Reporting Key Features in Cold-Adapted Bacteria
Abstract
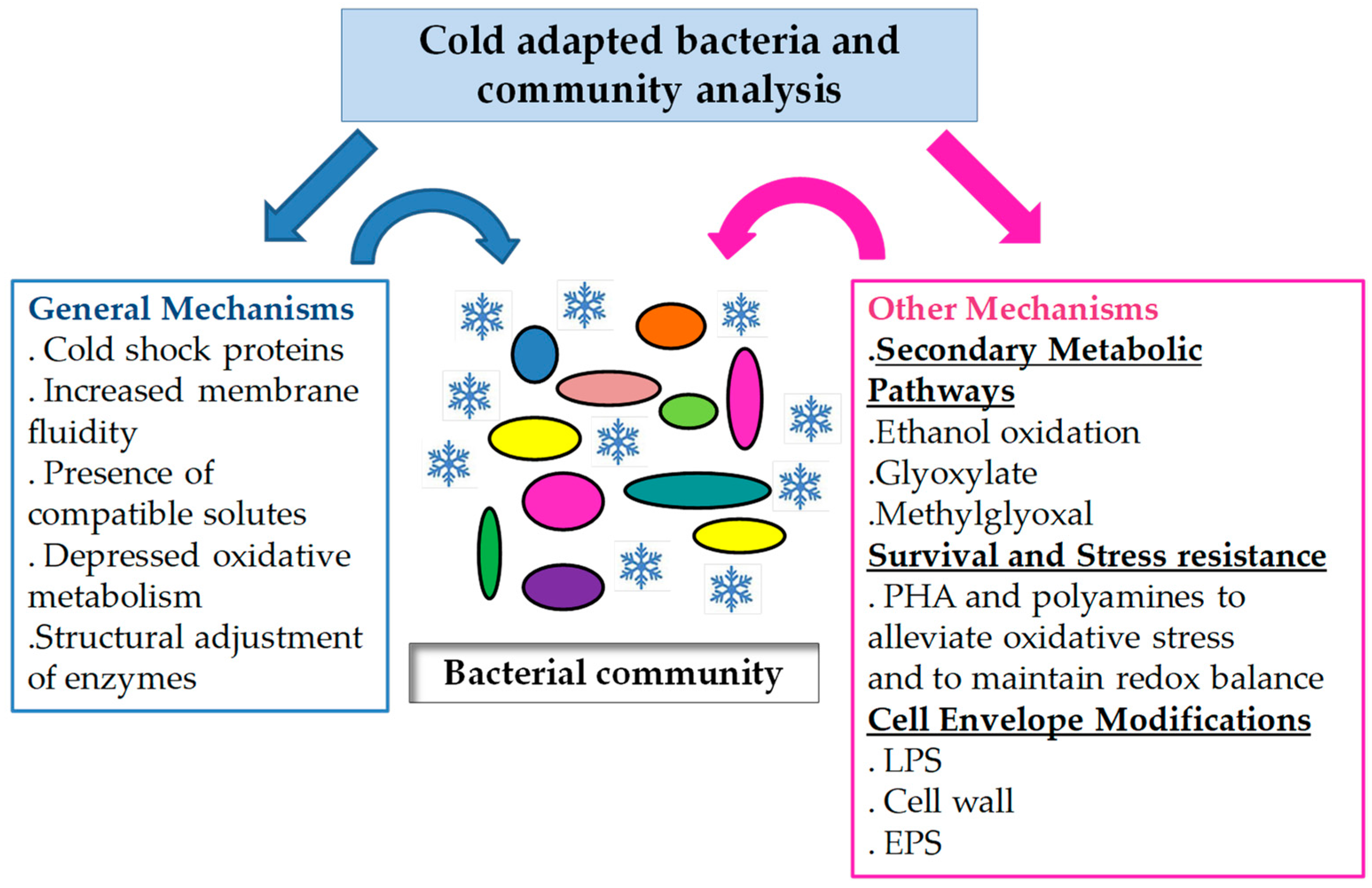
:1. Introduction
2. Metabolic Features Related to Energy Generation in Cold Environments
3. Compounds and Mechanisms Involved in Stress Resistance and Cold Adaptation
3.1. Compatible Solutes and Related Compounds
3.2. Polyhydroxyalkanoates Metabolism
4. Envelopes and Cold Adaptation
5. Functional Attributes of Cold-Adapted Bacterial Communities
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chattopadhyay, M.K.; Raghu, G.; Sharma, Y.V.R.K.; Biju, A.R.; Rajasekharan, M.V.; Shivaji, S. Increase in oxidative stress at low temperature in an Antarctic bacterium. Curr. Microbiol. 2011, 62, 544–546. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.F.; Tiedje, J.M. Coping with our cold planet. Appl. Environ. Microbiol. 2008, 74, 1677–1686. [Google Scholar] [CrossRef] [PubMed]
- Barria, C.; Malecki, M.; Arraiano, C.M. Bacterial adaptation to cold. Microbiology 2013, 159, 2437–2443. [Google Scholar] [CrossRef] [PubMed]
- De Maayer, P.; Anderson, D.; Cary, C.; Cowan, D.A. Some like it cold: Understanding the survival strategies of psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Maccario, L.; Sanguino, L.; Vogel, T.M.; Larose, C. Snow and ice ecosystems: Not so extreme. Res. Microbiol. 2015, 166, 782–795. [Google Scholar] [CrossRef] [PubMed]
- Cavicchioli, R. On the concept of a psychrophile. ISME J. 2016, 10, 793–795. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.Y. Psychrophilic bacteria. Bacteriol. Rev. 1975, 39, 144–167. [Google Scholar] [PubMed]
- Atlas, R.M.; Bartha, R. Microbial Ecology: Fundamentals and Applications, 4th ed.; Benjamin/Cummings Science Publishers: Menlo Park, CA, USA, 1998; pp. 1–704. [Google Scholar]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Microbiol. 2003, 1, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.F.; Ivanova, N.; He, Z.; Huebner, M.; Zhou, J.; Tiedje, J.M. Architecture of thermal adaptation in an Exiguobacterium sibiricum strain isolated from 3 million year old permafrost: A genome and transcriptome approach. BMC Genom. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Aliyu, H.; De Maayer, P.; Cowan, D. The genome of the Antarctic polyextremophile Nesterenkonia sp. AN1 reveals adaptive strategies for survival under multiple stress conditions. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [PubMed]
- Raymond-Bouchard, I.; Chourey, K.; Altshuler, I.; Iyer, R.; Hettich, R.L.; Whyte, L.G. Mechanisms of subzero growth in the cryophile Planococcus halocryophilus determined through proteomic analysis. Environ. Microbiol. 2017, 19, 4460–4479. [Google Scholar] [CrossRef] [PubMed]
- Mykytczuk, N.C.S.; Foote, S.J.; Omelon, C.R.; Southam, G.; Greer, C.W.; Whyte, L.G. Bacterial growth at −15 °C; molecular insights from the permafrost bacterium Planococcus halocryophilus Or1. ISME J. 2013, 7, 1211–1226. [Google Scholar] [CrossRef] [PubMed]
