Monitoring and Management of Imidazolinone-Resistant Red Rice (Oryza sativa L., var. sylvatica) in Clearfield® Italian Paddy Rice
Abstract
:1. Introduction
2. Results and Discussion
2.1. Field History of the Sampled Fields
| IBAF Code | Sampling Year | Location (province) | No. IMI treatments (*) | Rotation | Cyperaceae and Echinochloa not controlled | Phenotype | Amino acid |
|---|---|---|---|---|---|---|---|
| 010-1 | 2010 | Ottobiano (PV) | None | No | No | S | Ser |
| 010-2 | 2010 | Torre d'Isola (PV) | None | No | No | S | Ser |
| 010-3 | 2010 | Cergnago (PV) | None | No | No | S | - |
| 010-4 | 2010 | Rosate (MI) | 4-(2) | No | No | S | - |
| 010-5 | 2010 | Salussola (BI) | 2-(1) | No | No | R |  |
| 010-6 | 2010 | Zerbolò (PV) | 2-(1) | No | No | R | Asn |
| 010-7 | 2010 | Santhià (VC) | 6-(3) | No | No | S | - |
| 010-8 | 2010 | Robbio (PV) | 2-(1) | No | No | S | - |
| 010-9 | 2010 | Borgo Lavezzaro (NO) | 2-(1) | No | No | R |  |
| 010-10 | 2010 | Bianzè (VC) | 8-(4) | No | No | S | - |
| 010-11 | 2010 | Ottobiano (PV) | 6-(3) | No | No | R | Asn |
| 010-12 | 2010 | Lomello (PV) | 10-(5) | No | No | R | Asn |
| 010-13 | 2010 | Sannazzaro de Burgondi (PV) | 6-(3) | No | No | R |  |
| 010-14 | 2010 | Ferrera Erbognone (PV) | 10-(5) | No | No | R | Asn |
| 010-15 | 2010 | Pomaro Monferrato (AL) | 6-(3) | No | No | S | - |
| 010-17 | 2010 | Castelnovetto (PV) | 2-(1) | No | No | R | Asn |
| 010-18 | 2010 | Pomaro Monferrato (AL) | 8-(4) | No | No | S | - |
