Sad State of Phage Electron Microscopy. Please Shoot the Messenger
Abstract
:1. Introduction
2. Publications and Criteria for Analysis
2.1. General
2.2. Countries of Origin
| Country | Articles | |||
|---|---|---|---|---|
| Overall | Poor | Mediocre | Good | |
| Australia | 8 | 8 | - | - |
| Canada | 22 | 9 | 1 | 12 |
| China (P.R.) | 28 | 24 | 3 | 1 |
| Finland | 12 | 3 | - | 9 |
| India | 12 | 6 | 2 | 4 |
| Japan | 9 | 6 | 2 | 1 |
| Poland | 6 | 4 | - | 2 |
| Russia | 12 | 5 | 3 | 4 |
| South Korea | 29 | 28 | - | 1 |
| Switzerland | 7 | 3 | 1 | 3 |
| United Kingdom | 17 | 11 | 1 | 5 |
| USA | 37 | 26 | 2 | 9 |
2.3. Journals
| Articles | Journal |
|---|---|
| 33 | Applied and Environmental Microbiology |
| 27 | Archives of Virology |
| 17 | Journal of Virology |
| 12 | Journal of Bacteriology |
| 10 | Environmental Microbiology |
| 8 | Current Microbiology, Virological Journal |
| 7 | Microbial Ecology |
| 6 | Virology |
| 5 | Bacteriophage, Microbiology-UK |
2.4. Electron Microscopes
| Manufacturer | Model | Number | Model | Number | Model | Number |
|---|---|---|---|---|---|---|
| Hitachi | H-300 | 1 | H-7100 | 7 | H8100S | 1 |
| H-300TM | 1 | H-7500 | 5 | H8100 | 1 | |
| H-600 | 1 | H-7600 | 1 | 800 STEM | 2 | |
| H-7000 | 1 | H7650 | 1 | - | - | |
| JEOL | 7A | 1 | 1010 | 3 | 1230 | 4 |
| 100C | 1 | 1011 | 2 | 1400 | 5 | |
| 100CX | 3 | 1200 | 1 | 2000 | 3 | |
| 100S | 2 | 1200EX | 7 | 2000EX | 1 | |
| 100SX | 1 | 1200EXII | 14 | 2010 | 1 | |
| 100SXII | 2 | 1210 | 1 | 2010HC | 1 | |
| 200CX | 1 | 1220 | 1 | 2011 | 1 | |
| 210 | 1 | - | - | - | - | |
| Philips/FEI | 100 | 1 | 400T | 1 | T10 | 3 |
| 201 | 1 | 400HGM | 2 | T20 | 1 | |
| 201C | 1 | Morgagni 268 | 1 | T30 | 1 | |
| 205 | 1 | CM12 | 3 | G2 Spirit TWIN | 8 | |
| 208S | 1 | CM100 | 6 | |||
| 300 | 19 | CM120 | 2 | G2 Spirit Bio TWIN | 10 | |
| 400ST | 1 | F20 | 1 | |||
| Zeiss | EM10 | 2 | Leo 902 | 14 | Leo 912AB | 3 |
| EM109 | 1 | Leo 910 | 1 | Libra 120 | 1 | |
| Leo 900 | 1 | Leo 912 | 2 | