Kin Selection in the RNA World
Abstract
:1. Introduction
2. Results
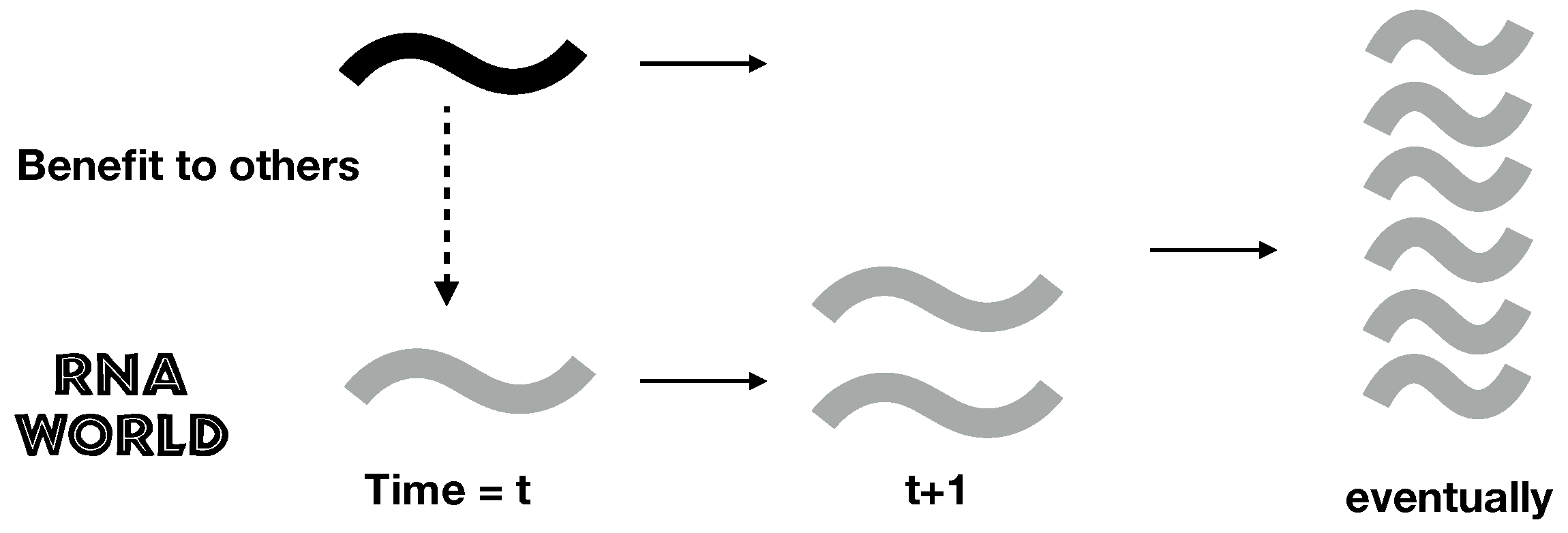
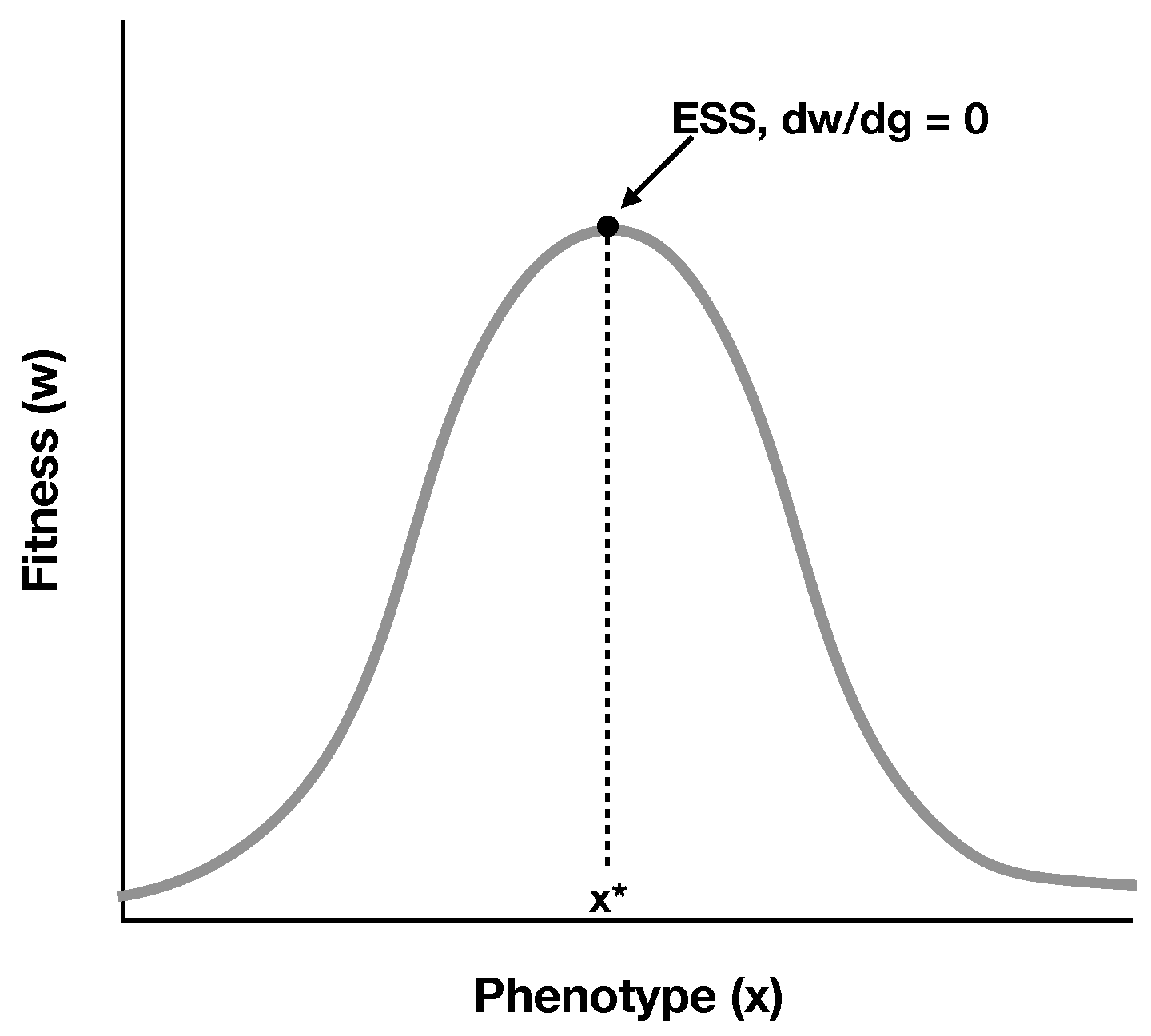
2.1. A Simple Model of RNA Cooperation
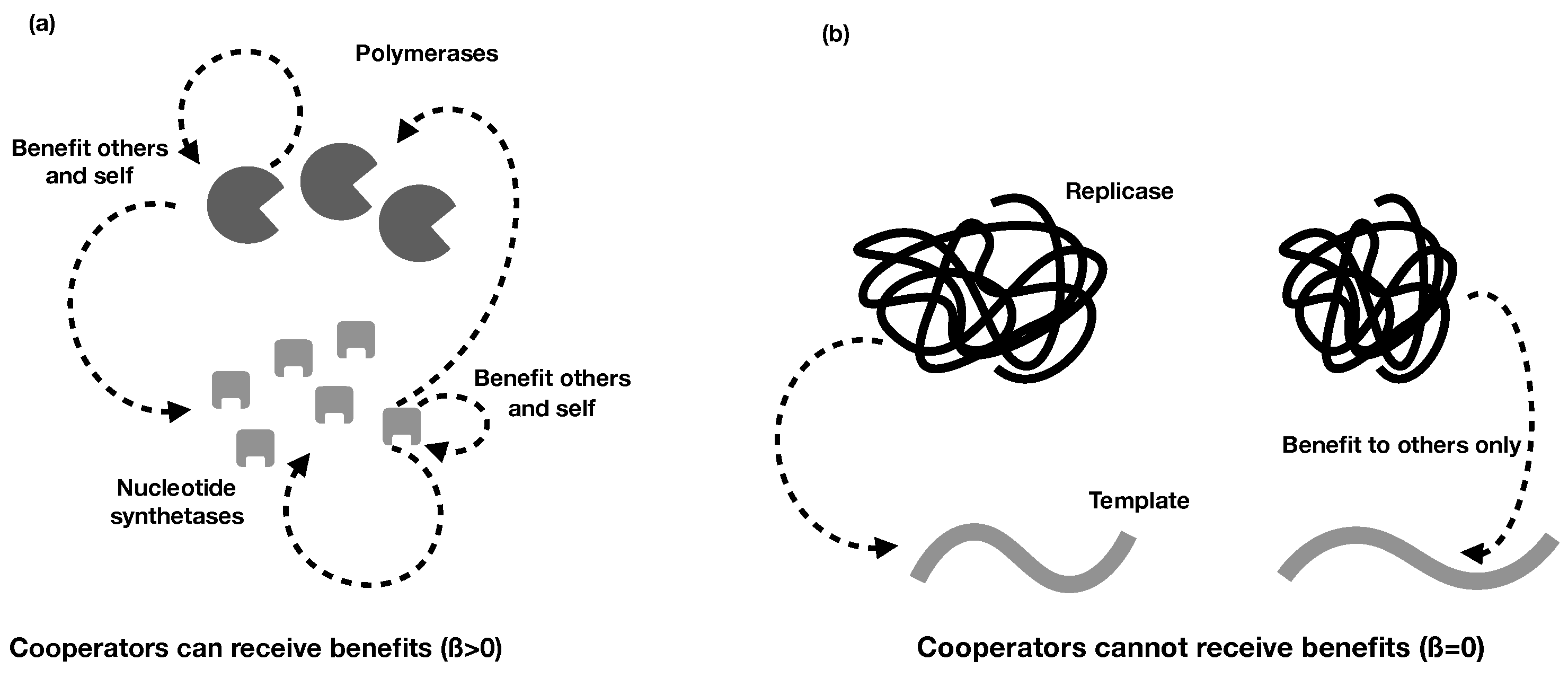
2.2. Different Types of RNA Cooperation