| 011-19 | 2011 | Ferrera Erbognone (PV) | 8-(4) | No | No | R | Asn |
| 011-20 | 2011 | Olevano Lomellina (PV) | 2-(1) | No | No | R | Asn |
| 011-21 | 2011 | Ceretto Lomellina (PV) | 10-(5) | No | No | R | Asn |
| 011-22 | 2011 | Castello d'Agogna (PV) | 4-(5) | Yes | No | S | - |
| 011-23 | 2011 | Sannazzaro de Burgondi (PV) | 6-(3) | No | No | R | Asn |
| 011-24 | 2011 | Bianzè (VC) | 4-(5) | Yes | No | S | Ser |
| 011-25 | 2011 | Vespolate (NO) | 2-(3) | Yes | No | R | Asn |
| 011-26 | 2011 | Santhià (VC) | 6-(3) | No | No | R | Asn |
| 011-27 | 2011 | Vercelli (VC) | 4-(3) | Yes | Yes | R | Asn |
| 011-28 | 2011 | Garbagna Novarese (NO) | 4-(4) | Yes | No | S | - |
| 011-29 | 2011 | Langosco (PV) | 6-(3) | No | No | S | - |
| 011-30 | 2011 | Robbio (PV) | 2-(2) | Yes | Yes | S | - |
| 011-31 | 2011 | Livorno Ferraris (VC) | 6-(3) | No | No | S | - |
| 011-32 | 2011 | Lomello (PV) | 12-(6) | No | No | R | Asn |
| 011-33 | 2011 | Ottobiano (PV) | 12-(6) | No | No | R | Asn |
| 011-34 | 2011 | Ferrera Erbognone (PV) | 10-(6) | No | No | R | Asn |
| 011-35 | 2011 | Sannazzaro de Burgondi (PV) | 8-(6) | No | No | R | Asn |
| 011-36 | 2011 | Pieve del Cairo (PV) | 12-(6) | No | No | R | Asn |
| 011-37 | 2011 | Gambarana (PV) | 4-(6) | Yes | No | S | - |
| 011-38 | 2011 | Olevano Lomellina (PV) | 8-(6) | Yes | No | R | Asn |
| 011-39 | 2011 | San Giorgio Lomellina (PV) | 6-(6) | No | No | S | - |
| 011-40 | 2011 | Semiana (PV) | 4-(2) | No | Yes | R | Asn |