- Ronholm, J.; Raymond-Bouchard, I.; Creskey, M.; Cyr, T.; Cloutis, E.A.; Whyte, L.G. Characterizing the surface-exposed proteome of Planococcus halocryophilus during cryophilic growth. Extremophiles 2015, 19, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Piette, F.; D’Amico, S.; Mazzucchelli, G.; Danchin, A.; Leprince, P.; Feller, G. Life in the cold: A proteomic study of cold-repressed proteins in the antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Appl. Environ. Microbiol. 2011, 77, 3881–3883. [Google Scholar] [CrossRef] [PubMed]
- Tribelli, P.M.; Venero, E.C.S.; Ricardi, M.M.; Gómez-Lozano, M.; Iustman, L.J.R.; Molin, S.; López, N.I. Novel essential role of ethanol oxidation genes at low temperature revealed by transcriptome analysis in the antarctic bacterium Pseudomonas extremaustralis. PLoS ONE 2015, 10, e0145353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayala-Del-Río, H.; Chain, P.S.; Grzymski, J.J.; Ponder, M.A.; Ivanova, N.; Bergholz, P.W.; Di Bartolo, G.; Hauser, L.; Land, M.; Bakermans, C.; et al. The genome sequence of Psychrobacter arcticus 273-4, a psychroactive siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl. Environ. Microbiol. 2010, 76, 2304–2312. [Google Scholar] [CrossRef] [PubMed]
- Ting, L.; Williams, T.J.; Cowley, M.J.; Lauro, F.M.; Guilhaus, M.; Raftery, M.J.; Cavicchioli, R. Cold adaptation in the marine bacterium, Sphingopyxis alaskensis, assessed using quantitative proteomics. Environ. Microbiol. 2010, 12, 2658–2676. [Google Scholar] [CrossRef] [PubMed]
- Piette, F.; Leprince, P.; Feller, G. Is there a cold shock response in the Antarctic psychrophile Pseudoalteromonas haloplanktis? Extremophiles 2012, 16, 681–683. [Google Scholar] [CrossRef] [PubMed]
- Koh, H.Y.; Park, H.; Lee, J.H.; Han, S.J.; Sohn, Y.C.; Lee, S.G. Proteomic and transcriptomic investigations on cold-responsive properties of the psychrophilic Antarctic bacterium Psychrobacter sp. PAMC 21119 at subzero temperatures. Environ. Microbiol. 2017, 19, 628–644. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, P.; Moreno, R.; Rojo, F. Growth of Pseudomonas putida at low temperature: Global transcriptomic and proteomic analyses. Environ. Microbiol. Rep. 2011, 3, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Textor, S.; Wendisch, V.F.; De Graaf, A.A.; Müller, U.; Linder, M.I.; Linder, D.; Buckel, W. Propionate oxidation in Escherichia coli: Evidence for operation of a methylcitrate cycle in bacteria. Arch. Microbiol. 1997, 168, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Horswill, A.R.; Escalante-Semerena, J.C. Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J. Bacteriol. 1999, 181, 5615–5623. [Google Scholar] [PubMed]
- Regenhardt, D.; Heuer, H.; Heim, S.; Fernandez, D.U.; Strömpl, C.; Moore, E.R.B.; Timmis, K.N. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ. Microbiol. 2002, 4, 912–915. [Google Scholar] [CrossRef] [PubMed]
- Reva, O.N.; Weinel, C.; Weinel, M.; Böhm, K.; Stjepandic, D.; Hoheisel, J.D.; Tümmler, B. Functional genomics of stress response in Pseudomonas putida KT2440. J. Bacteriol. 2006, 188, 4079–4092. [Google Scholar] [CrossRef] [PubMed]
