Starch Biosynthesis in the Developing Endosperms of Grasses and Cereals
Abstract
:1. Introduction
2. Endosperm Development
3. Provision of Carbon to the Endosperm
4. General Pathway of Starch Biosynthesis
5. Starch Deposition in the Endosperm
5.1. AGPase: The First Committed Step in Starch Biosynthesis
5.2. Elongation of α-Chains by Starch Synthases
5.2.1. Granule Bound Starch Synthase I
5.2.2. Soluble Starch Synthases
5.3. Branch Linkage Formation by Starch Branching Enzymes
5.4. Debranching Enzymes
5.5. Starch Phosphorylase
5.6. Disproportionating Enzyme
5.7. Reversible Phosphorylation of α-Linked Glucans
6. Starch Granule Growth, Size and Morphology
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comparot-Moss, S.; Denyer, K. The evolution of the starch biosynthetic pathway in cereals and other grasses. J. Exp. Bot. 2009, 60, 2481–2492. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, Q.G.; Dunwell, J.M.; Zhang, Y.M. Divergent evolutionary pattern of starch biosynthetic pathway genes in grasses and dicots. Mol. Biol. Evol. 2012, 29, 3227–3236. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhu, Z.; Ma, L.; Chen, M. The preferential retention of starch synthesis genes reveals the impact of whole-genome duplication on grass evolution. Mol. Biol. Evol. 2008, 25, 1003–1006. [Google Scholar] [CrossRef] [PubMed]
- Strömberg, C.A.E. Evolution of Grasses and Grassland Ecosystems. Annu. Rev. Earth Planet. Sci. 2011, 39, 517–544. [Google Scholar] [CrossRef]
- Borrill, M. Temperate grasses. In Evolution of Crop Plants; Simmonds, N.W., Ed.; Longman: London, UK, 1976. [Google Scholar]
- Piperno, D.R.; Weiss, E.; Holst, I.; Nadel, D. Processing of wild cereal grains in the Upper Palaeolithic revealed by starch grain analysis. Nature 2004, 430, 670–673. [Google Scholar] [CrossRef] [PubMed]
- Hardy, K.; Buckley, S.; Collins, M.J.; Estalrrich, A.; Brothwell, D.; Copeland, L.; García-Tabernero, A.; García-Vargas, S.; De La Rasilla, M.; Lalueza-Fox, C.; et al. Neanderthal medics? Evidence for food, cooking, and medicinal plants entrapped in dental calculus. Naturwissenschaften 2012, 99, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Prasad, V. Dinosaur Coprolites and the Early Evolution of Grasses and Grazers. Science 2005, 310, 1177–1180. [Google Scholar] [CrossRef] [PubMed]
- Mercader, J. Mozambican Grass Seed Consumption during the Middle Stone Age. Science 2009, 326, 1680–1683. [Google Scholar] [CrossRef] [PubMed]
- Henry, A.G.; Brooks, A.S.; Piperno, D.R. Microfossils in calculus demonstrate consumption of plants and cooked foods in Neanderthal diets (Shanidar III, Iraq; Spy I and II, Belgium). Proc. Natl. Acad. Sci. USA 2011, 108, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P. The genes of the Green Revolution. Trends Genet. 2003, 19, 5–9. [Google Scholar] [CrossRef]
- Siqueira, G.F.; Pierre, J.S.; El Tahchy, A.; Glassop, D.; Singh, S.; Bonnett, G.D.; Rae, A.L. Sugarcane seed composition and changes during artificial ageing. Crop Pasture Sci. 2015, 66, 1180–1189. [Google Scholar] [CrossRef]
- Langer, R.H.M.; Hill, G.D. Agricultural Plants; Cambridge University Press: Cambridge, UK, 1982. [Google Scholar]
- Venn, B.J.; Mann, J.I. Cereal grains, legumes and diabetes. Eur. J. Clin. Nutr. 2004, 58, 1443–1461. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, A.; Bingham, S.A.; Cummings, J.H. Starch intake and colorectal cancer risk: An international comparison. Br. J. Cancer 1994, 69, 937–942. [Google Scholar] [CrossRef] [PubMed]
- Noakes, M.; Clifton, P.; Nestel, P.; Le Leu, R.; McIntosh, G. Effect of high-amylose starch and oat bran on metabolic variables and bowel function in subjects with hypertriglyceridemia. Am. J. Clin. Nutr. 1996, 64, 944–951. [Google Scholar] [PubMed]
- Annison, G.; Topping, D.L. Nutritional Role of Resistant Starch: Chemical Structure vs Physiological Function Primary Structure of Starch Components. Annu. Rev. Nutr. 1994, 14, 297–320. [Google Scholar] [CrossRef] [PubMed]
- Godfray, C.; Beddington, J.; Crute, I.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenges of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Parry, M.A.J.; Hawkesford, M.J. Food security: Increasing yield and improving resource use efficiency. Proc. Nutr. Soc. 2010, 69, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Bihmidine, S.; Hunter, C.T.; Johns, C.E.; Koch, K.E.; Braun, D.M. Regulation of assimilate import into sink organs: Update on molecular drivers of sink strength. Front. Plant Sci. 2013, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- MacNeill, G.J.; Mehrpouyan, S.; Minow, M.A.A.; Patterson, J.A.; Tetlow, I.J.; Emes, M.J. Starch as a source, starch as a sink: The bifunctional role of starch in carbon allocation. J. Exp. Bot. 2017, 1, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Setter, T.L.; Parra, R. Relationship of carbohydrate and abscisic acid levels to kernel set in maize under postpollination water deficit. Crop Sci. 2010, 50, 980–988. [Google Scholar] [CrossRef]
- Slewinski, T. Non-structural carbohydrate partitioning in grass stems: A target to increase yield stability, stress tolerance, and biofuel production. J. Exp. Bot. 2012, 63, 4647–4670. [Google Scholar] [CrossRef] [PubMed]
- Thalmann, M.; Santelia, D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017, 214, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Areche, M.; Slafer, G. Grain weight response to increases in number of grains in wheat in a Mediterranean area. Field Crops Res. 2006, 98, 52–59. [Google Scholar] [CrossRef]
- Preston, J.C.; Kellogg, E.A. Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae). Genetics 2006, 174, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.H.; Bennetzen, J.L.; Messing, J. Dynamic gene copy number variation in collinear regions of grass genomes. Mol. Biol. Evol. 2012, 29, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.P.; Garvin, D.F.; Mockler, T.C.; Schmutz, J.; Rokhsar, D.; Bevan, M.W.; Barry, K.; Lucas, S.; Harmon-Smith, M.; Lail, K.; et al. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Pollock, C.; Chatterton, N. The Biochemistry of Plants: A Comprehensive Treatise; Preiss, J., Ed.; Academic Press Inc., Ltd.: London, UK, 1988; pp. 109–140. [Google Scholar]
- Chatterton, N.J.; Harrison, P.A.; Thornley, W.R.; Draper, E.A. Oligosaccharides in foliage of Agropyron, Bromus, Dactylis, Festuca, Lolium and Phleum. New Phytol. 1990, 114, 167–171. [Google Scholar] [CrossRef]
- Schnyder, H. The role of carbohydrate storage and redistribution in the source-sink relations of wheat and barley during grain filling—A review. New Phytol. 1993, 123, 233–245. [Google Scholar] [CrossRef]
- Graf, A.; Schlereth, A.; Stitt, M.; Smith, A.M. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. USA 2010, 107, 9458–9463. [Google Scholar] [CrossRef] [PubMed]
- De Souza, A.; Sandrin, C.Z.; Moraes, M.G.; de Cássia, L. Figueiredo-Ribeiro, R. Diurnal variations of non-structural carbohydrates in vegetative tissues of Melinis minutiflora, Echinolaena inflexa and Lolium multiflorum (Poaceae). Rev. Bras. Bot. 2005, 28, 755–763. [Google Scholar] [CrossRef]
- De Figueira, J.A.; Carvalho, P.H.; Sato, H.H. Sugarcane starch: Quantitative determination and characterization. Ciência Tecnol. Aliment. 2011, 31, 806–815. [Google Scholar] [CrossRef]
- Gordon, A.J.; Ryle, G.J.A.; Powell, C.E.; Mitchell, D. Export, mobilization, and respiration of assimilates in uniculm barley during light and darkness. J. Exp. Bot. 1980, 31, 461–473. [Google Scholar] [CrossRef]
- Gordon, A.J.; Ryle, G.J.A.; Webb, G. The relationship between sucrose and starch during “dark” export from leaves of uniculm barley. J. Exp. Bot. 1980, 31, 845–850. [Google Scholar] [CrossRef]
- Nie, G.; Hendrix, D.L.; Webber, A.N.; Kimball, B.A.; Long, S.P. Increased accumulation of carbohydrates and decreased photosynthetic gene transcript levels in wheat grown at an elevated CO2 concentration in the field. Plant Physiol. 1995, 108, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Farrar, S.; Farrar, F. Carbon fluxes in leaf blades of barley. New Phytol. 1985, 100, 271–283. [Google Scholar] [CrossRef]
- Pollock, C.J. Sucrose accumulation and the initiation of fructan biosynthesis in Lolium temulentum L. New Phytol. 1984, 96, 527–534. [Google Scholar] [CrossRef]
- Cairns, A.J.; Cookson, A.; Thomas, B.J.; Turner, L.B. Starch metabolism in the fructan-grasses: Patterns of starch accumulation in excised leaves of Lolium temulentum L. J. Plant Physiol. 2002, 159, 293–305. [Google Scholar] [CrossRef]
- Cairns, A. Starch accumulation in temperate forage grasses. IGER Innov. 2002, 6–9. [Google Scholar]
- Francis, S.; Chapman, D.; Doyle, P.; Leury, B.; Egan, A. Non-structural carbohydrate content of a perennial ryegrass cultivar bred for high sugar levels, compared to “normal” perennial ryegrass and white clover. Anim. Prod. Aust. 2002, 24, 73–76. [Google Scholar]
- Ciavarella, T.; Simpson, R.; Dove, H.; Leury, B.; Sims, I. Diurnal changes in the concentration of water-soluble carbohydrates in Phalaris aquatica L. pasture in spring, and the effect of short-term shading. Aust. J. Agric. Res. 2000, 51, 749–756. [Google Scholar] [CrossRef]
- Cairns, A.J.; Gallagher, J.A. Absence of turnover and futile cycling of sucrose in leaves of Lolium temulentum L.: Implications for metabolic compartmentation. Planta 2004, 219, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Lechtenberg, V.; Holt, D.; Youngberg, H. Diurnal variation in nonstructural carbohydrates of Festuca arundinacea (Schreb.) with and without N fertilizer. Agron. J. 1972, 64, 302–305. [Google Scholar] [CrossRef]
- Ruckle, M.; Meier, M.; Frey, L.; Eicke, S.; Kölliker, R.; Zeeman, S.; Studer, B. Diurnal Leaf Starch Content: An Orphan Trait in Forage Legumes. Agronomy 2017, 7, 16. [Google Scholar] [CrossRef]
- Lunn, J.E.; Hatch, M.D. Primary partitioning and storage of photosynthate in sucrose and starch in leaves of C4 plants. Planta 1995, 197, 385–391. [Google Scholar] [CrossRef]
- Sulpice, R.; Pyl, E.-T.; Ishihara, H.; Trenkamp, S.; Steinfath, M.; Witucka-Wall, H.; Gibon, Y.; Usadel, B.; Poree, F.; Piques, M.C.; et al. Starch as a major integrator in the regulation of plant growth. Proc. Natl. Acad. Sci. USA 2009, 106, 10348–10353. [Google Scholar] [CrossRef] [PubMed]
- Stitt, M.; Zeeman, S.C. Starch turnover: Pathways, regulation and role in growth. Curr. Opin. Plant Biol. 2012, 15, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, X.; Quan, Z.; Cheng, S.; Xu, X.; Pan, S.; Xie, M.; Zeng, P.; Yue, Z.; Wang, W.; et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 2012, 30, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Austin, R.; Morgan, C.; Ford, M.; Blackwell, R. Contributions to grain yield from pre-anthesis assimilation in tall and dwarf barley phenotypes in two contrasting seasons. Ann. Bot. 1980, 45, 309–319. [Google Scholar] [CrossRef]
- Austin, R.; Bingham, J.; Blackwell, R.; Evans, L.; Ford, M.; Morgan, C.; Taylor, M. Genetic improvements in winter wheat yields since 1900 and associated physiological changes. J. Agric. Sci. 1980, 94, 52–59. [Google Scholar] [CrossRef]
- Evans, L.T.; Dunstone, R.L. Some physiological aspects of evolution in wheat. Aust. J. Biol. Sci. 1970, 23, 725–741. [Google Scholar] [CrossRef]
- Weatherwax, P. The endosperm of Zea and Coix. Am. J. Bot. 1930, 17, 371–380. [Google Scholar] [CrossRef]
- Leroux, B.M.; Goodyke, A.J.; Schumacher, K.I.; Abbott, C.P.; Clore, A.M.; Yadegari, R.; Larkins, B.A.; Dannenhoffer, J.M. Maize early endosperm growth and development: From fertilization through cell type. Am. J. Bot. 2014, 101, 1259–1274. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.; Lemmon, B.; Olsen, O. Development of the endosperm in rice (Oryza sativa L.): Cellularization. J. Plant Res. 1996, 109, 301–313. [Google Scholar] [CrossRef]
- Briarty, L.G.; Hughes, C.E.; Evers, A.D. The Developing Endosperm of Wheat—A Stereological Analysis. Ann. Bot. 1979, 44, 641–658. [Google Scholar] [CrossRef]
- Bosnes, M.; Weideman, F.; Olsen, O.-A. Endosperm differentiation in barley wild-type and sex mutants. Plant J. 1992, 2, 661–674. [Google Scholar] [CrossRef]
- Shull, J.; Chandrashekar, A.; Kirleis, A.; Ejeta, G. Development of Sorghum (Sorghum bicolor) endosperm in varieties of varying hardness. Food Struct. 1990, 9, 253–267. [Google Scholar]