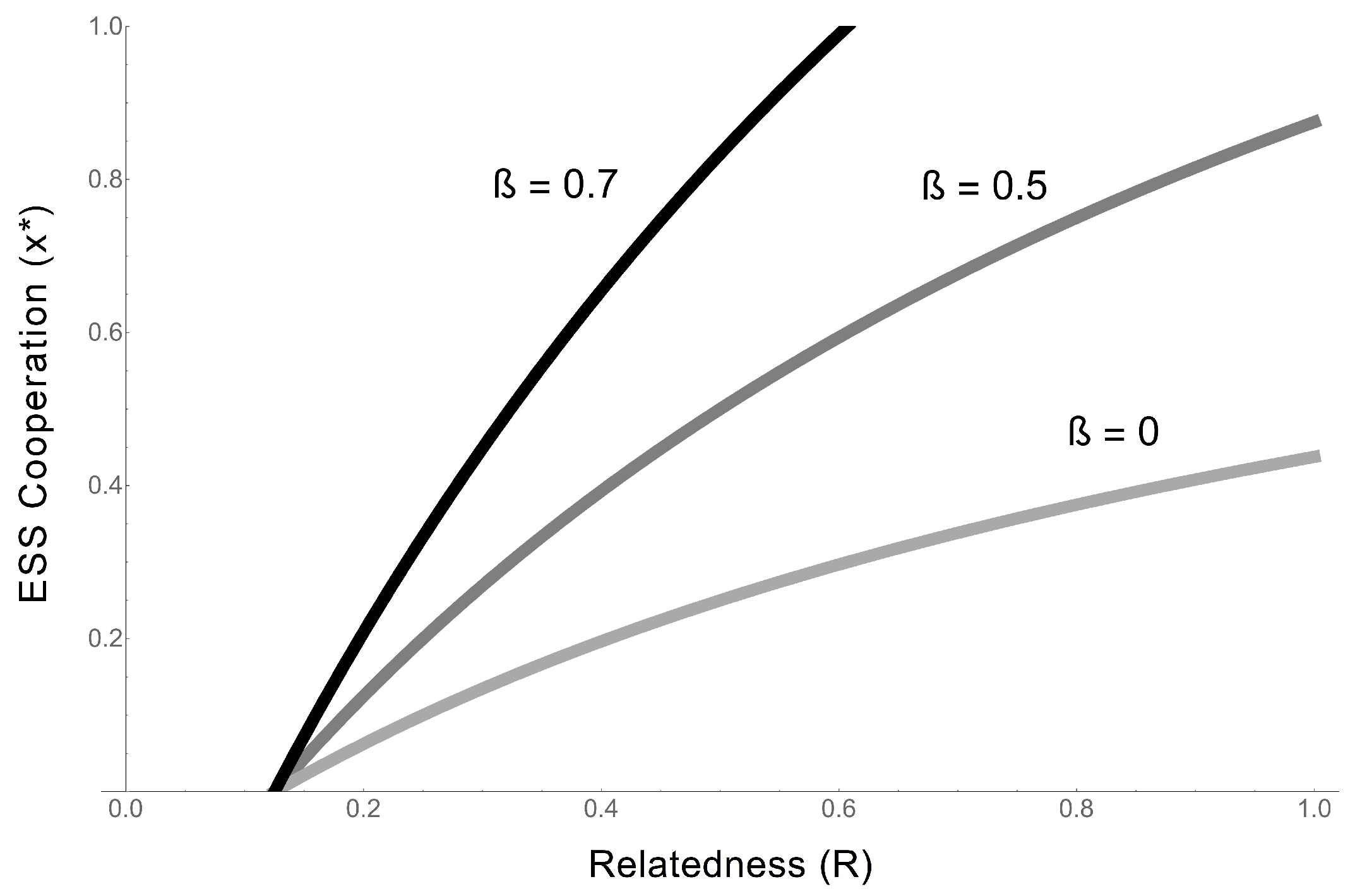
- Regardless of whether cooperative RNAs receive benefits (the value of ), the condition for cooperation to evolve () is . This tells us that the degree to which cooperators receive benefits has no effect on whether cooperation will evolve, although it impacts on the degree of cooperation. Regardless, higher benefits and relatedness and lower costs are favourable for RNA cooperation, confirming the more general model in the previous section.