| 011-41 | 2011 | Zeccone (PV) | 4-(2) | No | Yes | R | Asn |
| 011-42 | 2011 | Noviglio (MI) | 6-(3) | No | Yes | R | Asn |
| 011-43 | 2011 | Noviglio (MI) | 6-(3) | No | Yes | S | - |
| 011-44 | 2011 | Vernate (MI) | 4-(3) | Yes | Yes | S | - |
| 011-45 | 2011 | Ottobiano (PV) | 6-(3) | No | No | R | Asn |
| 011-46 | 2011 | Pomaro Monferrato (AL) | 6-(6) | Yes | No | R | Asn |
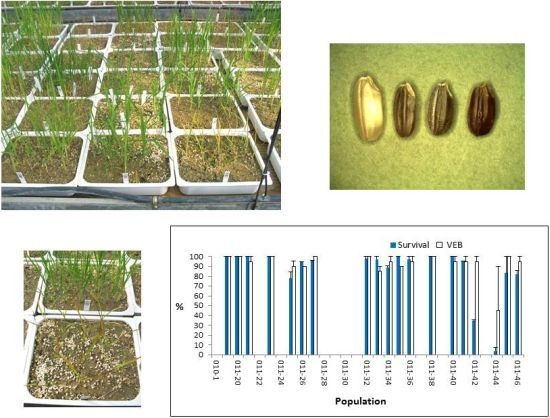
2.2. Whole-Plant Resistance Assessment
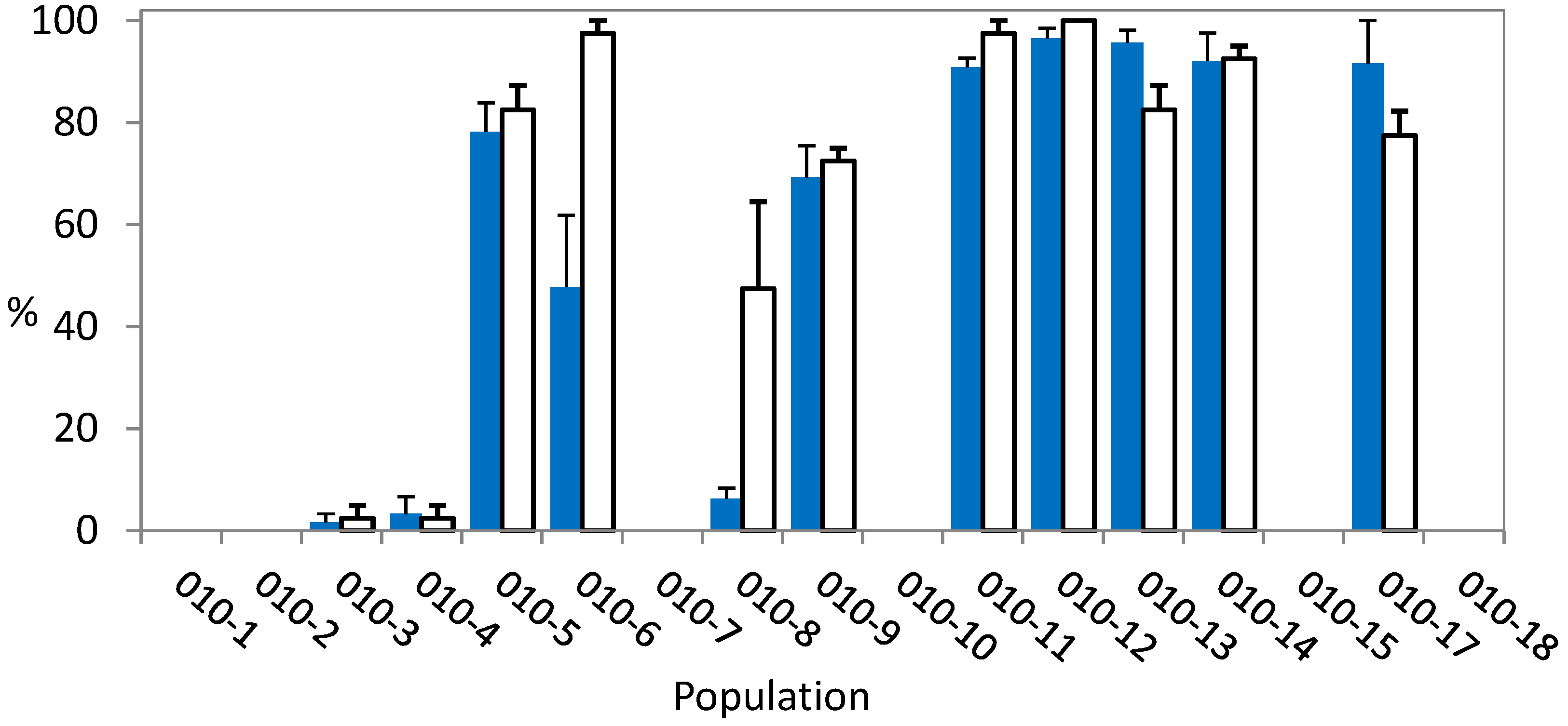
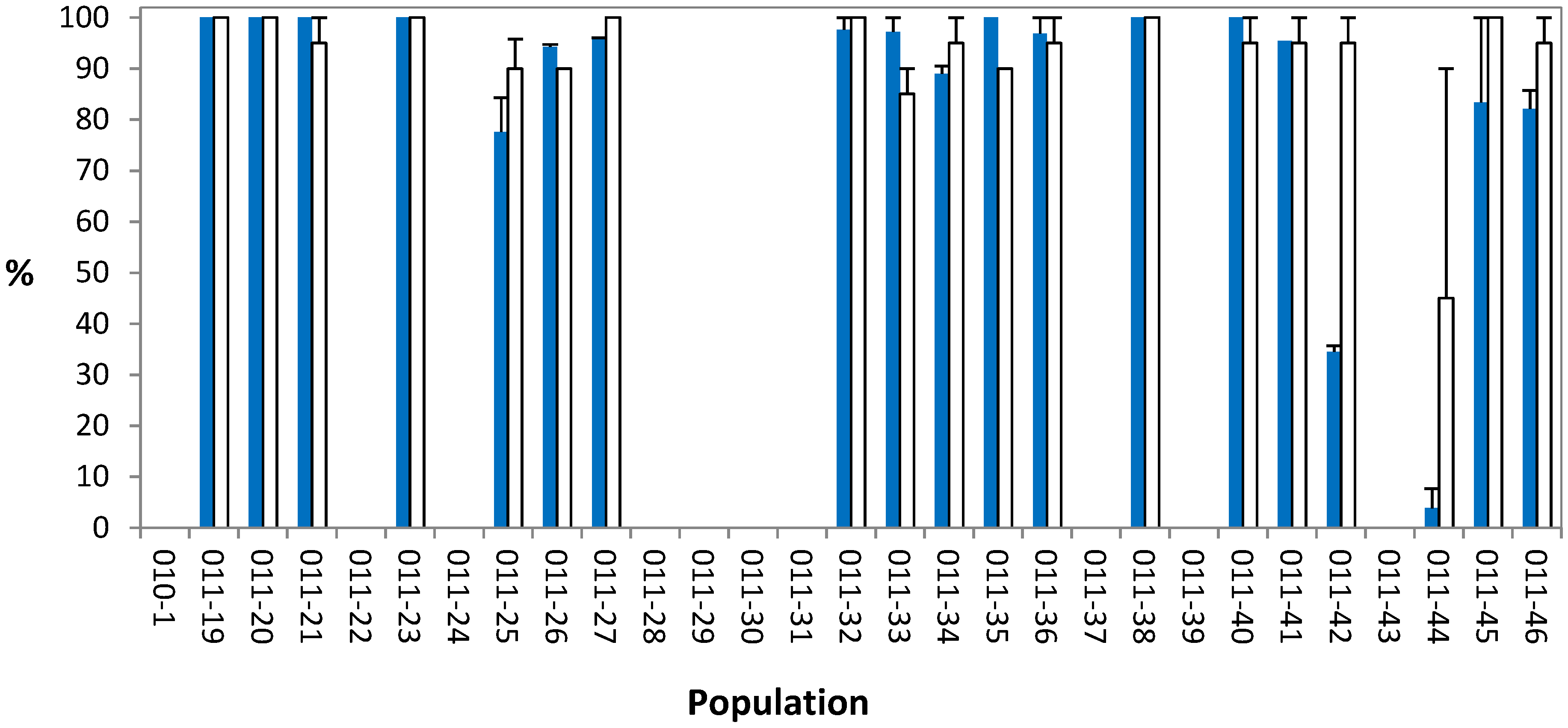
2.3. Resistance Mechanism
2.4. Sustainable Weed Management for Clearfield® Technology
3. Experimental Section
3.1. Plant Material
3.2. Whole-Plant Imazamox Sensitivity Assessment
3.3. DNA Extraction and Amplification
4. Conclusions
Acknowledgments
References
- Ferrero, A.; Vidotto, F.; Balsari, P.; Airoldi, G. Mechanical and chemical control of red rice (Oryza sativa L. var. sylvatica) in rice (Oryza sativa L.) pre planting. Crop Prot. 1999, 18, 245–251. [Google Scholar] [CrossRef]
- Craigmiles, J.P. troduction. In Red Rice: Research and Control; Easting, E.F., Ed.; Texas Agri. Exp. Stn. Bull; Texas A&M University: College Station, TX, USA, 1978; pp. 5–6. [Google Scholar]
- Kwon, S.L.; Smith, R.J.; Talbert, R.E. Comparative growth and development of red rice (Oryza sativa) and rice (O. sativa). Weed Sci. 1992, 40, 57–62. [Google Scholar]
- Fogliatto, S.; Vidotto, F.; Ferrero, A. Morphological characterisation of Italian weedy rice (Oryza sativa) populations. Weed Res. 2012, 52, 60–69. [Google Scholar] [CrossRef]