- Sabelli, P.A.; Larkins, B.A. The Development of Endosperm in Grasses. Plant Physiol. 2009, 149, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Opanowicz, M.; Hands, P.; Betts, D.; Parker, M.L.; Toole, G.A.; Mills, E.N.C.; Doonan, J.H.; Drea, S. Endosperm development in Brachypodium distachyon. J. Exp. Bot. 2011, 62, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Hands, P. Analysis of grain characters in temperate grasses reveals distinctive patterns of endosperm organisation associated with grain shape. J. Exp. Bot. 2012, 63, 6253–6266. [Google Scholar] [CrossRef] [PubMed]
- Olsen, O.-A. Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 2004, 16, S214–S227. [Google Scholar] [CrossRef] [PubMed]
- Buttrose, M. Ultrastructure of the developing aleurone cells of wheat grain. Aust. J. Biol. Sci. 1963, 16, 768–774. [Google Scholar] [CrossRef]
- Offler, C.E.; McCurdy, D.W.; Patrick, J.W.; Talbot, M.J. Transfer Cells: Cells Specialized for a Special Purpose. Annu. Rev. Plant Biol. 2003, 54, 431–454. [Google Scholar] [CrossRef] [PubMed]
- Ohdan, T.; Francisco, P.B.; Sawada, T.; Hirose, T.; Terao, T.; Satoh, H.; Nakamura, Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 2005, 56, 3229–3244. [Google Scholar] [CrossRef] [PubMed]
- Uhlmann, N.K.; Beckles, D.M. Storage products and transcriptional analysis of the endosperm of cultivated wheat and two wild wheat species. J. Appl. Genet. 2010, 51, 431–447. [Google Scholar] [CrossRef] [PubMed]
- Brocklehurst, P.A. Factors controlling grain weight in wheat. Nature 1977, 266, 348–349. [Google Scholar] [CrossRef]
- Chojecki, A.; Gale, M.; Bayliss, M.W. The number and sizes of starch granules in the wheat endosperm, and their association with grain weight. Ann. Bot. (Lond.) 1986, 58, 819–831. [Google Scholar] [CrossRef]
- Chojecki, A.; Bayliss, M.; Gale, M. Cell production and DNA accumulation in the wheat endosperm, and their association with grain weight. Ann. Bot. (Lond.) 1986, 58, 809–817. [Google Scholar] [CrossRef]
- Jones, R.J.; Schreiber, B.M.N.; Roessler, J.A. Kernel sink capacity in maize: Genotypic and maternal regulation. Crop Sci. 1996, 36, 301–306. [Google Scholar] [CrossRef]
- Beckles, D.M.; Thitisaksakul, M. How environmental stress affects starch composition and functionality in cereal endosperm. Starch/Staerke 2014, 66, 58–71. [Google Scholar] [CrossRef]
- Reddy, V.M.; Daynard, T.B. Endosperm characteristics associated with rate of grain filling and kernel size in corn. Maydica 1983, 28, 339–355. [Google Scholar]
- Commuri, P.D.; Jones, R.J. Ultrastructural characterization of maize (Zea mays L.) kernels exposed to high temperature during endosperm cell division. Plant Cell Environ. 1999, 22, 375–385. [Google Scholar] [CrossRef]
- Van Dongen, J.T.; Roeb, G.W.; Dautzenberg, M.; Froehlich, A.; Vigeolas, H.; Minchin, P.E.H.; Geigenberger, P.; Van Dongen, J.T.; Roeb, G.W.; Dautzenberg, M.; et al. Phloem Import and Storage Metabolism Are Highly Coordinated by the Low Oxygen Concentrations within Developing Wheat Seeds. Plant Physiol. 2004, 135, 1809–1821. [Google Scholar] [CrossRef] [PubMed]
- Rolletschek, H.; Koch, K.; Wobus, U.; Borisjuk, L. Positional cues for the starch/lipid balance in maize kernels and resource partitioning to the embryo. Plant J. 2005, 42, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Rolletschek, H.; Weschke, W.; Weber, H.; Wobus, U.; Borisjuk, L. Energy state and its control on seed development: Starch accumulation is associated with high ATP and steep oxygen gradients within barley grains. J. Exp. Bot. 2004, 55, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Radchuk, V.; Borisjuk, L. Physical, metabolic and developmental functions of the seed coat. Front. Plant Sci. 2014, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Rolletschek, H.; Melkus, G.; Grafahrend-Belau, E.; Fuchs, J.; Heinzel, N.; Schreiber, F.; Jakob, P.M.; Borisjuk, L. Combined Noninvasive Imaging and Modeling Approaches Reveal Metabolic Compartmentation in the Barley Endosperm. Plant Cell 2011, 23, 3041–3054. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N. Tocochromanols: Rancid lipids, seed longevity, and beyond. Proc. Natl. Acad. Sci. USA 2010, 107, 17857–17858. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C.; Kranner, I. Analyses of reactive oxygen species and antioxidants in relation to seed longevity and germination. Methods Mol. Biol. 2011, 773, 343–367. [Google Scholar] [PubMed]
- Hendry, G. Oxygen, free radical processes and seed longevity. Seed Sci. Res. 1993, 3, 141–153. [Google Scholar] [CrossRef]
- Alonso, A.P.; Val, D.L.; Shachar-Hill, Y. Central metabolic fluxes in the endosperm of developing maize seeds and their implications for metabolic engineering. Metab. Eng. 2011, 13, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Young, T.E.; Gallie, D.R. Programmed cell death during endosperm development. Plant Mol. Biol. 2000, 44, 283–301. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, L.; Li, J.; Song, X.; Yang, C. Study on programmed cell death and dynamic changes of starch accumulation in pericarp cells of Triticum aestivum L. Protoplasma 2009, 236, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Van Bel, A.J.E. Strategies of Phloem Loading. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993, 44, 253–281. [Google Scholar] [CrossRef]
- Van Bel, A.J.E.; Helariutta, Y.; Thompson, G.A.; Ton, J.; Dinant, S.; Ding, B.; Patrick, J.W. Phloem: The integrative avenue for resource distribution, signaling, and defense. Front. Plant Sci. 2013, 4, 471. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, R. The Role of Phloem Loading Reconsidered. Plant Physiol. 2010, 152, 1817–1823. [Google Scholar] [CrossRef] [PubMed]
- De Schepper, V.; De Swaef, T.; Bauweraerts, I.; Steppe, K. Phloem transport: A review of mechanisms and controls. J. Exp. Bot. 2013, 64, 4839–4850. [Google Scholar] [CrossRef] [PubMed]
- Jenner, C.F.; Rathjen, A.J. Factors limiting the supply of sucrose to the developing wheat grain. Ann. Bot. 1972, 36, 729–741. [Google Scholar] [CrossRef]
- Fisher, D.B.; Gifford, R.M. Accumulation and Conversion of Sugars by Developing Wheat Grains. Plant Physiol. 1987, 84, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Shannon, J.; Dougherty, C. Movement of 14C-Labeled assimilates into kernels of Zea mays L. II. Invertase activity of the pedicel and placento-chalazal tissues. Plant Physiol. 1972, 49, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, M.P. Morphology of the crease region in relation to assimilate uptake and water loss during caryopsis development in barley and wheat. Aust. J. Plant Physiol. 1983, 10, 473–491. [Google Scholar] [CrossRef]
- Thorne, J.H. Developing seeds. Annu. Rev. Plant Physiol. 1985, 36, 317–343. [Google Scholar] [CrossRef]
- Weschke, W.; Panitz, R.; Sauer, N.; Wang, Q.; Neubohn, B.; Weber, H.; Wobus, U. Sucrose transport into barley seeds: Molecular characterization of two transporters and implications for seed development and starch accumulation. Plant J. 2000, 21, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Dayanandan, P. Structural and histochemical studies on grain-filling in the caryopsis of rice (Oryza sativa L.). J. Biosci. 2003, 28, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Felker, F.C.; Shannon, J.C. Movement of 14C-labeled Assimilates into Kernels of Zea mays L: III. An anatomical examination and microautoradiographic study of assimilate transfer. Plant Physiol. 1980, 65, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Sosso, D.; Luo, D.; Li, Q.-B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; McCarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef] [PubMed]
- Vilhar, B.; Kladnik, A.; Blejec, A.; Chourey, P.S.; Dermastia, M. Cytometrical evidence that the loss of seed weight in the miniature1 seed mutant of maize is associated with reduced mitotic activity in the developing endosperm. Plant Physiol. 2002, 129, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Chourey, P.S.; Li, Q.B.; Pring, D.R. Expression of cell wall invertase and several other genes of sugar metabolism in relation to seed development in sorghum (Sorghum bicolor). J. Plant Physiol. 2008, 165, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Ou-Lee, T.M.; Setter, T.L. Enzyme activities of starch and sucrose pathways and growth of apical and basal maize kernels. Plant Physiol. 1985, 79, 848–851. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.E. Carbohydrate-Modulated Gene Expression in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 509–540. [Google Scholar] [CrossRef] [PubMed]
- Koch, K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 2004, 7, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L.; Jin, Y.; Yang, Y.J.; Li, G.J.; Boyer, J.S. Sugar input, metabolism, and signaling mediated by invertase: Roles in development, yield potential, and response to drought and heat. Mol. Plant 2010, 3, 942–955. [Google Scholar] [CrossRef] [PubMed]
- Barratt, D.H.P.; Derbyshire, P.; Findlay, K.; Pike, M.; Wellner, N.; Lunn, J.; Feil, R.; Simpson, C.; Maule, A.J.; Smith, A.M. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc. Natl. Acad. Sci. USA 2009, 106, 13124–13129. [Google Scholar] [CrossRef] [PubMed]
- Kato, T. Change of Sucrose Synthase Activity in Developing Endosperm of Rice Cultivars. Crop Sci. 1995, 35, 827–831. [Google Scholar] [CrossRef]
- Thévenot, C.; Simond-Côte, E.; Reyss, A.; Manicacci, D.; Trouverie, J.; Le Guilloux, M.; Ginhoux, V.; Sidicina, F.; Prioul, J.L. QTLs for enzyme activities and soluble carbohydrates involved in starch accumulation during grain filling in maize. J. Exp. Bot. 2005, 56, 945–958. [Google Scholar] [CrossRef] [PubMed]
- Chourey, P.S.; Taliercio, E.W.; Carlson, S.J.; Ruan, Y.L. Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol. Gen. Genet. 1998, 259, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Farré, E.M.; Geigenberger, P.; Willmitzer, L.; Trethewey, R.N. A possible role for pyrophosphate in the coordination of cytosolic and plastidial carbon metabolism within the potato tuber. Plant Physiol. 2000, 123, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Appeldoorn, N.J.G.; De Bruijn, S.M.; Koot-Gronsveld, E.A.M.; Visser, R.G.F.; Vreugdenhil, D.; Van Der Plas, L.H.W. Developmental changes in enzymes involved in the conversion of hexose phosphate and its subsequent metabolites during early tuberization of potato. Plant Cell Environ. 1999, 22, 1085–1096. [Google Scholar] [CrossRef]
- Zeng, Y.; Wu, Y.; Avigne, W.T.; Koch, K.E. Rapid repression of maize invertases by low oxygen. Invertase/sucrose synthase balance, sugar signaling potential, and seedling survival. Plant Physiol. 1999, 121, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Richard, B.; Rivoal, J.; Spiteri, A.; Pradet, A. Anaerobic Stress Induces the Transcription and Tranlslation of Sucrose Synthase in Rice. Plant Physiol. 1991, 95, 669–674. [Google Scholar] [CrossRef]
- Huber, S.C.; Akazawa, T. A novel sucrose synthase pathway for sucrose degradation in cultured sycamore cells. Plant Physiol. 1986, 81, 1008–1013. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.P.; Sung, S.J.; Loboda, T.; Kormanik, P.P.; Black, C.C. Characterization of Sucrolysis via the Uridine Diphosphate and Pyrophosphate-Dependent Sucrose Synthase Pathway. Plant Physiol. 1989, 90, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Guglielminetti, L.; Perata, P.; Alpi, A. Effect of Anoxia on Carbohydrate Metabolism in Rice Seedlings. Plant Physiol. 1995, 108, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Perata, P.; Pozueta-Romero, J.; Akazawa, T.; Yamaguchi, J. Effect of anoxia on starch breakdown in rice and wheat seeds. Planta 1992, 188, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Taliercio, E.; Chourey, P. The Miniature1 Seed Locus of Maize Encodes a Cell Wall Invertase Required for Normal Development of Endosperm and Maternal Cells in the Pedicel. Plant Cell 1996, 8, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A. Back to the drawing board: redefining starch synthesis in cereals. Trends Plant Sci. 1996, 1, 363–364. [Google Scholar] [CrossRef]
- Kleczkowski, L.A. Glucose activation and metabolism through UDP-glucose pyrophosphorylase in plants. Phytochemistry 1994, 37, 1507–1515. [Google Scholar] [CrossRef]