- In the extreme case in which cooperative RNAs receive no benefits (), the ESS value of cooperation is capped at 0.5, and only partial cooperation can evolve, because complete cooperation would mean that there were no individuals available to receive benefits. This applies, for example, in some RNA trans-replicases, in which becoming a replicase enzyme prevents individuals from being replicated by other replicases [16,44]. This result is analogous to the result obtained in Frank’s (1997) model of paired sibling suicide in animals [45]. This suggests that the evolution of the cooperative enzyme that cannot receive benefits (as in [16,24,44]) is analogous to the evolution of sterility in higher organisms.
2.3. Cooperation and Competition in RNA World
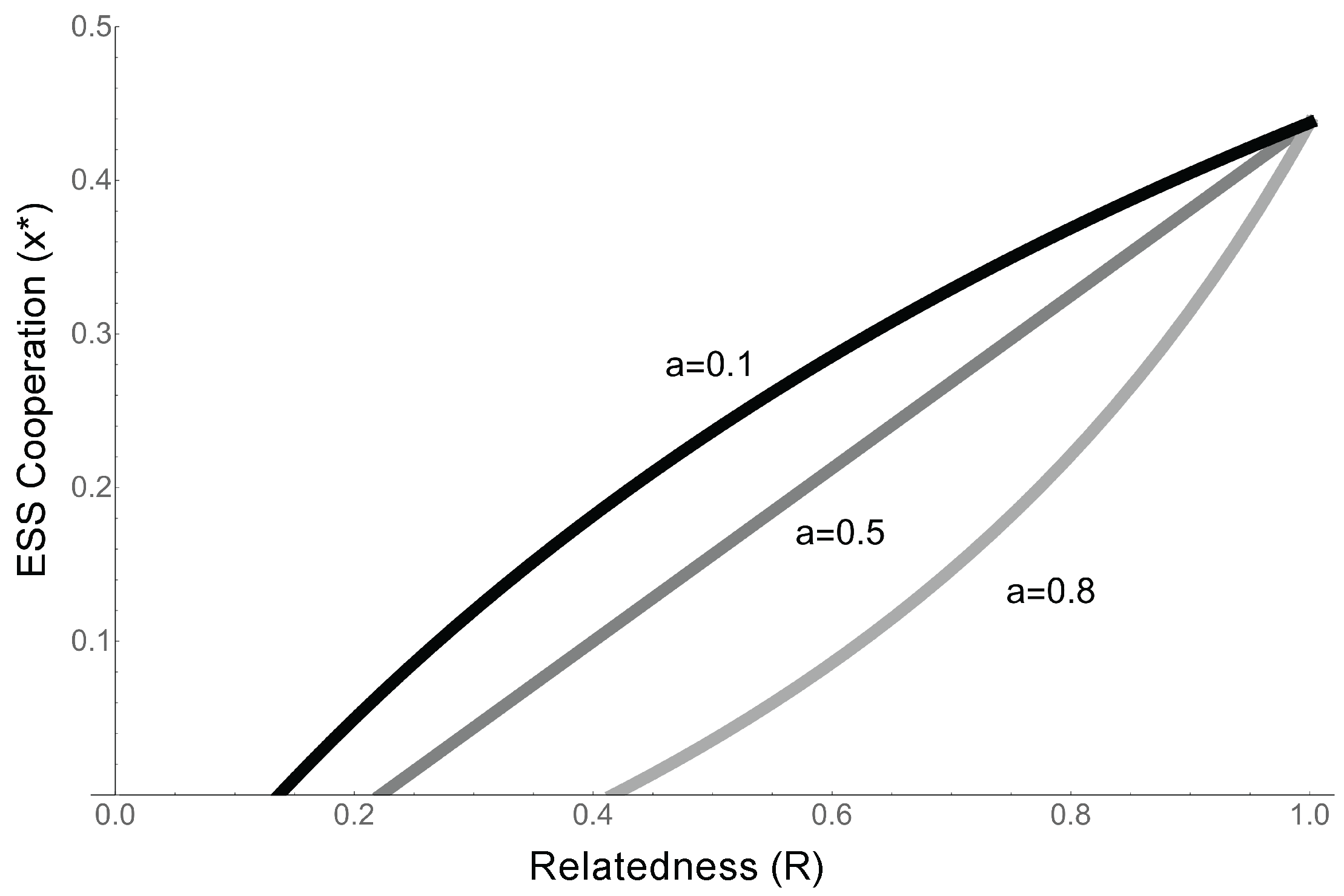
- When competition is completely global (), the ESS reverts to the that identified in the previous model (Equation (7), ), and the condition for cooperation to evolve is simply . This confirms our previous results, and is a qualitatively similar result to that found by Takeuchi et al., as in their model competition is relatively global and replicators cooperate about half the time (Equation (7), [24]).
