparatively more or less dense marsh vegetation, additional samples should be acquired to adapt the technique for those areas.
Future studies are advised to obtain larger biomass density sample sizes and synchronize the field data collection with the remote sensing and lidar acquisition times to the maximum extent possible. At a minimum, data should be synchronized on a seasonal basis. Furthermore, biomass growth rates can vary year to year due to climatic, weather, and environmental factors such as drought, severe storms, or a pollution event. Any evidence of abnormal growth patterns should be addressed.
Despite the uncertainty in the method proposed herein, it is still a substantial step in the right direction and a necessary attempt to address a problem with widespread implications. The elevation of the marsh platform is a key parameter in nearly every model set in coastal ecosystems. For example, the Marsh Equilibrium Model [
24] relies heavily on the initial estimation of the marsh platform elevation to project future accretion in response to sea level rise. The model is based on local bioassay field experiments that estimate above- and below-ground biomass production rates that also serve as inputs into the model.
The Sea Level Affecting Marshes Model (SLAMM) [
39] is another widely used tool that relies heavily on marsh platform elevation as an input. The technical documentation for this model specifically states that bare earth lidar should be used as the elevation dataset when available [
40]. If the mean elevation error in a bare earth lidar DEM is nearly 50 cm above its true elevation, regions with small tide ranges such as the northern Gulf of Mexico will not see any modeled inundation, as the erroneously high marsh platform often will exceed the elevation of high tide. With a tool as widely used as SLAMM, this could lead to the propagation of misinformation, with communities basing their future land planning decisions on highly inaccurate results. In fact, Freeman
et al. [
38] applied the SLAMM model to Apalachicola Bay in 2012 and used the same lidar DEM that served as the basis for adjustment in this paper (1.524 m (5 ft) resolution resampled to 30 m). Their results serve as guidance for adapting to projected impacts to coastal wetlands, infrastructure, and vulnerable species, thereby demonstrating the importance of addressing the issue of inaccuracies in lidar DEMs of coastal salt marshes.
6. Summary and Conclusions
A novel processing sequence that uses ASTER, IfSAR, and lidar data to estimate spatially variable biomass density classes is presented. Two biomass density classification schemes were tested: a three-class scheme (high, medium, and low) based on the 66th and 33rd percentiles, and a two-class scheme (high and low) based on a natural break at the 45th percentile in the field biomass density data. These biomass density classes were used to estimate the error in the lidar DEM of the marsh platform. To compute the lidar DEM adjustment value, the field biomass samples (n = 16) and their corresponding lidar DEM errors were analyzed. Within each classification scheme, two methods for computing the recommended adjustment values were investigated: a median approach, where the 50th percentile lidar DEM error within each class was selected, and a quartile approach, where the 75th percentile lidar DEM error within the high class, the 50th percentile within the medium class, and the 25th percentile with the low class were selected. All combinations of biomass density classification scheme and adjustment value were tested. All adjustment schemes were superior to the unadjusted lidar DEM by wide statistical margins. The schemes were also compared to a simple median adjustment based on the entire population of samples. Only the scenarios using the quartile adjustment selection were significantly better than simply using the overall median lidar DEM error. The two-class quartile adjustment method was most effective. The recommended adjustments associated with each of these classes are 0.30 m for high and 0.18 m for low. The performance of the method was tested on 229 spot elevation points in the lower Apalachicola River Marsh. The two-class quartile-adjusted DEM reduced the RMS error in elevation from 0.65 m to 0.40 m, a 38% improvement. The raw mean errors for the lidar DEM and the adjusted DEM were 0.61 ± 0.24 m and 0.32 ± 0.24 m, respectively, thereby reducing the high bias by approximately 49%.
Based on the findings of this study and the work by others cited herein, coastal researchers should avoid using unadjusted lidar DEMs in coastal marshes and wetlands dominated by emergent vegetation. Instead, they are advised to apply a spatially variable correction technique such as the one presented here. Future work in developing these techniques should address the major sources of uncertainty identified in this paper, namely the discrepancy in acquisition time between field and remote sensing datasets.