- Sung, S.J.S.; Xu, D.P.; Galloway, C.M.; Black, C.C. A reassessment of glycolysis and gluconeogenesis in higher plants. Physiol. Plant. 1988, 72, 650–654. [Google Scholar] [CrossRef]
- Ap Rees, T.; Morrell, S. Carbohydrate metabolism in developing potatoes. Am. Potato J. 1990, 67, 835–847. [Google Scholar] [CrossRef]
- Stitt, M. Pyrophosphate as an energy donor in the cytosol of plant cells: An enigmatic alternative to ATP. Bot. Acta 1998, 111, 167–175. [Google Scholar] [CrossRef]
- Zeng, Y.; Wu, Y.; Avigne, W.T.; Koch, K.E. Differential Regulation of Sugar-Sensitive Sucrose Synthases by Hypoxia and Anoxia Indicate Complementary Transcriptional and Posttranscriptional Responses1. Plant Physiol. 1998, 116, 1573–1583. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, D.J.; Turner, J.F. Pyruvate kinase of higher plants. Biochim. Biophys. Acta 1973, 329, 128–139. [Google Scholar] [CrossRef]
- Plaxton, W.C. The Organization and Regulation of Plant Glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 185–214. [Google Scholar] [CrossRef] [PubMed]
- Kurzawska, A.; Gõrecka, D.; Błaszczak, W.; Szwengiel, A.; Paukszta, D.; Lewandowicz, G. The molecular and supermolecular structure of common cattail (Typha latifolia) starch. Starch/Staerke 2014, 66, 849–856. [Google Scholar] [CrossRef]
- Wersal, R.M.; Madsen, J.D.; Cheshier, J.C. Seasonal Biomass and Starch Allocation of Common Reed (Phragmites australis) (Haplotype I) in Southern Alabama, USA. Invasive Plant Sci. Manag. 2013, 6, 140–146. [Google Scholar] [CrossRef]
- Chourey, P.S.; Nelson, O.E. The enzymatic deficiency conditioned by the shrunken-1 mutations in maize. Biochem. Genet. 1976, 14, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Chourey, P. The Maize Invertase-Deficient miniature-1 Seed Mutation Is Associated with Aberrant Pedicel and Endosperm Development. Plant Cell 1992, 4, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Fincher, G.B.; Stone, B.A. Cell walls and their components in cereal grain technology. In Advances in Cereal Science and Technology; Pomeranz, Y., Ed.; American Association of Cereal Chemists Inc.: St. Paul, MN, USA, 1986. [Google Scholar]
- Yu, X.; Li, B.; Wang, L.; Chen, X.; Wang, W.; Wang, Z.; Xiong, F. Systematic analysis of pericarp starch accumulation and degradation during wheat caryopsis development. PLoS ONE 2015, 10, e0138228. [Google Scholar] [CrossRef] [PubMed]
- Earp, C.F.; McDonough, C.M.; Rooney, L.W. Microscopy of pericarp development in the caryopsis of Sorghum bicolor (L.) Moench. J. Cereal Sci. 2004, 39, 21–27. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, Z.; Chen, J.; Zhao, G. The structure and function of pericarp in rice. Acta Agron. Sin. 2002, 28, 439–444. [Google Scholar]
- She, K.-C.; Kusano, H.; Koizumi, K.; Yamakawa, H.; Hakata, M.; Imamura, T.; Fukuda, M.; Naito, N.; Tsurumaki, Y.; Yaeshima, M.; et al. A Novel Factor FLOURY ENDOSPERM2 Is Involved in Regulation of Rice Grain Size and Starch Quality. Plant Cell 2010, 22, 3280–3294. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.G.; Park, S.; Matsuoka, M.; An, G. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C4-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J. 2005, 42, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zheng, X.; Yang, J.; Messing, J.; Wu, Y. Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc. Natl. Acad. Sci. USA 2016, 113, 10842–10847. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Xu, H.; Zhu, Y.; Liu, Q.Q.; Cai, X.L. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 2013, 64, 3453–3466. [Google Scholar] [CrossRef] [PubMed]
- Pérez, S.; Bertoft, E. The molecular structures of starch components and their contribution to the architecture of starch granules: A comprehensive review. Starch/Staerke 2010, 62, 389–420. [Google Scholar] [CrossRef]
- Tetlow, I.J. Starch biosynthesis in developing seeds. Seed Sci. Res. 2011, 21, 5–32. [Google Scholar] [CrossRef]
- Pfister, B.; Zeeman, S.C. Formation of starch in plant cells. Cell. Mol. Life Sci. 2016, 73, 2781–2807. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, J.R.; Kossmann, J. Transitory and storage starch metabolism: Two sides of the same coin? Curr. Opin. Biotechnol. 2015, 32, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Blennow, A.; Jensen, S.L.; Shaik, S.S.; Skryhan, K.; Carciofi, M.; Holm, P.B.; Hebelstrup, K.H.; Tanackovic, V. Future cereal starch bioengineering: Cereal ancestors encounter gene technology and designer enzymes. Cereal Chem. 2013, 90, 274–287. [Google Scholar] [CrossRef]
- Brozynska, M.; Furtado, A.; Henry, R.J. Genomics of crop wild relatives: Expanding the gene pool for crop improvement. Plant Biotechnol. J. 2016, 14, 1070–1085. [Google Scholar] [CrossRef] [PubMed]
- Hajjar, R.; Hodgkin, T. The use of wild relatives in crop improvement: A survey of developments over the last 20 years. Euphytica 2007, 156, 1–13. [Google Scholar] [CrossRef]
- Ghosh, H.; Preiss, J. Adenosine diphosphate glucose pyrophosphorylase: A regulatory enzyme in the biosynthesis of starch in spinach leaf chloroplasts. J. Biol. Chem. 1966, 241, 4491–4504. [Google Scholar] [PubMed]
- Thorbjørnsen, T.; Villand, P.; Denyer, K.; Olsen, O.A.; Smith, A.M. Distinct isoforms of ADPglucose pyrophosphorylase occur inside and outside the amyloplasts in barley endosperm. Plant J. 1996, 10, 243–250. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Davies, E.J.; Vardy, K.A.; Bowsher, C.G.; Burrell, M.M.; Emes, M.J. Subcellular localization of ADPglucose pyrophosphorylase in developing wheat endosperm and analysis of the properties of a plastidial isoform. J. Exp. Bot. 2003, 54, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Sikka, V.; Choi, B.; Kavakli, I.; Sakulsingharoj, C.; Gupta, S.; Ito, H.; Okita, T. Subcellular compartmentation and allosteric regulation of the rice endosperm ADPglucose pyrophosphorylase. Plant Sci. 2001, 161, 461–468. [Google Scholar] [CrossRef]
- Denyer, K.; Dunlap, F.; Thorbjørnsen, T.; Keeling, P.; Smith, A.M. The Major Form of ADP-Glucose Pyrophosphorylase in Maize Endosperm is Extra-Plastidial. Plant Physiol. 1996, 112, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-Y.; Nelson, O. Starch-deficient maize mutant lacking adenosine diphosphate glucose pyrophosphorylase activity. Science 1966, 151, 341–343. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Hwang, S.K.; Han, M.; Eom, J.S.; Kang, H.G.; Han, Y.; Choi, S.B.; Cho, M.H.; Bhoo, S.H.; An, G.; et al. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Mol. Biol. 2007, 65, 531–546. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.E.; Patron, N.J.; Bottrill, A.R.; Dinges, J.R.; Fahy, B.F.; Parker, M.L.; Waite, D.N.; Denyer, K.A. Low-starch barley mutant, Risø 16, lacking the cytosolic small subunit of ADP-glucose pyrophosphorylase, reveals the importance of the cytosolic isoform and the identity of the plastidial small subunit. Plant Physiol. 2003, 131, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Kirchberger, S.; Tjaden, J.; Ekkehard Neuhaus, H. Characterization of the Arabidopsis Brittle1 transport protein and impact of reduced activity on plant metabolism. Plant J. 2008, 56, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Bowsher, C.G.; Scrase-Field, E.F.A.L.; Esposito, S.; Emes, M.J.; Tetlow, I.J. Characterization of ADP-glucose transport across the cereal endosperm amyloplast envelope. J. Exp. Bot. 2007, 58, 1321–1332. [Google Scholar] [CrossRef] [PubMed]
- Cakir, B.; Shiraishi, S.; Tuncel, A.; Matsusaka, H.; Satoh, R.; Singh, S.; Crofts, N.; Hosaka, Y.; Fujita, N.; Hwang, S.-K.; et al. Analysis of the rice ADPglucose transporter (OsBT1) indicates the presence of regulatory processes in the amyloplast stroma that control ADPglucose flux into starch. Plant Physiol. 2016, 170, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Beckles, D.M.; Smith, A.M.; ap Rees, T. A cytosolic ADP-glucose pyrophosphorylase is a feature of graminaceous endosperms, but not of other starch-storing organs. Plant Physiol. 2001, 125, 818–827. [Google Scholar] [CrossRef] [PubMed]
- Gross, P.; ap Rees, T. Alkaline inorganic pyrophosphatase and starch synthesis in amyloplasts. Planta 1986, 167, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Tiessen, A.; Nerlich, A.; Faix, B.; Hümmer, C.; Fox, S.; Trafford, K.; Weber, H.; Weschke, W.; Geigenberger, P. Subcellular analysis of starch metabolism in developing barley seeds using a non-aqueous fractionation method. J. Exp. Bot. 2012, 63, 2071–2087. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Ballicora, M.A.; Leykam, J.F.; Preiss, J. Mechanism of reductive activation of potato tuber ADP-glucose pyrophosphorylase. J. Biol. Chem. 1998, 273, 25045–25052. [Google Scholar] [CrossRef] [PubMed]
- Tiessen, A.; Hendriks, J.H.M.; Stitt, M.; Branscheid, A.; Gibon, Y.; Farré, E.M.; Geigenberger, P. Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase. Am. Soc. Plant Biol. 2002, 14, 2191–2213. [Google Scholar] [CrossRef]
- Gómez-Casati, D.F.; Iglesias, A.A. ADP-glucose pyrophosphorylase from wheat endosperm. Purification and characterization of an enzyme with novel regulatory properties. Planta 2002, 214, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.; Villand, P.; Lüthi, E.; Olsen, O.; Preiss, J. Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate regulation. Plant Physiol. 1993, 101, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P. Regulation of Starch Biosynthesis in Response to a Fluctuating Environment. Plant Physiol. 2011, 155, 1566–1577. [Google Scholar] [CrossRef] [PubMed]
- Tuncel, A.; Okita, T.W. Improving starch yield in cereals by over-expression of ADPglucose pyrophosphorylase: Expectations and unanticipated outcomes. Plant Sci. 2013, 211, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Hannah, L.C.; Futch, B.; Bing, J.; Shaw, J.R.; Boehlein, S.; Stewart, J.D.; Beiriger, R.; Georgelis, N.; Greene, T. A shrunken-2 Transgene Increases Maize Yield by Acting in Maternal Tissues to Increase the Frequency of Seed Development. Plant Cell 2012, 24, 2352–2363. [Google Scholar] [CrossRef] [PubMed]
- Smidansky, E.D.; Meyer, F.D.; Blakeslee, B.; Weglarz, T.E.; Greene, T.W.; Giroux, M.J. Expression of a modified ADP-glucose pyrophosphorylase large subunit in wheat seeds stimulates photosynthesis and carbon metabolism. Planta 2007, 225, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Smidansky, E.D.; Martin, J.M.; Hannah, L.C.; Fischer, A.M.; Giroux, M.J. Seed yield and plant biomass increases in rice are conferred by deregulation of endosperm ADP-glucose pyrophosphorylase. Planta 2003, 216, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Smidansky, E.; Clancy, M.; Meyer, F.; Lanning, S.; Blake, N.; Talbert, L.; Giroux, M. Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. J. Plant Physiol. Pathol. 2002, 99, 1724–1729. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Smidansky, E.; Beecher, B.; Greene, T.; Giroux, M. The maize Sh2r6hs ADP-glucose pyrophosphorylase (AGP) large subunit confers enhanced AGP properties in transgenic wheat (Triticum aestivum). Plant Sci. 2004, 167, 899–911. [Google Scholar] [CrossRef]
- Oiestad, A.J.; Martin, J.M.; Giroux, M.J. Overexpression of ADP-glucose pyrophosphorylase in both leaf and seed tissue synergistically increase biomass and seed number in rice (Oryza sativa ssp. japonica). Funct. Plant Biol. 2016, 43, 1194–1204. [Google Scholar] [CrossRef]
- Greene, T.W.; Kavakli, I.H.; Kahn, M.L.; Okita, T.W. Generation of up-regulated allosteric variants of potato ADP-glucose pyrophosphorylase by reversion genetics. Proc. Natl. Acad. Sci. USA 1998, 95, 10322–10327. [Google Scholar] [CrossRef] [PubMed]
- Salamone, P.R.; Kavakli, I.H.; Slattery, C.J.; Okita, T.W. Directed molecular evolution of ADP-glucose pyrophosphorylase. Proc. Natl. Acad. Sci. USA 2002, 99, 1070–1075. [Google Scholar] [CrossRef] [PubMed]
- Halil Kavakli, I.; Kato, C.; Choi, S.B.; Kim, K.H.; Salamone, P.R.; Ito, H.; Okita, T.W. Generation, characterization, and heterologous expression of wild-type and up-regulated forms of Arabidopsis thaliana leaf ADP-glucose pyrophosphorylase. Planta 2002, 215, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Recondo, E.; Leloir, L.F. Adenosine diphosphate glucose and starch synthesis. Biochem. Biophys. Res. Commun. 1961, 6, 85–88. [Google Scholar] [CrossRef]