- As competition becomes more local (a increases), the condition for cooperation to evolve becomes more stringent. When competition (a) is high, the competitive effects of the extra offspring copies produced by, e.g., cooperative enzyme activity, are experienced locally. This means that in addition to benefiting from cooperation, relatives suffer increased competition from cooperation, making it harder for cooperation to evolve (Figure 6).
- When competition is completely local (), . Since the costs and benefits are both positive, cooperation cannot evolve.
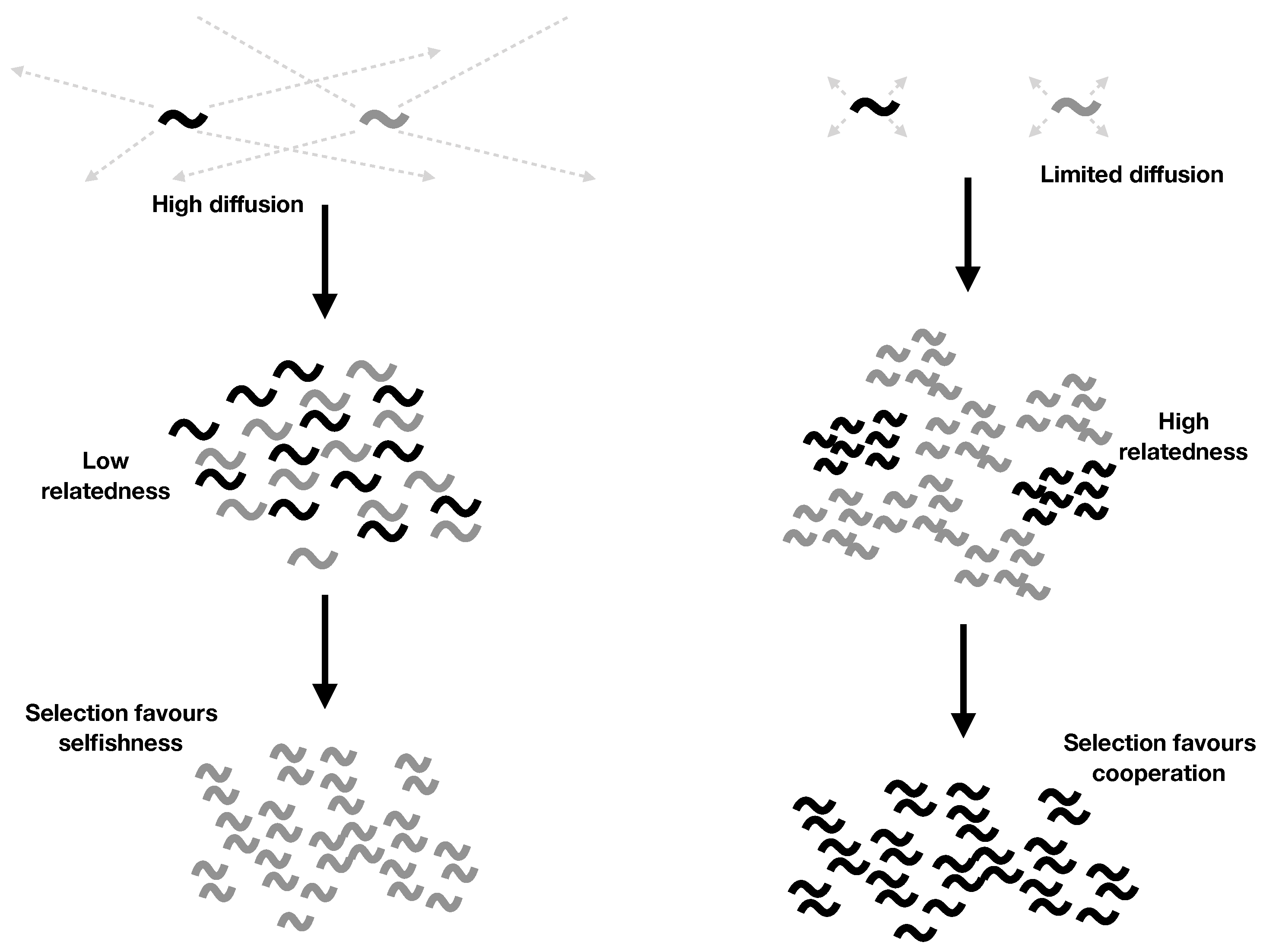
2.4. Explicit Population Structure: Closing the Model
- Cooperation is favoured by higher parent survival (larger k), limited diffusion (lower d), and smaller local group size (smaller N).