- Raiger Iustman, L.J.; Tribelli, P.M.; Ibarra, J.; Catone, M.V.; Solar Venero, E.C.; López, N.I. Genome sequence analysis of Pseudomonas extremaustralis provides new insights into environmental adaptability and extreme conditions resistance. Extremophiles 2015, 19, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Methe, B.A.; Nelson, K.E.; Deming, J.W.; Momen, B.; Melamud, E.; Zhang, X.; Moult, J.; Madupu, R.; Nelson, W.C.; Dodson, R.J.; et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl. Acad. Sci. USA 2005, 102, 10913–10918. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.E.; Deming, J.W. An inter-order horizontal gene transfer event enables the catabolism of compatible solutes by Colwellia psychrerythraea 34H. Extremophiles 2013, 17, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Mocali, S.; Chiellini, C.; Fabiani, A.; Decuzzi, S.; Pascale, D.; Parrilli, E.; Tutino, M.L.; Perrin, E.; Bosi, E.; Fondi, M.; et al. Ecology of cold environments: New insights of bacterial metabolic adaptation through an integrated genomic-phenomic approach. Sci. Rep. 2017, 7, 839. [Google Scholar] [CrossRef] [PubMed]
- Nunn, B.L.; Slattery, K.V.; Cameron, K.A.; Timmins-Schiffman, E.; Junge, K. Proteomics of Colwellia psychrerythraea at subzero temperatures—A life with limited movement, flexible membranes and vital DNA repair. Environ. Microbiol. 2015, 17, 2319–2335. [Google Scholar] [CrossRef] [PubMed]
- Ghobakhlou, A.F.; Johnston, A.; Harris, L.; Antoun, H.; Laberge, S. Microarray transcriptional profiling of Arctic Mesorhizobium strain N33 at low temperature provides insights into cold adaption strategies. BMC Genom. 2015, 16, 383. [Google Scholar] [CrossRef] [PubMed]
- López, N.I.; Pettinari, M.J.; Nikel, P.I.; Méndez, B.S. Polyhydroxyalkanoates: Much more than biodegradable plastics. Adv. Appl. Microbiol. 2015, 93, 73–106. [Google Scholar] [PubMed]
- Matin, A.; Veldhuis, C.; Stegeman, V.; Veenhuis, M. Selective advantage of a Spirillum sp. in a carbon-limited environment. Accumulation of poly-beta-hydroxybutyric acid and its role in starvation. J. Gen. Microbiol. 1979, 112, 349–355. [Google Scholar] [CrossRef] [PubMed]
- López, N.I.; Floccari, M.E.; Steinbüchel, A.; García, A.F.; Méndez, B.S. Effect of poly(3-hydroxybutyrate) (PHB) content on the starvation-survival of bacteria in natural waters. FEMS Microbiol. Ecol. 1995, 16, 95–101. [Google Scholar] [CrossRef]
- Handrick, R.; Reinhardt, S.; Jendrossek, D. Mobilization of poly(3-hydroxybutyrate) in Ralstonia eutropha. J. Bacteriol. 2000, 182, 5916–5918. [Google Scholar] [CrossRef] [PubMed]
- Kadouri, D.; Jurkevitch, E.; Okon, Y. Involvement of the reserve material poly-β-hydroxybutyrate in Azospirillum brasilense stress endurance and root colonization. Appl. Environ. Microbiol. 2003, 69, 3244–3250. [Google Scholar] [CrossRef] [PubMed]
- Mezzina, M.P.; Pettinari, M.J. Phasins, multifaceted polyhydroxyalkanoate granule-associated proteins. Appl. Environ. Microbiol. 2016, 82, 5060–5067. [Google Scholar] [CrossRef] [PubMed]
- Catone, M.V.; Ruiz, J.A.; Castellanos, M.; Segura, D.; Espin, G.; López, N.I. High polyhydroxybutyrate production in Pseudomonas extremaustralis is associated with differential expression of horizontally acquired and core genome polyhydroxyalkanoate synthase genes. PLoS ONE 2014, 9, e98873. [Google Scholar] [CrossRef] [PubMed]