- Parker, C.; Dean, M.L. Control of wild rice in rice. Pestic. Sci. 1976, 7, 403–416. [Google Scholar] [CrossRef]
- Català Forner, M. Chemical and cultural practices for red rice control in rice fields in Ebro Delta (Spain). Crop Prot. 1995, 14, 405–408. [Google Scholar] [CrossRef]
- Ferrero, A.; Vidotto, F. Shattering ability of red rice seeds in cultural conditions. In Proceedings of the 50th International Symposium on Crop Protaction, Gent, Belgium, 5 May 1998; 50, pp. 839–843.
- Webster, E.P.; Masson, J.A. Acetolactate synthase-inhibiting herbicides on imidazolinone-tolerant rice. Weed Sci. 2001, 49, 652–657. [Google Scholar] [CrossRef]
- Tan, S.; Evans, R.R.; Dahmer, M.L.; Singh, B.K.; Shaner, D.L. Imidazolinone-tolerant crops: History, current status and future. Pest Manag. Sci. 2005, 61, 246–257. [Google Scholar] [CrossRef]
- Gressel, J.; Valverde, B.E. A strategy to provide long-term control of weedy rice while mitigating herbicide resistance transgene flow, and its potential use for other crops with related weeds. Pest Manag. Sci. 2009, 65, 723–731. [Google Scholar] [CrossRef]
- Tranel, P.J.; Wright, T.R. Resistance of weeds to ALS-inhibiting herbicides: What have we learned? Weed Sci. 2002, 50, 700–712. [Google Scholar] [CrossRef]
- Tranel, P.J.; Wright, T.R.; Heap, I.N. ALS mutations from resistant weeds. Available online: http://www.weedscience.org (accessed on 20 October 2012).