- Both parent survival () and limited diffusion () are required for cooperation to evolve. While discrete generations () hold approximately for many higher organisms, we expect overlapping generations () to hold for most RNA systems, because we expect RNAs to survive well after their copies’ copies have replicated. This offers a potential explanation for why RNA molecules might have cooperated despite having a dispersal strategy that otherwise leads to high local competition.
3. Discussion
3.1. Why Bother?
3.1.1. Generality
3.1.2. Testability
3.1.3. Conceptual Links
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| RNA | Ribonucleic acid |
| ESS | Evolutionarily stable strategy |
References
- Crick, F.H. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Orgel, L.E. Evolution of the genetic apparatus. J. Mol. Biol. 1968, 38, 381–393. [Google Scholar] [CrossRef]
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Eigen, M.; Schuster, P. A principle of natural self-organization. Naturwissenschaften 1977, 64, 541–565. [Google Scholar] [CrossRef] [PubMed]
- Szathmáry, E.; Demeter, L. Group selection of early replicators and the origin of life. J. Theor. Biol. 1987, 128, 463–486. [Google Scholar] [CrossRef]
- Smith, J.M.; Szathmary, E. The Major Transitions in Evolution; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- Higgs, P.G.; Lehman, N. The RNA World: molecular cooperation at the origins of life. Nat. Rev. Genet. 2015, 16, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Boerlijst, M.C.; Hogeweg, P. Spiral wave structure in pre-biotic evolution: Hypercycles stable against parasites. Phys. D Nonlinear Phenom. 1991, 48, 17–28. [Google Scholar] [CrossRef]
- Frank, S.A. Kin selection and virulence in the evolution of protocells and parasites. Proc. R. Soc. Lond. B Biol. Sci. 1994, 258, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Boerlijst, M.C.; Hogeweg, P. Spatial gradients enhance persistence of hypercycles. Phys. D Nonlinear Phenom. 1995, 88, 29–39. [Google Scholar] [CrossRef]
- Cronhjort, M.B.; Blomberg, C. Cluster compartmentalization may provide resistance to parasites for catalytic networks. Phys. D Nonlinear Phenom. 1997, 101, 289–298. [Google Scholar] [CrossRef]
- McCaskill, J.S.; Füchslin, R.M.; Altmeyer, S. The stochastic evolution of catalysts in spatially resolved molecular systems. Biol. Chem. 2001, 382, 1343–1363. [Google Scholar] [CrossRef] [PubMed]
- Szabó, P.; Scheuring, I.; Czárán, T.; Szathmáry, E. In silico simulations reveal that replicators with limited dispersal evolve towards higher efficiency and fidelity. Nature 2002, 420, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Sardanyés, J.; Solé, R.V. Spatio-temporal dynamics in simple asymmetric hypercycles under weak parasitic coupling. Phys. D Nonlinear Phenom. 2007, 231, 116–129. [Google Scholar] [CrossRef]
- Bianconi, G.; Zhao, K.; Chen, I.A.; Nowak, M.A. Selection for replicases in protocells. PLoS Comput. Biol. 2013, 9, e1003051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shay, J.A.; Huynh, C.; Higgs, P.G. The origin and spread of a cooperative replicase in a prebiotic chemical system. J. Theor. Biol. 2015, 364, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Levin, S.R.; West, S.A. The evolution of cooperation in simple molecular replicators. Proc. R. Soc. Lond. B Biol. Sci. 2017, 284. [Google Scholar] [CrossRef] [PubMed]
- West, S.A.; Griffin, A.S.; Gardner, A. Evolutionary explanations for cooperation. Curr. Biol. 2007, 17, R661–R672. [Google Scholar] [CrossRef] [PubMed]
- Bourke, A.F. Principles of Social Evolution; Oxford University Press: Oxford, UK, 2011. [Google Scholar]
- Davies, N.; Krebs, J.; West, S. An Introduction to Behavioural Ecology, 4th ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2012. [Google Scholar]
- Hamilton, W.D. The genetical theory of social behavior. I and II. J. Theor. Biol. 1964, 7, 1–52. [Google Scholar] [CrossRef]
- Frank, S.A. Foundations of Social Evolution; Princeton University Press: Princeton, NJ, USA, 1998. [Google Scholar]