- Cuesta-Seijo, J.A.; Nielsen, M.M.; Ruzanski, C.; Krucewicz, K.; Beeren, S.R.; Rydhal, M.G.; Yoshimura, Y.; Striebeck, A.; Motawia, M.S.; Willats, W.G.T.; et al. In vitro biochemical characterization of all barley endosperm starch synthases. Front. Plant Sci. 2016, 6, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, K.; Tagaya, M.; Tanizawa, K.; Fukui, T. Role of the conserved Lys-X-Gly-Gly sequence at the ADP-glucose-binding site in Escherichia coli glycogen synthase. J. Biol. Chem. 1993, 268, 23837–23842. [Google Scholar] [PubMed]
- Furukawa, K.; Tagaya, M.; Preiss, J.; Fukui, Y. Identification of lysine 15 at the active site in E. coli glycogen synthase. J. Biol. Chem. 1990, 265, 2086–2090. [Google Scholar] [PubMed]
- Busi, M.; Palapoli, N.; Valdez, H.; Fornasari, M.; Wayllace, N.; Gomez-Casati, D.; Parisi, G.; Ugalde, R. Functional and structural characterization of the catalytic domain of the starch synthase III from Arabidopsis thaliana. Proteins 2008, 70, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Wanat, J.; Stinard, P.S.; James, M.G.; Myers, A.M. Characterization of DULL1, a maize gene coding for a novel starch synthase. Plant Cell 1998, 10, 399–412. [Google Scholar] [CrossRef] [PubMed]
- De Fekete, M.; Leloir, L.; Cardini, C. Mechanism of starch biosynthesis. Nature 1960, 187, 918–919. [Google Scholar] [CrossRef]
- Nelson, O.E.; Rines, H.W. The enzymatic deficiency in the waxy mutant of maize. Biochem. Biophys. Res. Commun. 1962, 9, 297–300. [Google Scholar] [CrossRef]
- Brink, R.; MacGillivray, J. Segregation for the waxy character in maize pollen and differential development of the male gametophyte. Am. J. Bot. 1924, 11, 465–469. [Google Scholar] [CrossRef]
- Denyer, K.; Waite, D.; Motawia, S.; Møller, B.L.; Smith, A.M. Granule-bound starch synthase I in isolated starch granules elongates malto-oligosaccharides processively. Biochem. J. 1999, 340, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Maruta, N.; Hizukuri, S. Structures of amylose subfractions with different molecular sizes. Carbohydr. Res. 1992, 226, 279–285. [Google Scholar] [CrossRef]
- Hanashiro, I.; Itoh, K.; Kuratomi, Y.; Yamazaki, M.; Igarashi, T.; Matsugasako, J.I.; Takeda, Y. Granule-bound starch synthase I is responsible for biosynthesis of extra-long unit chains of amylopectin in rice. Plant Cell Physiol. 2008, 49, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Hasegawa, H.; Taira, T. The isolation and characterization of a waxy mutant of diploid wheat (Triticum monococcum L.). Plant Sci. 2001, 160, 595–602. [Google Scholar] [CrossRef]
- Tester, R.F.; Morrison, W.R. Swelling and gelatinization of cereal starches. III. Some properties of waxy and normal nonwaxy barley starches. Cereal Chem. 1992, 69, 654–658. [Google Scholar]
- Yamanaka, S.; Nakamura, I.; Watanabe, K.N.; Sato, Y.I. Identification of SNPs in the waxy gene among glutinous rice cultivars and their evolutionary significance during the domestication process of rice. Theor. Appl. Genet. 2004, 108, 1200–1204. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.Y. The function of the Waxy locus in starch synthesis in maize endosperm. Biochem. Genet. 1974, 11, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Shure, M.; Wessler, S.; Fedoroff, N. Molecular identification and isolation of the Waxy locus in maize. Cell 1983, 35, 225–233. [Google Scholar] [CrossRef]
- Bettge, A.D.; Giroux, M.J.; Morris, C.F. Susceptibility of waxy starch granules to mechanical damage. Cereal Chem. 2000, 77, 750–753. [Google Scholar] [CrossRef]
- Yeh, J.Y.; Garwood, D.L.; Shannon, J.C. Characterization of Starch from Maize Endosperm Mutants. Starch Stärke 1981, 33, 222–230. [Google Scholar] [CrossRef]
- Ahuja, G.; Jaiswal, S.; Hucl, P.; Chibbar, R.N. Genome-specific granule-bound starch synthase i (GBSSI) influences starch biochemical and functional characteristics in near-isogenic wheat (Triticum aestivum L.) Lines. J. Agric. Food Chem. 2013, 61, 12129–12138. [Google Scholar] [CrossRef] [PubMed]
- Cooke, D.; Gidley, M.J. Loss of crystalline and molecular order during starch gelatinisation: Origin of the enthalpic transition. Carbohydr. Res. 1992, 227, 103–112. [Google Scholar] [CrossRef]
- Bogracheva, T.Y.; Cairns, P.; Noel, T.R.; Hulleman, S.; Wang, T.L.; Morris, V.J.; Ring, S.G.; Hedley, C.L. Effect of mutant genes at the r, rb, rug3, rug4, rug5 and lam loci on the granular structure and physico-chemical properties of pea seed starch. Carbohydr. Polym. 1999, 39, 303–314. [Google Scholar] [CrossRef]
- Inouchi, N.; Glover, D.V.; Fuwa, H. Chain Length Distribution of Amylopectins of Several Single Mutants and the Normal Counterpart, and Sugary-1 Phytoglycogen in Maize (Zea mays L.). Starch Stärke 1987, 39, 259–266. [Google Scholar] [CrossRef]
- Nakamura, T.; Vrinten, P.; Hayakawa, K.; Ikeda, J. Characterization of a granule-bound starch synthase isoform found in the pericarp of wheat. Plant Physiol. 1998, 118, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Taira, T. A 56-kDa protein is a novel granule-bound starch synthase existing in the pericarps, aleurone layers, and embryos of immature seed in diploid wheat (Triticum monococcum L.). Planta 1998, 207, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Vrinten, P.L.; Nakamura, T. Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues. Plant Physiol. 2000, 122, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Denyer, K.; Sidebottom, C.; Hylton, C.M.; Smith, A.M. Soluble isoforms of starch synthase and starch-branching enzyme also occur within starch granules in developing pea embryos. Plant J. 1993, 4, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Mu-forster, C.; Huang, R.; Powers, J.R.; Harriman, R.W.; Knight, M.; Singletary, C.W.; Keeling, P.; Wasserman, B.P. Physical Association. Plant Physiol. 1995, 111, 821–829. [Google Scholar] [CrossRef]
- Li, H.; Xiao, Q.; Zhang, C.; Du, J.; Li, X.; Huang, H.; Wei, B.; Li, Y.; Yu, G.; Liu, H.; et al. Identification and characterization of transcription factor ZmEREB94 involved in starch synthesis in maize. J. Plant Physiol. 2017, 216, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.; Borthakur, A.; Bornemann, S.; Venail, J.; Denyer, K.; Waite, D.; Fulton, D.; Smith, A.; Martin, C. Specificity of starch synthase isoforms from potato. Eur. J. Biochem. 1999, 266, 724–736. [Google Scholar] [CrossRef] [PubMed]
- Denyer, K.; Clarke, B.; Hylton, C.; Tatge, H.; Smith, A.M. The elongation of amylose and amylopectin chains in isolated starch granules. Plant J. 1996, 10, 1135–1143. [Google Scholar] [CrossRef]
- Clarke, B.R.; Denyer, K.; Jenner, C.F.; Smith, A.M. The relationship between the rate of starch synthesis, the adenosine 5′-diphosphoglucose concentration and the amylose content of starch in developing pea embryos. Planta 1999, 209, 324–329. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.R.; Huang, W.X.; Cai, X.L. Oligomerization of rice granule-bound starch synthase 1 modulates its activity regulation. Plant Sci. 2013, 210, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Grimaud, F.; Rogniaux, H.; James, M.G.; Myers, A.M.; Planchot, V. Proteome and phosphoproteome analysis of starch granule-associated proteins from normal maize and mutants affected in starch biosynthesis. J. Exp. Bot. 2008, 59, 3395–3406. [Google Scholar] [CrossRef] [PubMed]
- Seung, D.; Soyk, S.; Coiro, M.; Maier, B.A.; Eicke, S.; Zeeman, S.C. PROTEIN TARGETING TO STARCH Is Required for Localising GRANULE-BOUND STARCH SYNTHASE to Starch Granules and for Normal Amylose Synthesis in Arabidopsis. PLoS Biol. 2015, 13, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Commuri, P.D.; Keeling, P.L. Chain-length specificities of maize starch synthase I enzyme: Studies of glucan affinity and catalytic properties. Plant J. 2001, 25, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Yoshida, M.; Asakura, N.; Ohdan, T.; Miyao, A.; Hirochika, H.; Nakamura, Y. Function and Characterization of Starch Synthase I Using Mutants in Rice. Plant Physiol. 2006, 140, 1070–1084. [Google Scholar] [CrossRef] [PubMed]
- Imparl-Radosevich, J.M.; Li, P.; Zhang, L.; McKean, A.L.; Keeling, P.L.; Guan, H. Purification and characterization of maize starch synthase I and its truncated forms. Arch. Biochem. Biophys. 1998, 353, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Wilkens, C.; Cuesta-Seijo, J.; Palcic, M.; Svensson, B. Selectivity of the surface binding site (SBS) on barley starch synthase I. Biologia (Bratisl.) 2014, 69, 1118–1121. [Google Scholar] [CrossRef]
- Mcmaugh, S.J.; Thistleton, J.L.; Anschaw, E.; Luo, J.; Konik-Rose, C.; Wang, H.; Huang, M.; Larroque, O.; Regina, A.; Jobling, S.A.; et al. Suppression of starch synthase I expression affects the granule morphology and granule size and fine structure of starch in wheat endosperm. J. Exp. Bot. 2014, 65, 2189–2201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Colleoni, C.; Ratushna, V.; Sirghie-colleoni, M.; James, M.; Myers, A. Molecular characterization demonstrates that the Zea mays gene sugary2 codes for the starch synthase isoform SSIIa. Plant Mol. Biol. 2004, 54, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Imparl-Radosevich, J.; Gameon, J.; McKean, A.; Wetterberg, D.; Keeling, P.; Guan, H. Understanding catalytic properties and functions of maize starch synthase isozymes. J. Appl. Glycosci. 2003, 50, 177–182. [Google Scholar] [CrossRef]
- Cao, H.; Imparl-Radosevich, J.; Guan, H.; Keeling, P.L.; James, M.G.; Myers, A.M. Identification of the soluble starch synthase activities of maize endosperm. Plant Physiol. 1999, 120, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Waters, D.L.E.; Henry, R.J.; Reinke, R.F.; Fitzgerald, M.A. Gelatinization temperature of rice explained by polymorphisms in starch synthase. Plant Biotechnol. J. 2006, 4, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Sakurai, A.; Inaba, Y.; Kimura, K.; Iwasawa, N.; Nagamine, T. The fine structure of amylopectin in endosperm from Asian cultivated rice can be largely classified into two classes. Starch/Staerke 2002, 54, 117–131. [Google Scholar] [CrossRef]
- Nakamura, Y.; Francisco, B., Jr.; Hosaka, Y.; Satoh, A.; Sawada, T.; Kubo, A.; Fujita, N. Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice cultivars. Starch Stärke 2005, 58, 213–227. [Google Scholar]
- Umemoto, T.; Aoki, N. Single-nucleotide polymorphisms in rice starch synthase IIa that alter starch gelatinisation and starch association of the enzyme. Funct. Plant Biol. 2005, 32, 763–768. [Google Scholar] [CrossRef]
- Umemoto, T.; Aoki, N.; Lin, H.; Nakamura, Y.; Inouchi, N.; Sato, Y.; Yano, M.; Hirabayashi, H.; Maruyama, S. Natural variation in rice starch synthase IIa affects enzyme and starch properties. Funct. Plant Biol. 2004, 31, 671–684. [Google Scholar] [CrossRef]
- Umemoto, T.; Terashima, K.; Nakamura, Y.; Satoh, H. Differences in Amylopectin Structure between Two Rice Varieties in Relation to the Effects of Temperature during Grain-Filling. Starch Stärke 1999, 51, 58–62. [Google Scholar] [CrossRef]
- Umemoto, T.; Yano, M.; Satoh, H.; Shomura, A.; Nakamura, Y. Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor. Appl. Genet. 2002, 104, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Morell, M.K.; Kosar-Hashemi, B.; Cmiel, M.; Samuel, M.S.; Chandler, P.; Rahman, S.; Buleon, A.; Batey, I.L.; Li, Z. Barley sex6 mutants lack starch synthase lla activity and contain a starch with novel properties. Plant J. 2003, 34, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Hennen-Bierwagen, T.A.; Liu, F.; Marsh, R.S.; Kim, S.; Gan, Q.; Tetlow, I.J.; Emes, M.J.; James, M.G.; Myers, A.M. Starch Biosynthetic Enzymes from Developing Maize Endosperm Associate in Multisubunit Complexes. Plant Physiol. 2008, 146, 1892–1908. [Google Scholar] [CrossRef] [PubMed]
- Hennen-Bierwagen, T.A.; Lin, Q.; Grimaud, F.; Planchot, V.; Keeling, P.L.; James, M.G.; Myers, A.M. Proteins from Multiple Metabolic Pathways Associate with Starch Biosynthetic Enzymes in High Molecular Weight Complexes: A Model for Regulation of Carbon Allocation in Maize Amyloplasts. Plant Physiol. 2009, 149, 1541–1559. [Google Scholar] [CrossRef] [PubMed]
- Tetlow, I.; Liu, F.; Emes, M. Protein-protein interactions during starch biosynthesis. In Starch Metabolism and Structure; Springer: Tokyo, Japan, 2015; pp. 291–313. [Google Scholar]