- Ayub, N.D.; Tribelli, P.M.; López, N.I. Polyhydroxyalkanoates are essential for maintenance of redox state in the Antarctic bacterium Pseudomonas sp. 14–3 during low temperature adaptation. Extremophiles 2009, 13, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Tribelli, P.M.; López, N.I. Poly(3-hydroxybutyrate) influences biofilm formation and motility in the novel Antarctic species Pseudomonas extremaustralis under cold conditions. Extremophiles 2011, 15, 541. [Google Scholar] [CrossRef] [PubMed]
- Ayub, N.D.; Pettinari, M.J.; Méndez, B.S.; López, N.I. The polyhydroxyalkanoate genes of a stress resistant Antarctic Pseudomonas are situated within a genomic island. Plasmid 2007, 58, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.A.; López, N.I.; Fernández, R.O.; Méndez, B.S. Polyhydroxyalkanoate degradation is associated with nucleotide accumulation and enhances stress resistance and survival of Pseudomonas oleovorans in natural water microcosms. Appl. Environ. Microbiol. 2001, 67, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.A.; López, N.I.; Méndez, B.S. rpoS gene expression in carbon-starved cultures of the polyhydroxyalkanoate-accumulating species Pseudomonas oleovorans. Curr. Microbiol. 2004, 48, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Obruca, S.; Sedlacek, P.; Krzyzanek, V.; Mravec, F.; Hrubanova, K.; Samek, O.; Kucera, D.; Benesova, P.; Marova, I. Accumulation of poly(3-hydroxybutyrate) helps bacterial cells to survive freezing. PLoS ONE 2016, 11, e0157778. [Google Scholar] [CrossRef] [PubMed]
- Obruca, S.; Sedlacek, P.; Mravec, F.; Samek, O.; Marova, I. Evaluation of 3-hydroxybutyrate as an enzyme-protective agent against heating and oxidative damage and its potential role in stress response of poly(3-hydroxybutyrate) accumulating cells. Appl. Microbiol. Biotechnol. 2016, 100, 1365–1376. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, G.V.; Zakirova, O.N.; Oktyabrskii, O.N. The role of antioxidant systems in the cold stress response of Escherichia coli. Microbiology 2001, 70, 45–50. [Google Scholar] [CrossRef]
- Zhang, L.; Onda, K.; Imai, R.; Fukuda, R.; Horiuchi, H.; Ohta, A. Growth temperature downshift induces antioxidant response in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2003, 307, 308–314. [Google Scholar] [CrossRef]
- Margesin, R.; Miteva, V. Diversity and ecology of psychrophilic microorganisms. Res. Microbiol. 2011, 162, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Cabiscol, E.; Tamarit, J.; Ros, J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 2000, 3, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Tkachenko, A.G.; Pshenichnov, M.R.; Nesterova, L.Y. Putrescine as a factor protecting Escherichia coli against oxidative stress. Microbiology 2001, 70, 422–428. [Google Scholar] [CrossRef]
- Zhu, X.; Li, Q.; Yin, C.; Fang, X.; Xu, X. Role of spermidine in overwintering of cyanobacteria. J. Bacteriol. 2015, 197, 2325–2334. [Google Scholar] [CrossRef] [PubMed]