- Sales, M.A.; Shivrain, V.K.; Burgos, N.R.; Kuk, Y.I. Amino acid substitutions in the acetolactate synthase gene of red rice (Oryza sativa) confer resistance to imazethapyr. Weed Sci. 2008, 56, 485–489. [Google Scholar] [CrossRef]
- Gealy, D.R.; Mitten, D.H.; Rutger, J.N. Gene flow between red rice (Oryza sativa) and Herbicide-resistant rice (O. sativa): implications for weed management. Weed Technol. 2003, 17, 627–645. [Google Scholar] [CrossRef]
- Chaleff, R.S.; Day, E.B. Herbicide resistant mutants from tabacco cell cultures. Science 1984, 223, 1148–1151. [Google Scholar]
- Olofsdotter, M.; Valverde, B.E.; Madsen, K.H. Herbicide resistant rice (Oryza sativa L.): Global implications for weedy rice and weed management. Ann. Appl. Biol. 2000, 137, 279–295. [Google Scholar] [CrossRef]
- Scarabel, L.; Locascio, A.; Furini, A.; Sattin, M.; Varotto, S. Characterisation of ALS genes in the polyploid species Schoenoplectus mucronatus and implications for resistance management. Pest Manag. Sci. 2009, 66, 337–344. [Google Scholar]
- Panozzo, S.; Scarabel, L.; Tranel, P.J.; Sattin, M. Target-site resistance to ALS inhibitors in the polyploid species Echinochloa crus-galli. Pestic. Biochem. Phys. 2012. [Google Scholar]
- Busconi, M.; Rossi, D.; Lorenzoni, C.; Baldi, G.; Fogher, C. Spread of herbicide-resistant weedy rice (red rice, Oryza sativa L.) after 5 years of Clearfield rice cultivation in Italy. Plant Biol. 2012, 14, 751–759. [Google Scholar] [CrossRef]
- Sattin, M.; Berto, D.; Zanin, G.; Tabacchi, M. Resistance to ALS inhibitors in weeds of rice in north-western Italy. In Proceedings of International Brighton Conference on Weeds, Brighton, UK, 15–18 November 1999; British Crop Protection Council: Surrey, UK, 1999; 1–3, pp. 783–790. [Google Scholar]
- Gealy, D.R. Gene movement between rice (Oryza sativa) and weedy rice (Oryza sativa)—A US temperate rice perspective. In Crop Ferality and Volunteerism; Gressel, J., Ed.; CRC Press Taylor and Francis Group: Boca Raton, FL, USA, 2005; pp. 323–354. [Google Scholar]
- Shivrain, V.K.; Burgos, N.R.; Anders, M.M.; Rajguru, S.N.; Moore, J.; Sales, M.A. Gene flow between ClearfieldTM rice and red rice. Crop Prot. 2007, 26, 349–356. [Google Scholar] [CrossRef]
- Fogliatto, S.; Vidotto, F.; Ferrero, A. Germination of weedy rice in response to field conditions during winter. Weed Technol. 2011, 25, 252–261. [Google Scholar] [CrossRef]
- Fogliatto, S.; Vidotto, F.; Ferrero, A. Effects of winter flooding on weedy rice (Oryza sativa L.). Crop Prot. 2010, 29, 1232–1240. [Google Scholar] [CrossRef]
- Software Statistica 6.1; StatSoft: Tulsa, OK, USA, 2004. Available online: http://www.statsoft.com (accessed on 25 September 2012).
- Doyle, J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Scarabel, L.; Cenghialta, C.; Manuello, D.; Sattin, M. Monitoring and Management of Imidazolinone-Resistant Red Rice (Oryza sativa L., var. sylvatica) in Clearfield® Italian Paddy Rice. Agronomy 2012, 2, 371-383. https://doi.org/10.3390/agronomy2040371
Scarabel L, Cenghialta C, Manuello D, Sattin M. Monitoring and Management of Imidazolinone-Resistant Red Rice (Oryza sativa L., var. sylvatica) in Clearfield® Italian Paddy Rice. Agronomy. 2012; 2(4):371-383. https://doi.org/10.3390/agronomy2040371
Chicago/Turabian StyleScarabel, Laura, Cesare Cenghialta, Dario Manuello, and Maurizio Sattin. 2012. "Monitoring and Management of Imidazolinone-Resistant Red Rice (Oryza sativa L., var. sylvatica) in Clearfield® Italian Paddy Rice" Agronomy 2, no. 4: 371-383. https://doi.org/10.3390/agronomy2040371