- Nee, S. The evolutionary ecology of molecular replicators. R. Soc. Open Sci. 2016, 3, 160235. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, N.; Hogeweg, P.; Kaneko, K. The origin of a primordial genome through spontaneous symmetry breaking. Nat. Commun. 2017, 8, 250. [Google Scholar] [CrossRef] [PubMed]
- West, S.A.; Buckling, A. Cooperation, virulence and siderophore production in bacterial parasites. Proc. R. Soc. Lond. B Biol. Sci. 2003, 270, 37–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, S.A.; Diggle, S.P.; Buckling, A.; Gardner, A.; Griffin, A.S. The social lives of microbes. Annu. Rev. Ecol. Evol. Syst. 2007, 38, 53–77. [Google Scholar] [CrossRef]
- Frank, S.A. A general model of the public goods dilemma. J. Evolut. Biol. 2010, 23, 1245–1250. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.; West, S.A.; Wild, G. The genetical theory of kin selection. J. Evolut. Biol. 2011, 24, 1020–1043. [Google Scholar] [CrossRef] [PubMed]
- Biernaskie, J.M.; West, S.A. Cooperation, clumping and the evolution of multicellularity. Proc. R. Soc. B R. Soc. 2015, 282, 1075. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M.; Price, G. The Logic of Animal Conflict. Nature 1973, 246, 15–18. [Google Scholar] [CrossRef]
- Taylor, P.D.; Frank, S.A. How to make a kin selection model. J. Theor. Biol. 1996, 180, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, W.D. Selfish and spiteful behaviour in an evolutionary model. Nature 1970, 228, 1218–1220. [Google Scholar] [CrossRef] [PubMed]
- Grafen, A. A geometric view of relatedness. Oxf. Surv. Evolut. Biol. 1985, 2, 28–89. [Google Scholar]
- West, S.A.; Pen, I.; Griffin, A.S. Cooperation and competition between relatives. Science 2002, 296, 72–75. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, L.; Rousset, F. How life history and demography promote or inhibit the evolution of helping behaviours. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 2599–2617. [Google Scholar] [CrossRef] [PubMed]
- Hughes, W.O.; Oldroyd, B.P.; Beekman, M.; Ratnieks, F.L. Ancestral monogamy shows kin selection is key to the evolution of eusociality. Science 2008, 320, 1213–1216. [Google Scholar] [CrossRef] [PubMed]
- Cornwallis, C.; West, S.; Griffin, A. Routes to indirect fitness in cooperatively breeding vertebrates: Kin discrimination and limited dispersal. J. Evolut. Biol. 2009, 22, 2445–2457. [Google Scholar] [CrossRef] [PubMed]
- Cornwallis, C.K.; West, S.A.; Davis, K.E.; Griffin, A.S. Promiscuity and the evolutionary transition to complex societies. Nature 2010, 466, 969–972. [Google Scholar] [CrossRef] [PubMed]
- Griffin, A.S.; West, S.A.; Buckling, A. Cooperation and competition in pathogenic bacteria. Nature 2004, 430, 1024–1027. [Google Scholar] [CrossRef] [PubMed]
- Diggle, S.P.; Gardner, A.; West, S.A.; Griffin, A.S. Evolutionary theory of bacterial quorum sensing: When is a signal not a signal? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007, 362, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Kuzdzal-Fick, J.J.; Fox, S.A.; Strassmann, J.E.; Queller, D.C. High relatedness is necessary and sufficient to maintain multicellularity in Dictyostelium. Science 2011, 279, 1548–1551. [Google Scholar] [CrossRef] [PubMed]
- Lukas, D.; Clutton-Brock, T. Cooperative breeding and monogamy in mammalian societies. Proc. R. Soc. B R. Soc. 2012, 334, 2151–2156. [Google Scholar] [CrossRef] [PubMed]
- West, S.A.; Griffin, A.S.; Gardner, A. Social semantics: altruism, cooperation, mutualism, strong reciprocity and group selection. J. Evolut. Biol. 2007, 20, 415–432. [Google Scholar] [CrossRef] [PubMed]
- Von der Dunk, S.H.; Colizzi, E.S.; Hogeweg, P. Evolutionary Conflict Leads to Innovation: Symmetry Breaking in a Spatial Model of RNA-Like Replicators. Life 2017, 7, 43. [Google Scholar] [CrossRef] [PubMed]