- Goh, Y.S.; Tan, I.K.P. Polyhydroxyalkanoate production by antarctic soil bacteria isolated from Casey Station and Signy Island. Microbiol. Res. 2012, 167, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Ciesielski, S.; Górniak, D.; Możejko, J.; Świątecki, A.; Grzesiak, J.; Zdanowski, M. The diversity of bacteria isolated from Antarctic freshwater reservoirs possessing the ability to produce polyhydroxyalkanoates. Curr. Microbiol. 2014, 69, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Luhtanen, A.M.; Eronen-Rasimus, E.; Kaartokallio, H.; Rintala, J.M.; Autio, R.; Roine, E. Isolation and characterization of phage-host systems from the Baltic Sea ice. Extremophiles 2014, 18, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Parnanen, K.; Karkman, A.; Virta, M.; Eronen-Rasimus, E.; Kaartokallio, H. Discovery of bacterial polyhydroxyalkanoate synthase (PhaC)-encoding genes from seasonal Baltic Sea ice and cold estuarine waters. Extremophiles 2015, 19, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Médigue, C.; Krin, E.; Pascal, G.; Barbe, V.; Bernsel, A.; Bertin, P.N.; Cheung, F.; Cruveiller, S.; D’Amico, S.; Duilio, A.; et al. Coping with cold: The genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 2005, 15, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Bergholz, P.W.; Bakermans, C.; Tiedje, J.M. Psychrobacter arcticus 273-4 Uses resource efficiency and molecular motion adaptations for subzero temperature growth. J. Bacteriol. 2009, 191, 2340–2352. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.S.; Jagannadham, M.V.; Ray, M.K. Low-temperature-induced changes in composition and fluidity of lipopolysaccharides in the antarctic psychrotrophic bacterium Pseudomonas syringae. J. Bacteriol. 2002, 184, 6746–6749. [Google Scholar] [CrossRef] [PubMed]
- Benforte, F.C.; Colonnella, M.A.; Ricardi, M.M.; Venero, E.C.S.; Lizarraga, L.; López, N.I.; Tribelli, P.M. Novel role of the LPS core glycosyltransferase WapH for cold adaptation in the Antarctic bacterium Pseudomonas extremaustralis. PLoS ONE 2018, 13, e0192559. [Google Scholar] [CrossRef] [PubMed]
- Mykytczuk, N.C.S.; Lawrence, J.R.; Omelon, C.R.; Southam, G.; Whyte, L.G. Microscopic characterization of the bacterial cell envelope of Planococcus halocryophilus Or1 during subzero growth at −15 °C. Polar Biol. 2016, 39, 701–712. [Google Scholar] [CrossRef]
- Mancuso Nichols, C.A.; Garon, S.; Bowman, J.P.; Raguénès, G.; Guézennec, J. Production of exopolysaccharides by Antarctic marine bacterial isolates. J. Appl. Microbiol. 2004, 96, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Corsaro, M.M.; Lanzetta, R.; Parrilli, E.; Parrilli, M.; Tutino, M.L.; Ummarino, S. Influence of growth temperature on lipid and phosphate contents of surface polysaccharides from the antarctic bacterium Pseudoalteromonas haloplanktis TAC 125. J. Bacteriol. 2004, 186, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Nichols, C.M.; Lardière, S.G.; Bowman, J.P.; Nichols, P.D.; Gibson, J.A.E.; Guézennec, J. Chemical characterization of exopolysaccharides from Antarctic marine bacteria. Microb. Ecol. 2005, 49, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Caruso, C.; Rizzo, C.; Mangano, S.; Poli, A.; Di Donato, P.; Nicolaus, B.; Di Marco, G.; Michaud, L.; Lo Giudice, A. Extracellular polymeric substances with metal adsorption capacity produced by Pseudoalteromonas sp. MER144 from Antarctic seawater. Environ. Sci. Pollut. Res. Int. 2017, 25, 4667–4677. [Google Scholar] [CrossRef] [PubMed]