- Yamamori, M.; Fujita, S.; Hayakawa, K.; Matsuki, J.; Yasui, T. Genetic elimination of a starch granule protein, SGP-1, of wheat generates an altered starch with apparent high amylose. Theor. Appl. Genet. 2000, 101, 21–29. [Google Scholar] [CrossRef]
- Luo, J.; Ahmed, R.; Kosar-Hashemi, B.; Larroque, O.; Butardo, V.M.; Tanner, G.J.; Colgrave, M.L.; Upadhyaya, N.M.; Tetlow, I.J.; Emes, M.J.; et al. The different effects of starch synthase IIa mutations or variation on endosperm amylose content of barley, wheat and rice are determined by the distribution of starch synthase I and starch branching enzyme IIb between the starch granule and amyloplast. Theor. Appl. Genet. 2015, 128, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Romanova, N.; Lee, E.A.; Ahmed, R.; Evans, M.; Gilbert, E.P.; Morell, M.K.; Emes, M.J.; Tetlow, I.J. Glucan affinity of starch synthase IIa determines binding of starch synthase I and starch-branching enzyme IIb to starch granules. Biochem. J. 2012, 448, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, D.; Du, X.; Wang, H.; Larroque, O.; Jenkins, C.L.D.; Jobling, S.A.; Morell, M.K. The barley amo1 locus is tightly linked to the starch synthase IIIa gene and negatively regulates expression of granule-bound starch synthetic genes. J. Exp. Bot. 2011, 62, 5217–5231. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Terao, T. A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 2004, 220, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Dian, W.; Jiang, H.; Wu, P. Evolution and expression analysis of starch synthase III and IV in rice. J. Exp. Bot. 2005, 56, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Yoshida, M.; Kondo, T.; Saito, K.; Utsumi, Y.; Tokunaga, T.; Nishi, A.; Satoh, H.; Park, J.-H.; Jane, J.-L.; et al. Characterization of SSIIIa-Deficient Mutants of Rice: The Function of SSIIIa and Pleiotropic Effects by SSIIIa Deficiency in the Rice Endosperm. Plant Physiol. 2007, 144, 2009–2023. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Huang, B.; Zhang, M.; Zhang, X.; Rivenbark, J.; Lappe, R.L.; James, M.G.; Myers, A.M.; Hennen-Bierwagen, T.A. Functional Interactions between Starch Synthase III and Isoamylase-Type Starch-Debranching Enzyme in Maize Endosperm. Plant Physiol. 2012, 158, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Mangelsdorf, P.C. The Inheritance of Amylaceous Sugary Endosperm and Its Derivatives in Maize. Genetics 1947, 32, 448–458. [Google Scholar] [PubMed]
- Inouchi, N.; Glover, D.V.; Takaya, T.; Fuwa, H. Development Changes in Fine Structure of Starches of Several Endosperm Mutants of Maize. Starch Stärke 1983, 35, 371–376. [Google Scholar] [CrossRef]
- Ryoo, N.; Yu, C.; Park, C.S.; Baik, M.Y.; Park, I.M.; Cho, M.H.; Bhoo, S.H.; An, G.; Hahn, T.R.; Jeon, J.S. Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep. 2007, 26, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; James, M.; Myers, A. Purification and characterization of soluble starch synthases from maize endosperm. Arch. Biochem. Biophys. 2000, 373, 135–146. [Google Scholar] [CrossRef] [PubMed]
- James, M.G.; Denyer, K.; Myers, A.M. Starch synthesis in the cereal endosperm. Curr. Opin. Plant Biol. 2003, 6, 215–222. [Google Scholar] [CrossRef]
- Szydlowski, N.; Ragel, P.; Raynaud, S.; Lucas, M.M.; Roldan, I.; Montero, M.; Munoz, F.J.; Ovecka, M.; Bahaji, A.; Planchot, V.; et al. Starch Granule Initiation in Arabidopsis Requires the Presence of Either Class IV or Class III Starch Synthases. Plant Cell Online 2009, 21, 2443–2457. [Google Scholar] [CrossRef] [PubMed]
- Crofts, N.; Abe, K.; Aihara, S.; Itoh, R.; Nakamura, Y.; Itoh, K.; Fujita, N. Lack of starch synthase IIIa and high expression of granule-bound starch synthase I synergistically increase the apparent amylose content in rice endosperm. Plant Sci. 2012, 193–194, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Boyer, C.D.; Preiss, J. Evidence for independent genetic control of the multiple forms of maize endosperm branching enzymes and starch synthases. Plant Physiol. 1981, 67, 1141–1145. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; White, P.; Pollak, L.; Jane, J. Characterization of starch structures of 17 maize endosperm mutant genotypes with Oh43 inbred line background. Cereal Chem. 1993, 70, 171–179. [Google Scholar]
- Valdez, H.A.; Busi, M.V.; Wayllace, N.Z.; Parisi, G.; Ugalde, R.A.; Gomez-casati, D.F. Role of the N-Terminal starch-binding domains in the kinetic properties of Starch Synthase III from Arabidopsis thaliana. Biochemistry 2008, 47, 3026–3032. [Google Scholar] [CrossRef] [PubMed]
- Wayllace, N.Z.; Valdez, H.A.; Ugalde, R.A.; Busi, M.V.; Gomez-Casati, D.F. The starch-binding capacity of the noncatalytic SBD2 region and the interaction between the N- and C-terminal domains are involved in the modulation of the activity of starch synthase III from Arabidopsis thaliana: Enzymes and catalysis. FEBS J. 2010, 277, 428–440. [Google Scholar] [CrossRef] [PubMed]
- Leterrier, M.; Holappa, L.D.; Broglie, K.E.; Beckles, D.M. Cloning, characterisation and comparative analysis of a starch synthase IV gene in wheat: Functional and evolutionary implications. BMC Plant Biol. 2008, 8, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roldán, I.; Wattebled, F.; Mercedes Lucas, M.; Delvallé, D.; Planchot, V.; Jiménez, S.; Pérez, R.; Ball, S.; D’Hulst, C.; Mérida, Á. The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J. 2007, 49, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Crumpton-Taylor, M.; Grandison, S.; Png, K.M.Y.; Bushby, A.J.; Smith, A.M. Control of Starch Granule Numbers in Arabidopsis Chloroplasts. Plant Physiol. 2012, 158, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Ragel, P.; Streb, S.; Feil, R.; Sahrawy, M.; Annunziata, M.G.; Lunn, J.E.; Zeeman, S.; Merida, A. Loss of Starch Granule Initiation Has a Deleterious Effect on the Growth of Arabidopsis Plants Due to an Accumulation of ADP-Glucose. Plant Physiol. 2013, 163, 75–85. [Google Scholar] [CrossRef] [PubMed]
- D’Hulst, C.; Mérida, Á. The priming of storage glucan synthesis from bacteria to plants: Current knowledge and new developments. New Phytol. 2010, 188, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Malinova, I.; Mahlow, S.; Alseekh, S.; Orawetz, T.; Fernie, A.R.; Baumann, O.; Steup, M.; Fettke, J. Double Knockout Mutants of Arabidopsis Grown under Normal Conditions Reveal that the Plastidial Phosphorylase Isozyme Participates in Transitory Starch Metabolism. Plant Physiol. 2014, 164, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Malinova, I.; Alseekh, S.; Feil, R.; Fernie, A.R.; Baumann, O.; Schöttler, M.A.; Lunn, J.E.; Fettke, J. Starch Synthase 4 and Plastidal Phosphorylase Differentially Affect Starch Granule Number and Morphology. Plant Physiol. 2017, 174, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Toyosawa, Y.; Kawagoe, Y.; Matsushima, R.; Crofts, N.; Ogawa, M.; Fukuda, M.; Kumamaru, T.; Okazaki, Y.; Kusano, M.; Saito, K.; et al. Deficiency of Starch Synthase IIIa and IVb Alters Starch Granule Morphology from Polyhedral to Spherical in Rice Endosperm. Plant Physiol. 2016, 170, 1255–1270. [Google Scholar] [CrossRef] [PubMed]
- Gámez-Arjona, F.M.; Raynaud, S.; Ragel, P.; Mérida, Á. Starch synthase 4 is located in the thylakoid membrane and interacts with plastoglobule-associated proteins in Arabidopsis. Plant J. 2014, 80, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Raynaud, S.; Ragel, P.; Rojas, T.; Mérida, Á. The N-terminal part of Arabidopsis thaliana starch synthase 4 determines the localization and activity of the enzyme. J. Biol. Chem. 2016, 291, 10759–10771. [Google Scholar] [CrossRef] [PubMed]
- Yun, M.S.; Kawagoe, Y. Septum formation in amyloplasts produces compound granules in the rice endosperm and is regulated by plastid division proteins. Plant Cell Physiol. 2010, 51, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Drummond, G.S.; Smith, E.E.; Whelan, W.J. Purification and Properties of Potato: α-1,4-Glucan 6-Glycosyltransferase (Q-Enzyme). Eur. J. Biochem. 1972, 26, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Hawker, J.S.; Ozbun, J.L.; Ozaki, H.; Greenberg, E.; Preiss, J. Interaction of spinach leaf adenosine diphosphate glucose α-1,4-glucan α-4-glucosyl transferase and α-1,4-glucan, α-1,4-glucan-6-glycosyl transferase in synthesis of branched α-glucan. Arch. Biochem. Biophys. 1974, 160, 530–551. [Google Scholar] [CrossRef]
- Brust, H.; Lehmann, T.; D’Hulst, C.; Fettke, J. Analysis of the functional interaction of arabidopsis starch synthase and branching enzyme isoforms reveals that the cooperative action of SSI and BEs results in glucans with polymodal chain length distribution similar to amylopectin. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wu, A.C.; Go, R.M.; Malouf, J.; Turner, M.S.; Malde, A.K.; Mark, A.E.; Gilbert, R.G. The characterization of modified starch branching enzymes: Toward the control of starch chain-length distributions. PLoS ONE 2015, 10, e0125507. [Google Scholar] [CrossRef] [PubMed]
- Pfister, B.; Lu, K.-J.; Eicke, S.; Feil, R.; Lunn, J.E.; Streb, S.; Zeeman, S.C. Genetic Evidence That Chain Length and Branch Point Distributions Are Linked Determinants of Starch Granule Formation in Arabidopsis. Plant Physiol. 2014, 165, 1457–1474. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, A.; Perez, S. The relationship between internal chain length of amylopectin and crystallinity. Biopolymers 1999, 50, 381–390. [Google Scholar] [CrossRef]
- Boyer, L.; Roussel, X.; Courseaux, A.; Ndjindji, O.M.; Lancelon-Pin, C.; Putaux, J.L.; Tetlow, I.J.; Emes, M.J.; Pontoire, B.; D’Hulst, C.; et al. Expression of Escherichia coli glycogen branching enzyme in an Arabidopsis mutant devoid of endogenous starch branching enzymes induces the synthesis of starch-like polyglucans. Plant Cell Environ. 2016, 39, 1432–1447. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.-J.; Streb, S.; Meier, F.; Pfister, B.; Zeeman, S.C. Molecular genetic analysis of glucan branching enzymes from plants and bacteria in Arabidopsis reveals marked differences in their functions and capacity to mediate starch granule formation. Plant Physiol. 2015, 169, 1638–1655. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, H.M.; Ann MacGregor, E.; Henrissat, B.; Sierks, M.R.; Svensson, B. Starch- and glycogen-debranching and branching enzymes: Prediction of structural features of the catalytic (β/α)8-barrel domain and evolutionary relationship to other amylolytic enzymes. J. Protein Chem. 1993, 12, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Borovsky, D.; Smith, E.E.; Whelan, W.J. On The Mechanism of Amylose Branching by Potato Q-Enzyme. Eur. J. Biochem. 1976, 62, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Borovsky, D.; Smith, E.E.; Whelan, W.J.; French, D.; Kikumoto, S. The mechanism of Q-enzyme action and its influence on the structure of amylopectin. Arch. Biochem. Biophys. 1979, 198, 627–631. [Google Scholar] [CrossRef]
- Abad, M.C.; Binderup, K.; Rios-Steiner, J.; Arni, R.K.; Preiss, J.; Geiger, J.H. The x-ray crystallographic structure of Escherichia coli branching enzyme. J. Biol. Chem. 2002, 277, 42164–42170. [Google Scholar] [CrossRef] [PubMed]
- Devillers, C.H.; Piper, M.E.; Ballicora, M.A.; Preiss, J. Characterization of the branching patterns of glycogen branching enzyme truncated on the N-terminus. Arch. Biochem. Biophys. 2003, 418, 34–38. [Google Scholar] [CrossRef]
- Palomo, M.; Pijning, T.; Booiman, T.; Dobruchowska, J.M.; Van Der Vlist, J.; Kralj, S.; Planas, A.; Loos, K.; Kamerling, J.P.; Dijkstra, B.W.; et al. Thermus thermophilus glycoside hydrolase family 57 branching enzyme: Crystal structure, mechanism of action, and products formed. J. Biol. Chem. 2011, 286, 3520–3530. [Google Scholar] [CrossRef] [PubMed]
- Kuriki, T.; Stewart, D.C.; Chem, J.B.; Kuriki, T.; Stewart, D.C.; Preiss, J. Construction of Chimeric Enzymes out of Maize Endosperm Branching Enzymes I and II: Activity and Properties. J. Biol. Chem. 1997, 272, 28999–29004. [Google Scholar] [CrossRef] [PubMed]