- Frank, S.A. Multivariate analysis of correlated selection and kin selection, with an ESS maximization method. J. Theor. Biol. 1997, 189, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.E.; Higgs, P.G. Co-operation between Polymerases and Nucleotide Synthetases in the RNA World. PLoS Comput. Biol. 2016, 12, e1005161. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.F.; Szostak, J.W. Coupled growth and division of model protocell membranes. J. Am. Chem. Soc. 2009, 131, 5705–5713. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.; West, S.A. Spite and the scale of competition. J. Evolut. Biol. 2004, 17, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- West, S.A.; Murray, M.G.; Machado, C.A.; Griffin, A.S.; Herre, E.A. Testing Hamilton’s rule with competition between relatives. Nature 2001, 409, 510. [Google Scholar] [CrossRef] [PubMed]
- Kümmerli, R.; Gardner, A.; West, S.A.; Griffin, A.S. Limited dispersal, budding dispersal, and cooperation: An experimental study. Evolution 2009, 63, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.; West, S.A.; Buckling, A. Bacteriocins, spite and virulence. Proc. R. Soc. B R. Soc. 2004, 271, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.; Hardy, I.C.; Taylor, P.D.; West, S.A. Spiteful soldiers and sex ratio conflict in polyembryonic parasitoid wasps. Am. Nat. 2007, 169, 519–533. [Google Scholar] [CrossRef] [PubMed]
- West, S.A.; Gardner, A. Altruism, spite, and greenbeards. Science 2010, 327, 1341–1344. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.D. Altruism in viscous populations—An inclusive fitness model. Evolut. Ecol. 1992, 6, 352–356. [Google Scholar] [CrossRef]
- Queller, D.C. Genetic relatedness in viscous populations. Evolut. Ecol. 1994, 8, 70–73. [Google Scholar] [CrossRef]
- Gardner, A.; West, S. Demography, altruism, and the benefits of budding. J. Evolut. Biol. 2006, 19, 1707–1716. [Google Scholar] [CrossRef] [PubMed]
- Comins, H.N.; Hamilton, W.D.; May, R.M. Evolutionarily stable dispersal strategies. J. Theor. Biol. 1980, 82, 205–230. [Google Scholar] [CrossRef]
- Grafen, A. The hawk-dove game played between relatives. Anim. Behav. 1979, 27, 905–907. [Google Scholar] [CrossRef]
- West, S. Sex Allocation; Princeton University Press: Princeton, NJ, USA, 2009. [Google Scholar]
- Fisher, R.M.; Cornwallis, C.K.; West, S.A. Group formation, relatedness, and the evolution of multicellularity. Curr. Biol. 2013, 23, 1120–1125. [Google Scholar] [CrossRef] [PubMed]
- Bourke, A.F. Hamilton’s rule and the causes of social evolution. Phil. Trans. R. Soc. B 2014, 369, 20130362. [Google Scholar] [CrossRef] [PubMed]
- Frank, S.A. Hierarchical selection theory and sex ratios. II. On applying the theory, and a test with fig wasps. Evolution 1985, 39, 949–964. [Google Scholar] [CrossRef] [PubMed]
- West, S.A.; Griffin, A.S.; Gardner, A.; Diggle, S.P. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 2006, 4, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Wild, G.; Gardner, A.; West, S.A. Adaptation and the evolution of parasite virulence in a connected world. Nature 2009, 459, 983. [Google Scholar] [CrossRef] [PubMed]
| Example Model System | Relatedness Is Generated by | Do Benefits Return to Cooperators? () | Scale of Competition (a) | Testable Predictions? | |||||
|---|---|---|---|---|---|---|---|---|---|
| Limited Diffusion | Proto-Cells | Other Spatial Clustering | Yes (High ) | No (Low ) | Global (Low a) | Local (High a) | Comparative | Experimental | |
| Replicases in protocells [5,9,15] | X | X | X | X | |||||
| Replicases on surfaces [12,13] | X | X | X | X | |||||
| Trans-replicases [16] | X | X | X | X | X | ||||
| Nucleatase and polymerase [46] | X | X | X | X | X | ||||
| Origin of genome-like molecules [24] | X | X | X | X | |||||
| Hyper-cycles [8,11] | X | X | X | X | |||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Levin, S.R.; West, S.A. Kin Selection in the RNA World. Life 2017, 7, 53. https://doi.org/10.3390/life7040053
Levin SR, West SA. Kin Selection in the RNA World. Life. 2017; 7(4):53. https://doi.org/10.3390/life7040053
Chicago/Turabian StyleLevin, Samuel R., and Stuart A. West. 2017. "Kin Selection in the RNA World" Life 7, no. 4: 53. https://doi.org/10.3390/life7040053