- Caruso, C.; Rizzo, C.; Mangano, S.; Poli, A.; Di Donato, P.; Finore, I.; Nicolaus, B.; Di Marco, G.; Michaud, L.; Lo Giudice, A. Production and biotechnological potential of extracellular polymeric substances from sponge-sssociated antarctic bacteria. Appl. Environ. Microbiol. 2018, 84, e01624-17. [Google Scholar] [CrossRef] [PubMed]
- Varin, T.; Lovejoy, C.; Jungblut, A.D.; Vincent, W.F.; Corbeil, J. Metagenomic analysis of stress genes in microbial mat communities from Antarctica and the high Arctic. Appl. Environ. Microbiol. 2012, 78, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Hakim, J.A.; Fisher, P.R.E.; Grueneberg, A.; Andersen, D.T.; Bej, A.K. Distribution of cold adaptation proteins in microbial mats in Lake Joyce, Antarctica: Analysis of metagenomic data by using two bioinformatics tools. J. Microbiol. Methods 2016, 120, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Liljeqvist, M.; Ossandon, F.J.; González, C.; Rajan, S.; Stell, A.; Valdes, J.; Holmes, D.S.; Dopson, M. Metagenomic analysis reveals adaptations to a cold-adapted lifestyle in a low-temperature acid mine drainage stream. FEMS Microbiol. Ecol. 2015, 91. [Google Scholar] [CrossRef] [PubMed]
- MacKelprang, R.; Burkert, A.; Haw, M.; Mahendrarajah, T.; Conaway, C.H.; Douglas, T.A.; Waldrop, M.P. Microbial survival strategies in ancient permafrost: Insights from metagenomics. ISME J. 2017, 11, 2305–2318. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.J.; Long, E.; Evans, F.; Demaere, M.Z.; Lauro, F.M.; Raftery, M.J.; Ducklow, H.; Grzymski, J.J.; Murray, A.E.; Cavicchioli, R. A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J. 2012, 6, 1883–1900. [Google Scholar] [CrossRef] [PubMed]
- Coolen, M.J.L.; Orsi, W.D. The transcriptional response of microbial communities in thawing Alaskan permafrost soils. Front. Microbiol. 2015, 6, 197. [Google Scholar] [CrossRef] [PubMed]
| Bacterial Species | Metabolic Features | Increase of Membrane Fluidity | References | ||
|---|---|---|---|---|---|
| TCA Repression or Shortened | Cytochrome Repression | Presence of Alternative Pathways | |||
| Exiguobacterium sibiricum | NI | X | X a | X | [10] |
| Nesterenkonia sp. AN1 | NI | NI | X a | X | [11] |
| Planococcus halocryophilus Or1 | X | X | X a | Decrease | [12,13,14] |
| Pseudoalteromonas haloplanktis | X | X | NI | X | [15] |
| Pseudomonas extremaustralis | X | X | X c | NI | [16] |
| Psychrobacter arcticus 273-4 | X | NI | X a | X | [17] |
| Sphingopyxis alaskensis | X | X | X b | X | [18] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tribelli, P.M.; López, N.I. Reporting Key Features in Cold-Adapted Bacteria. Life 2018, 8, 8. https://doi.org/10.3390/life8010008
Tribelli PM, López NI. Reporting Key Features in Cold-Adapted Bacteria. Life. 2018; 8(1):8. https://doi.org/10.3390/life8010008
Chicago/Turabian StyleTribelli, Paula M., and Nancy I. López. 2018. "Reporting Key Features in Cold-Adapted Bacteria" Life 8, no. 1: 8. https://doi.org/10.3390/life8010008