- Tetlow, I.J.; Emes, M.J. A review of starch-branching enzymes and their role in amylopectin biosynthesis. IUBMB Life 2014, 66, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Blauth, S.L.; Kim, K.N.; Klucinec, J.; Shannon, J.C.; Thompson, D.; Guiltinan, M. Identification of mutator insertional mutants of starch-branching enzyme 1 (sbe1) in Zea mays L. Plant Mol. Biol. 2002, 48, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Gouy, M.; Yang, Y.W.; Sharp, P.M.; Li, W.H. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 1989, 86, 6201–6205. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Fisher, D.K.; Kim, K.-N.; Shannon, J.C.; Guiltinan, M.J. Evolutionary conservation and expression patterns of maize starch branching enzyme I and IIb genes suggests isoform specialization. Plant Mol. Biol. 1996, 30, 1223–1232. [Google Scholar] [CrossRef] [PubMed]
- Yamanouchi, H.; Nakamura, Y. Organ specificity of isoforms of starch branching enzyme (Q-enzyme) in rice. Plant Cell Physiol. 1992, 33, 985–991. [Google Scholar] [CrossRef]
- Morell, M.K.; Blennow, A.; Kosar-Hashemi, B.; Samuel, M.S. Differential Expression and Properties of Starch Branching Enzyme Isoforms in Developing Wheat Endosperm. Plant Physiol. 1997, 113, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S. Comparison of Starch-Branching Enzyme Genes Reveals Evolutionary Relationships Among Isoforms. Characterization of a Gene for Starch-Branching Enzyme IIa from the Wheat D Genome Donor Aegilops tauschii. Plant Physiol. 2001, 125, 1314–1324. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Li, P.; Imparl-Radosevich, J.; Preiss, J.; Keeling, P. Comparing the properties of Escherichia coli branching enzyme and maize branching enzyme. Arch. Biochem. Biophys. 1997, 342, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Guan, H.P.; Preiss, J. Branching of amylose by the branching isoenzymes of maize endosperm. Carbohydr. Res. 1993, 240, 253–263. [Google Scholar] [CrossRef]
- Guan, H.P.; Preiss, J. Differentiation of the Properties of the Branching Isozymes from Maize (Zea mays). Plant Physiol. 1993, 102, 1269–1273. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Utsumi, Y.; Sawada, T.; Aihara, S.; Utsumi, C.; Yoshida, M.; Kitamura, S. Characterization of the reactions of starch branching enzymes from rice endosperm. Plant Cell Physiol. 2010, 51, 776–794. [Google Scholar] [CrossRef] [PubMed]
- Regina, A.; Kosar-Hashemi, B.; Li, Z.; Rampling, L.; Cmiel, M.; Gianibelli, M.C.; Konik-Rose, C.; Larroque, O.; Rahman, S.; Morell, M.K. Multiple isoforms of starch branching enzyme-I in wheat: Lack of the major SBE-I isoform does not alter starch phenotype. Funct. Plant Biol. 2004, 31, 591–601. [Google Scholar] [CrossRef]
- Satoh, H.; Nishi, A.; Yamashita, K.; Takemoto, Y.; Tanaka, Y.; Hosaka, Y.; Sakurai, A.; Fujita, N.; Nakamura, Y. Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol. 2003, 133, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Thompson, D.B.; Guiltinan, M.J. Maize starch-branching enzyme isoforms and amylopectin structure. In the absence of starch-branching enzyme IIb, the further absence of starch-branching enzyme Ia leads to increased branching. Plant Physiol. 2004, 136, 3515–3523. [Google Scholar] [CrossRef] [PubMed]
- Tetlow, I.J.; Wait, R.; Lu, Z.; Akkasaeng, R.; Bowsher, C.G.; Esposito, S.; Kosar-Hashemi, B.; Morell, M.K.; Emes, M.J. Protein Phosphorylation in Amyloplasts Regulates Starch Branching Enzyme Activity and Protein—Protein Interactions. Plant Cell 2004, 16, 694–708. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Ahmed, Z.; Lee, E.A.; Donner, E.; Liu, Q.; Ahmed, R.; Morell, M.K.; Emes, M.J.; Tetlow, I.J. Allelic variants of the amylose extender mutation of maize demonstrate phenotypic variation in starch structure resulting from modified protein-protein interactions. J. Exp. Bot. 2012, 63, 1167–1183. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z.; Tetlow, I.J.; Ahmed, R.; Morell, M.K.; Emes, M.J. Protein-protein interactions among enzymes of starch biosynthesis in high-amylose barley genotypes reveal differential roles of heteromeric enzyme complexes in the synthesis of A and B granules. Plant Sci. 2015, 233, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Crofts, N.; Abe, N.; Oitome, N.F.; Matsushima, R.; Hayashi, M.; Tetlow, I.J.; Emes, M.J.; Nakamura, Y.; Fujita, N. Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J. Exp. Bot. 2015, 66, 4469–4482. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Yandeau-Nelson, M.; Thompson, D.B.; Guiltinan, M.J. Deficiency of maize starch-branching enzyme i results in altered starch fine structure, decreased digestibility and reduced coleoptile growth during germination. BMC Plant Biol. 2011, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Fisher, D.K.; Gao, M.; Kim, K.N.; Boyer, C.D.; Guiltinan, M.J. Allelic Analysis of the Maize amylose-extender Locus Suggests That Independent Genes Encode Starch-Branching Enzymes IIa and IIb. Plant Physiol. 1996, 110, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Fisher, D.K.; Kim, K.N.; Shannon, J.C.; Guiltinan, M.J. Independent genetic control of maize starch-branching enzymes IIa and IIb. Isolation and characterization of a Sbe2a cDNA. Plant Physiol. 1997, 114, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Tyler, L.; Bragg, J.N.; Wu, J.; Yang, X.; Tuskan, G.A.; Vogel, J.P. Annotation and comparative analysis of the glycoside hydrolase genes in Brachypodium distachyon. BMC Genom. 2010, 11, 600. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Kawasaki, T.; Shimada, H.; Satoh, H.; Kobayashi, E.; Okumura, S.; Arai, Y.; Baba, T. Alteration of the structural properties of starch components by the lack of an isoform of starch branching enzyme in rice seeds. J. Biol. Chem. 1993, 268, 19084–19091. [Google Scholar] [PubMed]
- Regina, A.; Bird, A.; Topping, D.; Bowden, S.; Freeman, J.; Barsby, T.; Kosar-Hashemi, B.; Li, Z.; Rahman, S.; Morell, M. High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc. Natl. Acad. Sci. USA 2006, 103, 3546–3551. [Google Scholar] [CrossRef] [PubMed]
- Regina, A.; Kosar-Hashemi, B.; Li, Z.; Pedler, A.; Mukai, Y.; Yamamoto, M.; Gale, K.; Sharp, P.J.; Morell, M.K.; Rahman, S. Starch branching enzyme IIb in wheat is expressed at low levels in the endosperm compared to other cereals and encoded at a non-syntenic locus. Planta 2005, 222, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Mu, H.H.; Wasserman, B.P.; Carman, G.M. Identification of the maize amyloplast stromal 112-kD protein as a plastidic starch phosphorylase. Plant Physiol. 2001, 125, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Sathish, P.; Ahlandsberg, S.; Jansson, C. The two genes encoding starch-branching enzymes IIa and IIb are differentially expressed in barley. Plant Physiol. 1998, 118, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Regina, A.; Kosar-Hashemi, B.; Ling, S.; Li, Z.; Rahman, S.; Morell, M. Control of starch branching in barley defined through differential RNAi suppression of starch branching enzyme IIa and IIb. J. Exp. Bot. 2010, 61, 1469–1482. [Google Scholar] [CrossRef] [PubMed]
- Cuesta-Seijo, J.A.; Ruzanski, C.; Krucewicz, K.; Meier, S.; Hägglund, P.; Svensson, B.; Palcic, M.M. Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development. PLoS ONE 2017, 12, e0175488. [Google Scholar] [CrossRef] [PubMed]
- Banks, W.; Greenwood, C.T.; Muir, D. Studies on starches of high amylose-content. Die Stärke 1974, 9, 289–328. [Google Scholar] [CrossRef]
- Blauth, S.L. Identification of Mutator Insertional Mutants of Starch-Branching Enzyme 2a in Corn. Plant Physiol. 2001, 125, 1396–1405. [Google Scholar] [CrossRef] [PubMed]
- Butardo, V.M.; Fitzgerald, M.A.; Bird, A.R.; Gidley, M.J.; Flanagan, B.M.; Larroque, O.; Resurreccion, A.P.; Laidlaw, H.K.C.; Jobling, S.A.; Morell, M.K.; et al. Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA-and hairpin RNA-mediated RNA silencing. J. Exp. Bot. 2011, 62, 4927–4941. [Google Scholar] [CrossRef] [PubMed]
- Klucinec, J.D.; Thompson, D.B. Structure of amylopectins from ae -containing maize starches. Cereal Chem. 2002, 79, 19–23. [Google Scholar] [CrossRef]
- Wellner, N.; Georget, D.M.R.; Parker, M.L.; Morris, V.J. In situ Raman microscopy of starch granule structures in wild type and ae mutant maize kernels. Starch/Staerke 2011, 63, 128–138. [Google Scholar] [CrossRef]
- Shaik, S.S.; Obata, T.; Hebelstrup, K.H.; Schwahn, K.; Fernie, A.R.; Mateiu, R.V.; Blennow, A. Starch granule re-structuring by starch branching enzyme and glucan water dikinase modulation affects caryopsis physiology and metabolism. PLoS ONE 2016, 11, e0149613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slade, A.J.; McGuire, C.; Loeffler, D.; Mullenberg, J.; Skinner, W.; Fazio, G.; Holm, A.; Brandt, K.M.; Steine, M.N.; Goodstal, J.F.; et al. Development of high amylose wheat through TILLING. BMC Plant Biol. 2012, 12, 69. [Google Scholar] [CrossRef] [PubMed]
- Sparla, F.; Falini, G.; Botticella, E.; Pirone, C.; Talamè, V.; Bovina, R.; Salvi, S.; Tuberosa, R.; Sestili, F.; Trost, P. New starch phenotypes produced by TILLING in barley. PLoS ONE 2014, 9, e0107779. [Google Scholar] [CrossRef] [PubMed]
- Hazard, B.; Zhang, X.; Colasuonno, P.; Uauy, C.; Beckles, D.M.; Dubcovsky, J. Induced Mutations in the Starch Branching Enzyme II (SBEII) Genes Increase Amylose and Resistant Starch Content in Durum Wheat. Crop Sci. 2012, 52, 1754–1766. [Google Scholar] [CrossRef] [PubMed]
- Denyer, K.; Hylton, C.M.; Jenner, C.F.; Smith, A.M. Identification of multiple isoforms of soluble and granule-bound starch synthase in developing wheat endosperm. Planta Int. J. Plant Biol. 1995, 196, 256–265. [Google Scholar] [CrossRef]
- Rahman, S.; Kosar-Hashemi, B.; Samuel, M.; Hill, A.; Abbott, D.; Skerritt, J.; Preiss, J.; Appels, R.; Morell, M. The Major Proteins of Wheat Endosperm Starch Granules. Aust. J. Plant Physiol. 1995, 22, 793–803. [Google Scholar] [CrossRef]
- Borén, M.; Larsson, H.; Falk, A.; Jansson, C. The barley starch granule proteome—Internalized granule polypeptides of the mature endosperm. Plant Sci. 2004, 166, 617–626. [Google Scholar] [CrossRef]
- Liu, F.; Makhmoudova, A.; Lee, E.A.; Wait, R.; Emes, M.J.; Tetlow, I.J. The amylose extender mutant of maize conditions novel protein-protein interactions between starch biosynthetic enzymes in amyloplasts. J. Exp. Bot. 2009, 60, 4423–4440. [Google Scholar] [CrossRef] [PubMed]
- Makhmoudova, A.; Williams, D.; Brewer, D.; Massey, S.; Patterson, J.; Silva, A.; Vassall, K.A.; Liu, F.; Subedi, S.; Harauz, G.; et al. Identification of multiple phosphorylation sites on maize endosperm starch branching enzyme IIb, a key enzyme in amylopectin biosynthesis. J. Biol. Chem. 2014, 289, 9233–9246. [Google Scholar] [CrossRef] [PubMed]
- Crofts, N.; Nakamura, Y.; Fujita, N. Critical and speculative review of the roles of multi-protein complexes in starch biosynthesis in cereals. Plant Sci. 2017, 262, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tetlow, I.J.; Beisel, K.G.; Cameron, S.; Makhmoudova, A.; Liu, F.; Bresolin, N.S.; Wait, R.; Morell, M.K.; Emes, M.J. Analysis of Protein Complexes in Wheat Amyloplasts Reveals Functional Interactions among Starch Biosynthetic Enzymes. Plant Physiol. 2008, 146, 1878–1891. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Fujita, N.; Nishi, A.; Satoh, H.; Hosaka, Y.; Ugaki, M.; Kawasaki, S.; Nakamura, Y. The structure of starch can be manipulated by changing the expression levels of starch branching enzyme IIb in rice endosperm. Plant Biotechnol. J. 2004, 2, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Facon, M.; Putaux, J.L.; Dinges, J.R.; Wattebled, F.; D’Hulst, C.; Hennen-Bierwagen, T.A.; Myers, A.M. Function of isoamylase-type starch debranching enzymes ISA1 and ISA2 in the Zea mays leaf. New Phytol. 2013, 200, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Dinges, J.R.; Colleoni, C.; James, M.G.; Myers, A.M. Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism. Plant Cell 2003, 15, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Wattebled, F.; Dong, Y.; Dumez, S.; Delvallé, D.; Planchot, V.; Berbezy, P.; Vyas, D.; Colonna, P.; Chatterjee, M.; Ball, S.; et al. Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol. 2005, 138, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Delatte, T.; Umhang, M.; Trevisan, M.; Eicke, S.; Thorneycroft, D.; Smith, S.M.; Zeeman, S.C. Evidence for distinct mechanisms of starch granule breakdown in plants. J. Biol. Chem. 2006, 281, 12050–12059. [Google Scholar] [CrossRef] [PubMed]
- Yun, M.S.; Umemoto, T.; Kawagoe, Y. Rice debranching enzyme isoamylase3 facilitates starch metabolism and affects plastid morphogenesis. Plant Cell Physiol. 2011, 52, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.; Morris, C. Glycogen in the seed of Zea mays. J. Biol. Chem. 1939, 130, 535–544. [Google Scholar]
- James, M.G.; Robertson, D.S.; Myers, A.M.; James, M.G.; Robertson, D.S.; Myers, A.N. Characterization of the Maize Gene sugary1, a Determinant Characterization in Kernels of Starch Composition in kernels. Plant Cell 2015, 7, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.A.; Jenner, H.; Carrangis, L.; Fahy, B.; Fincher, G.B.; Hylton, C.; Laurie, D.A.; Parker, M.; Waite, D.; Van Wegen, S.; et al. Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J. 2002, 31, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Kubo, A.; Rahman, S.; Utsumi, Y.; Li, Z.; Mukai, Y.; Yamamoto, M.; Ugaki, M.; Harada, K.; Satoh, H.; Konik-Rose, C.; et al. Complementation of sugary-1 phenotype in rice endosperm with the wheat isoamylase1 gene supports a direct role for isoamylase1 in amylopectin biosynthesis. Plant Physiol. 2005, 137, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Dinges, J.R.; Colleoni, C.; Myers, A.M.; James, M.G. Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol. 2001, 125, 1406–1418. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Nakamura, Y.; Li, Z.; Clarke, B.; Fujita, N.; Mukai, M.; Yamamoto, M.; Regina, A.; Tan, Z.; Kawasaki, S.; et al. The sugary-type isoamylase gene from rice and Aegilops tauschii: Characterization and comparison with maize and Arabidopsis. Genome 2003, 46, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, T.H.; Baunsgaard, L.; Blennow, A. Intermediary glucan structures formed during starch granule biosynthesis are enriched in short side chains, a dynamic pulse labeling approach. J. Biol. Chem. 2002, 277, 20249–20255. [Google Scholar] [CrossRef] [PubMed]
- Gidley, M.; Bulpin, P. Crystallisation of malto-oligosaccharides as models of the crystalline forms of starch: Minimum chain-length requirement for the formation of double helices. Carbohydr. Res. 1987, 161, 291–300. [Google Scholar] [CrossRef]
- Ball, S.; Guan, H.; James, M.; Myers, A.; Keeling, P.; Mouille, G.; Colonna, P.; Preiss, J. From Glycogen to Amylopectin: A Model for the Biogenesis of the Plant Starch Granule. Cell 1996, 86, 349–352. [Google Scholar] [CrossRef]
- Mouille, G. Preamylopectin Processing: A Mandatory Step for Starch Biosynthesis in Plants. Plant Cell Online 1996, 8, 1353–1366. [Google Scholar] [CrossRef] [PubMed]
- Sim, L.; Beeren, S.R.; Findinier, J.; Dauville, D.; Ball, S.G.; Henriksen, A.; Palcic, M.M. Crystal structure of the Chlamydomonas starch debranching enzyme isoamylase ISA1 reveals insights into the mechanism of branch trimming and complex assembly. J. Biol. Chem. 2014, 289, 22991–23003. [Google Scholar] [CrossRef] [PubMed]
- Zeeman, S.C.; Umemoto, T.; Lue, W.L.; Au-Yeung, P.; Martin, C.; Smith, A.M.; Chen, J. A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell 1998, 10, 1699–1712. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Hanashiro, I.; Suzuki, S.; Higuchi, T.; Toyosawa, Y.; Utsumi, Y.; Itoh, R.; Aihara, S.; Nakamura, Y. Elongated phytoglycogen chain length in transgenic rice endosperm expressing active starch synthase IIa affects the altered solubility and crystallinity of the storage α-glucan. J. Exp. Bot. 2012, 63, 5859–5872. [Google Scholar] [CrossRef] [PubMed]
- Streb, S.; Delatte, T.; Umhang, M.; Eicke, S.; Schorderet, M.; Reinhardt, D.; Zeeman, S.C. Starch Granule Biosynthesis in Arabidopsis Is Abolished by Removal of All Debranching Enzymes but Restored by the Subsequent Removal of an Endoamylase. Plant Cell Online 2008, 20, 3448–3466. [Google Scholar] [CrossRef] [PubMed]
- Kubo, A.; Fujita, N.; Harada, K.; Matsuda, T.; Satoh, H.; Nakamura, Y. The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol. 1999, 121, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Toyosawa, Y.; Utsumi, Y.; Higuchi, T.; Hanashiro, I.; Ikegami, A.; Akuzawa, S.; Yoshida, M.; Mori, A.; Inomata, K.; et al. Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa L.) and the function of PUL on starch biosynthesis in the developing rice endosperm. J. Exp. Bot. 2009, 60, 1009–1023. [Google Scholar] [CrossRef] [PubMed]
- Delatte, T.; Trevisan, M.; Parker, M.L.; Zeeman, S.C. Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J. 2005, 41, 815–830. [Google Scholar] [CrossRef] [PubMed]
- Kawagoe, Y.; Kubo, A.; Satoh, H.; Takaiwa, F.; Nakamura, Y. Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm. Plant J. 2005, 42, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, M.; Pfister, B.; Fulton, D.; Bischof, S.; Delatte, T.; Eicke, S.; Stettler, M.; Smith, S.M.; Streb, S.; Zeeman, S.C. The Heteromultimeric Debranching Enzyme Involved in Starch Synthesis in Arabidopsis Requires Both Isoamylase1 and Isoamylase2 Subunits for Complex Stability and Activity. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Facon, M.; Lin, Q.; Azzaz, A.M.; Hennen-Bierwagen, T.A.; Myers, A.M.; Putaux, J.-L.; Roussel, X.; D’Hulst, C.; Wattebled, F. Distinct Functional Properties of Isoamylase-Type Starch Debranching Enzymes in Monocot and Dicot Leaves. Plant Physiol. 2013, 163, 1363–1375. [Google Scholar] [CrossRef] [PubMed]
- Hussain, H.; Mant, A.; Seale, R.; Zeeman, S.; Hinchliffe, E.; Edwards, A.; Hylton, C.; Bornemann, S.; Smith, A.M.; Martin, C.; et al. Three isoforms of isoamylase contribute different catalytic properties for the debranching of potato glucans. Plant Cell 2003, 15, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Kubo, A.; Colleoni, C.; Dinges, J.R.; Lin, Q.; Lappe, R.R.; Rivenbark, J.G.; Meyer, A.J.; Ball, S.G.; James, M.G.; Hennen-Bierwagen, T.A.; et al. Functions of Heteromeric and Homomeric Isoamylase-Type Starch-Debranching Enzymes in Developing Maize Endosperm. Plant Physiol. 2010, 153, 956–969. [Google Scholar] [CrossRef] [PubMed]
- Utsumi, Y.; Utsumi, C.; Sawada, T.; Fujita, N.; Nakamura, Y. Functional Diversity of Isoamylase Oligomers: The ISA1 Homo-Oligomer Is Essential for Amylopectin Biosynthesis in Rice Endosperm. Plant Physiol. 2011, 156, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Utsumi, Y.; Nakamura, Y. Structural and enzymatic characterization of the isoamylase1 homo-oligomer and the isoamylase1-isoamylase2 hetero-oligomer from rice endosperm. Planta 2006, 225, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Hanes, C. The reversible formation of starch from glucose-1-phosphate catalysed by potato phosphorylase. Proc. R. Soc. Lond. B Biol. Sci. 1940, 129, 174–208. [Google Scholar] [CrossRef]
- Dauvillée, D.; Chochois, V.; Steup, M.; Haebel, S.; Eckermann, N.; Ritte, G.; Ral, J.P.; Colleoni, C.; Hicks, G.; Wattebled, F.; et al. Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J. 2006, 48, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Burr, B.; Nelson, O.E.; Phosphorylase, M. Maize α-Glucan Phosphorylase. Eur. J. Biochem. 1975, 546, 539–546. [Google Scholar] [CrossRef]
- Yang, Y.; Steup, M. Polysaccharide Fraction from Higher Plants which Strongly Interacts with the Cytosolic Phosphorylase Isozyme: I. Isolation and Characterization. Plant Physiol. 1990, 94, 960–969. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.Y.; Salamini, F.; Nelson, O.E. Enzymes of Carbohydrate Metabolism in the Developing Endosperm of Maize. Plant Physiol. 1970, 46, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.E.; Kosar-Hashemi, B.; Li, Z.; Howitt, C.A.; Larroque, O.; Flanagan, B.; Morell, M.K.; Rahman, S. Characterization of starch phosphorylases in barley grains. J. Sci. Food Agric. 2013, 93, 2137–2145. [Google Scholar] [CrossRef] [PubMed]
- Satoh, H.; Shibahara, K.; Tokunaga, T.; Nishi, A.; Tasaki, M.; Hwang, S.-K.; Okita, T.W.; Kaneko, N.; Fujita, N.; Yoshida, M.; et al. Mutation of the Plastidial α-Glucan Phosphorylase Gene in Rice Affects the Synthesis and Structure of Starch in the Endosperm. Plant Cell Online 2008, 20, 1833–1849. [Google Scholar] [CrossRef] [PubMed]
- Sonnewald, U.; Basner, A.; Greve, B.; Steup, M. A second L-type isozyme of potato glucan phosphorylase: Cloning, antisense inhibition and expression analysis. Plant Mol. Biol. 1995, 27, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Zeeman, S.C.; Thorneycroft, D.; Schupp, N.; Chapple, A.; Weck, M.; Dunstan, H.; Haldimann, P.; Bechtold, N.; Smith, A.M.; Smith, S.M. Plastidial alpha-glucan phosphorylase is not required for starch degradation in Arabidopsis leaves but has a role in the tolerance of abiotic stress. Plant Physiol. 2004, 135, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Tetlow, I.J.; Blissett, K.J.; Emes, M.J. Metabolite pools during starch synthesis and carbohydrate oxidation in amyloplasts isolated from wheat endosperm. Planta 1998, 204, 100–108. [Google Scholar] [CrossRef]
- Hwang, S.K.; Nishi, A.; Satoh, H.; Okita, T.W. Rice endosperm-specific plastidial α-glucan phosphorylase is important for synthesis of short-chain malto-oligosaccharides. Arch. Biochem. Biophys. 2010, 495, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Radchuk, V.V.; Borisjuk, L.; Sreenivasulu, N.; Merx, K.; Mock, H.-P.; Rolletschek, H.; Wobus, U.; Weschke, W. Spatiotemporal Profiling of Starch Biosynthesis and Degradation in the Developing Barley Grain. Plant Physiol. 2009, 150, 190–204. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-López, M.; Baroja-Fernández, E.; Zandueta-Criado, A.; Pozueta-Romero, J. Adenosine diphosphate glucose pyrophosphatase: A plastidial phosphodiesterase that prevents starch biosynthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 8705–8710. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Ono, M.; Utsumi, C.; Steup, M. Functional interaction between plastidial starch phosphorylase and starch branching enzymes from rice during the synthesis of branched maltodextrins. Plant Cell Physiol. 2012, 53, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Subasinghe, R.M.; Liu, F.; Polack, U.C.; Lee, E.A.; Emes, M.J.; Tetlow, I.J. Multimeric states of starch phosphorylase determine protein-protein interactions with starch biosynthetic enzymes in amyloplasts. Plant Physiol. Biochem. 2014, 83, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Fukui, T. The complete amino acid sequence of potato alpha-glucan phosphorylase. J. Biol. Chem. 1986, 261, 8230–8236. [Google Scholar] [PubMed]
- Albrecht, T.; Greve, B.; Pusch, K.; Kossmann, J.; Buchner, P.; Wobus, U.; Steup, M. Homodimers and heterodimers of Pho1-type phosphorylase isoforms in Solanum tuberosum L. as revealed by sequence-specific antibodies. Eur. J. Biochem. 1998, 251, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Buchbinder, J.L.; Rath, V.L.; Fletterick, R.J. Structural relationships among regulated and unregulated phosphorylases. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 191–209. [Google Scholar] [CrossRef] [PubMed]
- Takaha, T.; Yanase, M.; Okada, S.; Smith, S.M. Disproportionating enzyme (4-α-glucanotransferase; EC 2.4.1.25) of potato. Purification, molecular cloning, and potential role in starch metabolism. J. Biol. Chem. 1993, 268, 1391–1396. [Google Scholar] [PubMed]
- Colleoni, C.; Dauville, D.; Mouille, G.; Morell, M.; Samuel, M.; Slomiany, M.C.; Li nard, L.; Wattebled, F.; d’Hulst, C.; Ball, S. Biochemical characterization of the Chlamydomonas reinhardtii alpha-1,4 glucanotransferase supports a direct function in amylopectin biosynthesis. Plant Physiol. 1999, 120, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Colleoni, C.; Dauville, D.; Mouille, G.; Bulon, A.; Gallant, D.; Bouchet, B.; Morell, M.; Samuel, M.; Delrue, B.; d’Hulst, C.; et al. Genetic and biochemical evidence for the involvement of alpha-1,4 glucanotransferases in amylopectin synthesis. Plant Physiol. 1999, 120, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.K.; Koper, K.; Satoh, H.; Okita, T.W. Rice endosperm starch phosphorylase (Pho1) assembles with disproportionating enzyme (Dpe1) to form a protein complex that enhances synthesis of malto-oligosaccharides. J. Biol. Chem. 2016, 291, 19994–20007. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.P.; Preiss, J. Characterization of d-Enzyme (4-alpha-Glucanotransferase) in Arabidopsis Leaf. Plant Physiol. 1988, 86, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.P.M.; Schwacke, R.; Flügge, U.-I. Solute Transporters of the Plastid Envelope Membrane. Annu. Rev. Plant Biol. 2005, 56, 133–164. [Google Scholar] [CrossRef] [PubMed]
- Niittyla, T. A Previously Unknown Maltose Transporter Essential for Starch Degradation in Leaves. Science 2004, 303, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Critchley, J.H.; Zeeman, S.C.; Takaha, T.; Smith, A.M.; Smith, S.M. A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J. 2001, 26, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Bresolin, N.S.; Li, Z.; Kosar-Hashemi, B.; Tetlow, I.J.; Chatterjee, M.; Rahman, S.; Morell, M.K.; Howitt, C.A. Characterisation of disproportionating enzyme from wheat endosperm. Planta 2006, 224, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Zhang, D.; Liu, J.; Liu, Q.Q.; Liu, H.; Tian, L.; Jiang, L.; Qule, Q. Plastidial Disproportionating Enzyme Participates in Starch Synthesis in Rice Endosperm by Transferring Maltooligosyl Groups from Amylose and Amylopectin to Amylopectin. Plant Physiol. 2015, 169, 2496–2512. [Google Scholar] [CrossRef] [PubMed]
- Hizukuri, S.; Tabata, S.; Kagoshima; Nikuni, Z. Studies on Starch Phosphate Part 1. Estimation of glucose-6-phosphate residues in starch and the presence of other bound phosphate(s). Starch Stärke 1970, 22, 338–343. [Google Scholar] [CrossRef]
- Ritte, G.; Lloyd, J.R.; Eckermann, N.; Rottmann, A.; Kossmann, J.; Steup, M. The starch-related R1 protein is an alpha-glucan, water dikinase. Proc. Natl. Acad. Sci. USA 2002, 99, 7166–7171. [Google Scholar] [CrossRef] [PubMed]
- Ritte, G.; Heydenreich, M.; Mahlow, S.; Haebel, S.; Kötting, O.; Steup, M. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 2006, 580, 4872–4876. [Google Scholar] [CrossRef] [PubMed]
- Baunsgaard, L.; Lütken, H.; Mikkelsen, R.; Glaring, M.A.; Pham, T.T.; Blennow, A. A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated α-glucans and is involved in starch degradation in Arabidopsis. Plant J. 2005, 41, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Blennow, A.; Engelsen, S.B.; Munck, L.; Møller, B.L. Starch molecular structure and phosphorylation investigated by a combined chromatographic and chemometric approach. Carbohydr. Polym. 2000, 41, 163–174. [Google Scholar] [CrossRef]
- Kotting, O.; Santelia, D.; Edner, C.; Eicke, S.; Marthaler, T.; Gentry, M.S.; Comparot-Moss, S.; Chen, J.; Smith, A.M.; Steup, M.; et al. STARCH-EXCESS4 Is a Laforin-Like Phosphoglucan Phosphatase Required for Starch Degradation in Arabidopsis thaliana. Plant Cell Online 2009, 21, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Ral, J.P.; Bowerman, A.F.; Li, Z.; Sirault, X.; Furbank, R.; Pritchard, J.R.; Bloemsma, M.; Cavanagh, C.R.; Howitt, C.A.; Morell, M.K. Down-regulation of Glucan, Water-Dikinase activity in wheat endosperm increases vegetative biomass and yield. Plant Biotechnol. J. 2012, 10, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sun, X.; Zhou, X.; Hebelstrup, K.H.; Blennow, A.; Bao, J. Highly phosphorylated functionalized rice starch produced by transgenic rice expressing the potato GWD1 gene. Sci. Rep. 2017, 7, 3339. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, G.R.; Creek, J.A.; Runt, J. Spherulitic crystallization in starch as a model for starch granule initiation. Biomacromolecules 2005, 6, 1547–1554. [Google Scholar] [CrossRef] [PubMed]
- Puteaux, J.; Potocki-Veronese, G.; Remaud-Simeon, M.; Buléon, A. Alpha-D-glucan-based dendritic nanoparticles prepared by in vitro enzymatic chain extension of glycogen. Biomacromolecules 2006, 7, 1720–1728. [Google Scholar] [CrossRef] [PubMed]
- Fannon, J.E.; Hauber, R.J.; Bemiller, J.N. Surface pores of starch granules. Cereal Chem. 1992, 69, 284–288. [Google Scholar]
- Fannon, J.E.; Gray, J.A.; Gunawan, N.; Huber, K.C.; BeMiller, J.N. Heterogeneity of starch granules and the effect of granule channelization on starch modification. Cellulose 2004, 11, 247–254. [Google Scholar] [CrossRef]
- Glaring, M.A.; Koch, C.B.; Blennow, A. Genotype-specific spatial distribution of starch molecules in the starch granule: A combined CLSM and SEM approach. Biomacromolecules 2006, 7, 2310–2320. [Google Scholar] [CrossRef] [PubMed]
- Huber, K.C.; BeMiller, J.N. Visualization of channels and cavities of corn and sorghum starch granules. Cereal Chem. 1997, 74, 537–541. [Google Scholar] [CrossRef]
- Benmoussa, M.; Hamaker, B.R.; Huang, C.P.; Sherman, D.M.; Weil, C.F.; BeMiller, J.N. Elucidation of maize endosperm starch granule channel proteins and evidence for plastoskeletal structures in maize endosperm amyloplasts. J. Cereal Sci. 2010, 52, 22–29. [Google Scholar] [CrossRef]
- Jane, J.-L.; Kasemsuwan, T.; Leas, S.; Zobel, H.; Robyt, J.F. Anthology of Starch Granule Morphology by Scanning Electron Microscopy. Starch Stärke 1994, 46, 121–129. [Google Scholar] [CrossRef]
- Ellis, R.P.; Cochrane, M.P.; Dale, M.F.B.; Duþus, C.M.; Lynn, A.; Morrison, I.M.; Prentice, R.D.M.; Swanston, J.S.; Tiller, S.A. Starch Production and Industrial Use. J. Sci. Food Agric. 1998, 77, 289–311. [Google Scholar] [CrossRef]
- Lindeboom, N.; Chang, P.R.; Tyler, R.T. Analytical, Biochemical and Physicochemical Aspects of Starch Granule Size, with Emphasis on Small Granule Starches: A Review. Starch Stärke 2004, 56, 89–99. [Google Scholar] [CrossRef]
- Jobling, S. Improving starch for food and industrial applications. Curr. Opin. Plant Biol. 2004, 7, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Ai, Y.; Gong, L.; Reed, M.; Huang, J.; Zhang, Y.; Jane, J.L. Characterization of starch from bamboo seeds. Starch/Staerke 2016, 68, 131–139. [Google Scholar] [CrossRef]
- Buléon, A.; Colonna, P.; Planchot, V.; Ball, S. Starch granules: Structure and biosynthesis. Int. J. Biol. Macromol. 1998, 23, 85–112. [Google Scholar] [CrossRef]
- Chen, G.; Zhu, J.; Zhou, J.; Subburaj, S.; Zhang, M.; Han, C.; Hao, P.; Li, X.; Yan, Y. Dynamic development of starch granules and the regulation of starch biosynthesis in Brachypodium distachyon: Comparison with common wheat and Aegilops peregrina. BMC Plant Biol. 2014, 14, 198. [Google Scholar] [CrossRef] [PubMed]
- Meredith, P. Large and Small Starch Granules in Wheat—Are They Really Different? Starch Stärke 1981, 33, 40–44. [Google Scholar] [CrossRef]
- May, L.; Buttrose, M. Physiology of cereal grain. II. Starch granule formation in the developing barley kernel. Aust. J. Biol. Sci. 1959, 12, 146–159. [Google Scholar] [CrossRef]
- Bechtel, D.B.; Zayas, I.; Kaleikau, L.; Pomeranz, Y. Size-distribution of wheat starch granules during endosperm development. Cereal Chem. 1990, 67, 59–63. [Google Scholar]
- Shapter, F.M.; Henry, R.J.; Lee, L.S. Endosperm and starch granule morphology in wild cereal relatives. Plant Genet. Resour. 2008, 6, 85–97. [Google Scholar] [CrossRef]
- Shapter, F.M.; Eggler, P.; Lee, L.S.; Henry, R.J. Variation in Granule Bound Starch Synthase I (GBSSI) loci amongst Australian wild cereal relatives (Poaceae). J. Cereal Sci. 2009, 49, 4–11. [Google Scholar] [CrossRef]
- Tateoka, T. Starch grains of endosperm in grass systematics. Bot. Mag. Tokyo 1962, 75, 377–383. [Google Scholar] [CrossRef]
- Draper, J.; Mur, L.A.J.; Jenkins, G.; Ghosh-Biswas, G.C.; Bablak, P.; Hasterok, R.; Routledge, A.P.M. Brachypodium distachyon. A New Model System for Functional Genomics in Grasses1. Plant Physiol. 2001, 127, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Tanackovic, V.; Svensson, J.T.; Jensen, S.L.; Buléon, A.; Blennow, A. The deposition and characterization of starch in Brachypodium distachyon. J. Exp. Bot. 2014, 65, 5179–5192. [Google Scholar] [CrossRef] [PubMed]
- Evers, A.D. The Size Distriubution Among Starch Granules in Wheat Endosperm. Starch Stärke 1973, 25, 304–308. [Google Scholar] [CrossRef]
- Parker, M.L. The relationship between A-type and B-type starch granules in the developing endosperm of wheat. J. Cereal Sci. 1985, 3, 271–278. [Google Scholar] [CrossRef]
- Dengate, H.; Meredith, P. Variation in size distribution of starch granules from wheat grain. J. Cereal Sci. 1984, 2, 83–90. [Google Scholar] [CrossRef]
- Wilson, J.; Bechtel, D.; Todd, T.; Seib, P. Measurement of wheat starch granule size distribution using image analysis and laser diffraction technology. Cereal Chem. 2006, 83, 259–268. [Google Scholar] [CrossRef]
- Stoddard, F.L.; Sarker, R. Characterization of starch in Aegilops species. Cereal Chem. 2000, 77, 445–447. [Google Scholar] [CrossRef]
- Wei, C.; Zhang, J.; Chen, Y.; Zhou, W.; Xu, B.; Wang, Y.; Chen, J. Physicochemical properties and development of wheat large and small starch granules during endosperm development. Acta Physiol. Plant. 2010, 32, 905–916. [Google Scholar] [CrossRef]
- Salman, H.; Blazek, J.; Lopez-Rubio, A.; Gilbert, E.P.; Hanley, T.; Copeland, L. Structure-function relationships in A and B granules from wheat starches of similar amylose content. Carbohydr. Polym. 2009, 75, 420–427. [Google Scholar] [CrossRef]
- Ahmed, Z.; Tetlow, I.J.; Falk, D.E.; Liu, Q.; Emes, M.J. Resistant starch content is related to granule size in barley. Cereal Chem. 2016, 93, 618–630. [Google Scholar] [CrossRef]
- Langeveld, S.M.; van Wijk, R.; Stuurman, N.; Kijne, J.W.; de Pater, S. B-type granule containing protrusions and interconnections between amyloplasts in developing wheat endosperm revealed by transmission electron microscopy and GFP expression. J. Exp. Bot. 2000, 51, 1357–1361. [Google Scholar] [CrossRef] [PubMed]
- Bechtel, D.B.; Wilson, J.D. Amyloplast formation and starch granule development in hard red winter wheat. Cereal Chem. 2003, 80, 175–183. [Google Scholar] [CrossRef]
- Stahl, Y.; Coates, S.; Bryce, J.H.; Morris, P.C. Antisense downregulation of the barley limit dextrinase inhibitor modulates starch granule size distribution, starch composition and amylopectin structure. Plant J. 2004, 39, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Howard, T.; Rejab, N.A.; Griffiths, S.; Leigh, F.; Leverington-Waite, M.; Simmonds, J.; Uauy, C.; Trafford, K. Identification of a major QTL controlling the content of B-type starch granules in Aegilops. J. Exp. Bot. 2011, 62, 2217–2228. [Google Scholar] [CrossRef] [PubMed]
- Chia, T.; Adamski, N.M.; Saccomanno, B.; Greenland, A.; Nash, A.; Uauy, C.; Trafford, K.; Lunn, J. Transfer of a starch phenotype from wild wheat to bread wheat by deletion of a locus controlling B-type starch granule content. J. Exp. Bot. 2017. [Google Scholar] [CrossRef] [PubMed]
- De Pater, S.; Caspers, M.; Kottenhagen, M.; Meima, H.; Ter Stege, R.; De Vetten, N. Manipulation of starch granule size distribution in potato tubers by modulation of plastid division. Plant Biotechnol. J. 2006, 4, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Wang, Y.; Liu, F.; Ren, Y.; Zhou, K.; Lv, J.; Zheng, M.; Zhao, S.; Zhang, L.; Wang, C.; et al. FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J. 2014, 77, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ren, Y.; Lu, B.; Yang, C.; Feng, Z.; Liu, Z.; Chen, J.; Ma, W.; Wang, Y.; Yu, X.; et al. FLOURY ENDOSPERM7 encodes a regulator of starch synthesis and amyloplast development essential for peripheral endosperm development in rice. J. Exp. Bot. 2016, 67, 633–647. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, R.; Maekawa, M.; Kusano, M.; Kondo, H.; Fujita, N.; Kawagoe, Y.; Sakamoto, W. Amyloplast-Localized SUBSTANDARD STARCH GRAIN4 Protein Influences the Size of Starch Grains in Rice Endosperm. Plant Physiol. 2014, 164, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Hebelstrup, K.H.; Sagnelli, D.; Blennow, A. The future of starch bioengineering: GM microorganisms or GM plants? Front. Plant Sci. 2015, 6, 247. [Google Scholar] [CrossRef] [PubMed]
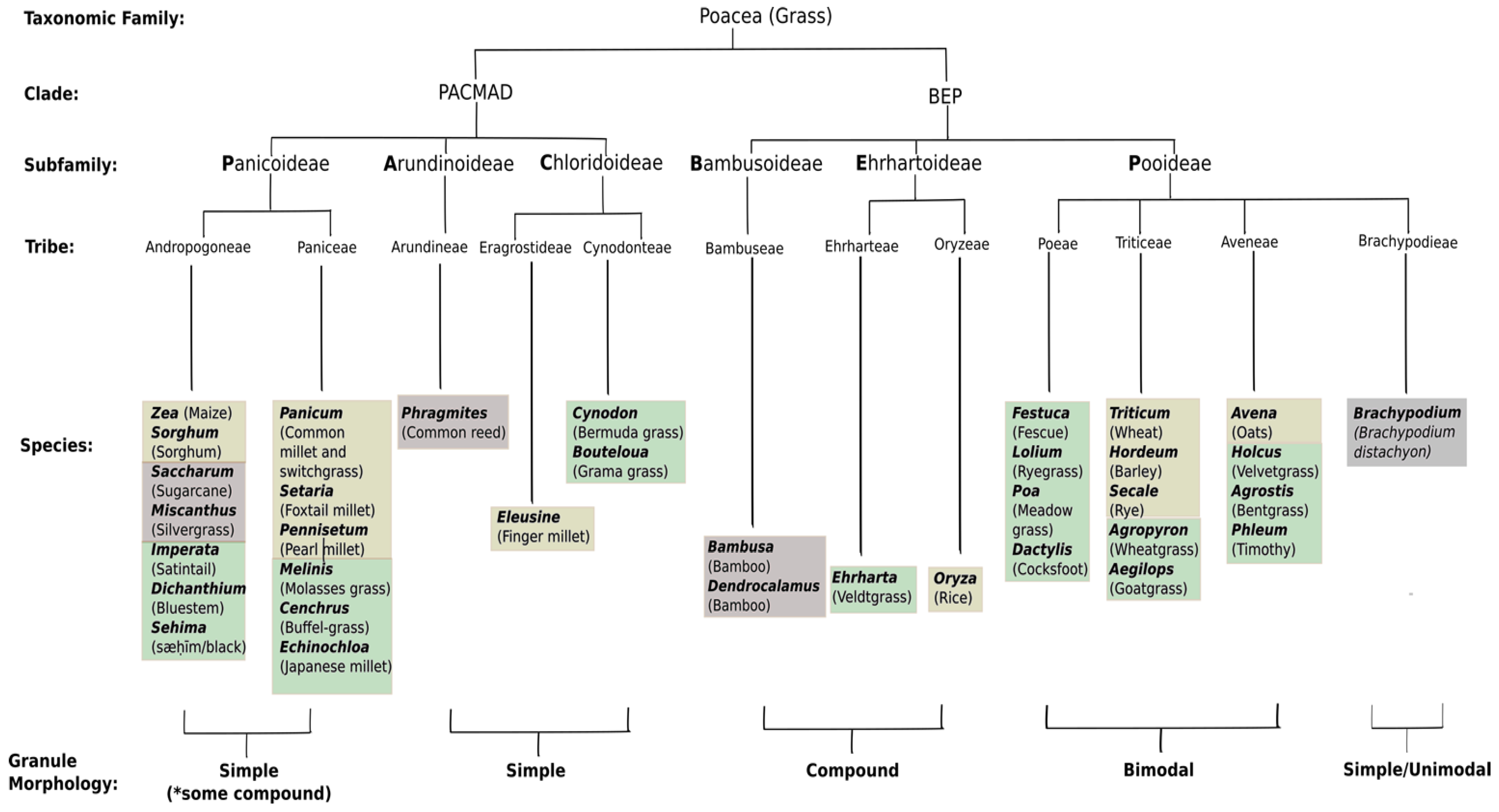
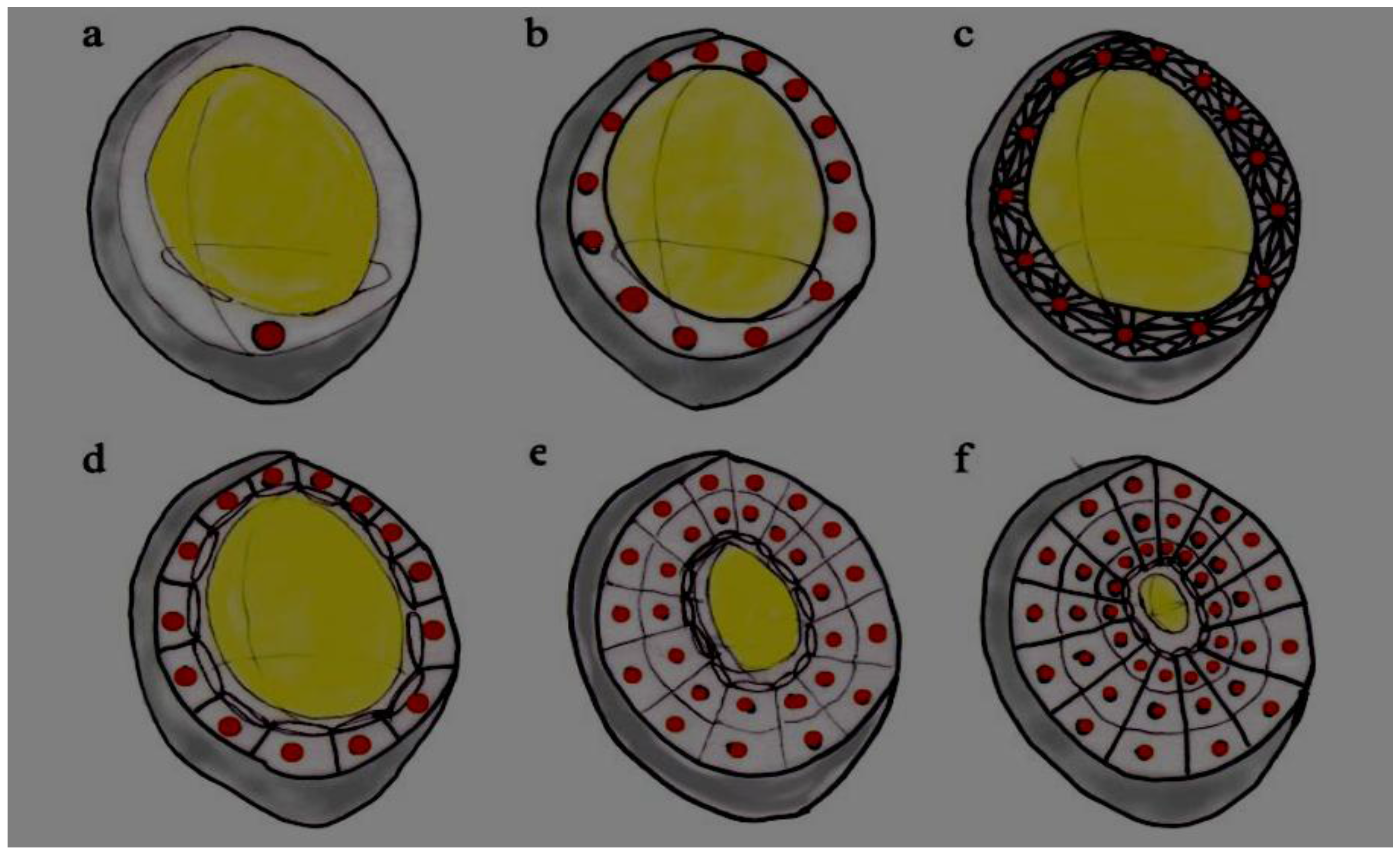
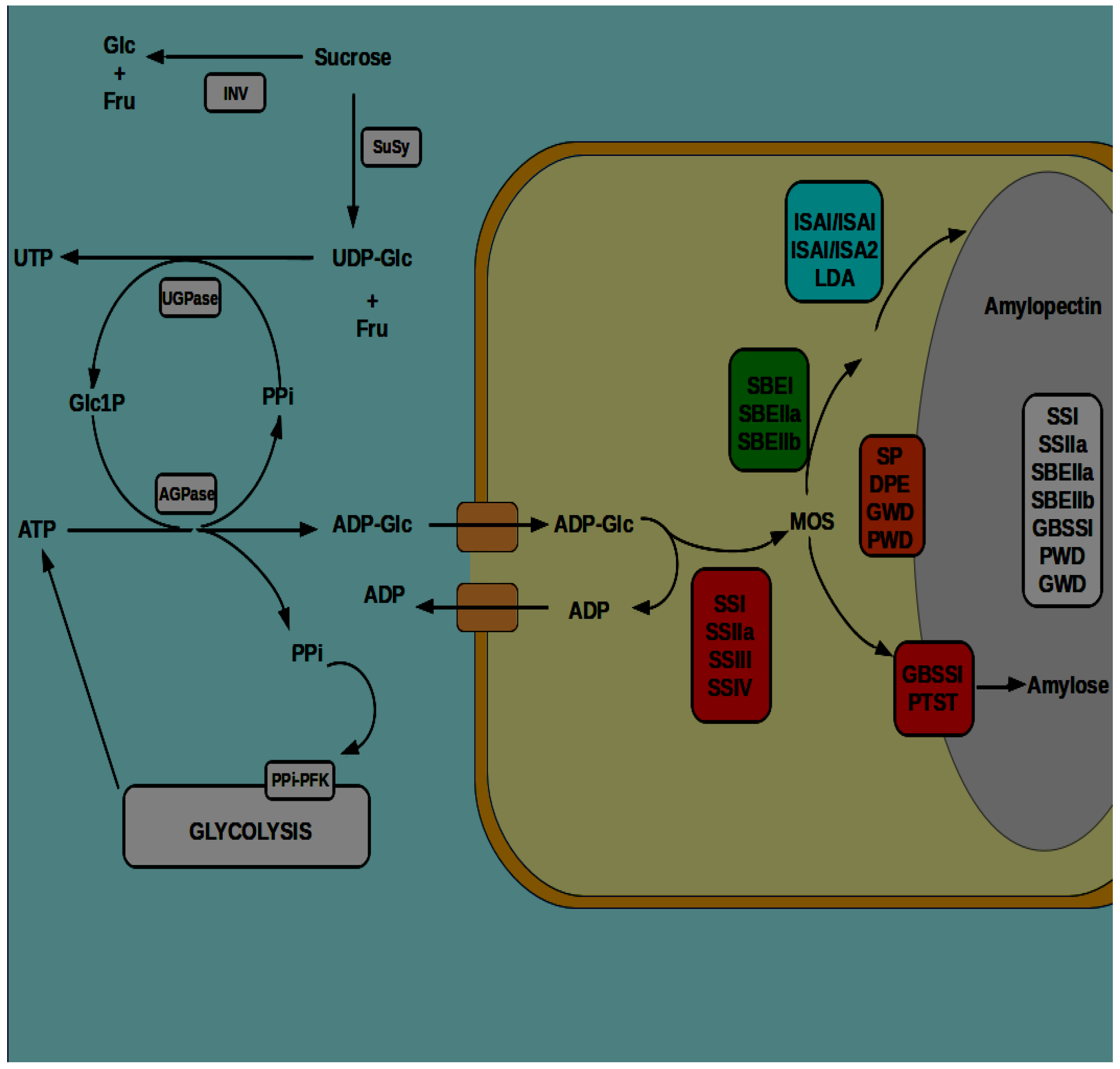
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tetlow, I.J.; Emes, M.J. Starch Biosynthesis in the Developing Endosperms of Grasses and Cereals. Agronomy 2017, 7, 81. https://doi.org/10.3390/agronomy7040081
Tetlow IJ, Emes MJ. Starch Biosynthesis in the Developing Endosperms of Grasses and Cereals. Agronomy. 2017; 7(4):81. https://doi.org/10.3390/agronomy7040081
Chicago/Turabian StyleTetlow, Ian J., and Michael J. Emes. 2017. "Starch Biosynthesis in the Developing Endosperms of Grasses and Cereals" Agronomy 7, no. 4: 81. https://doi.org/10.3390/agronomy7040081





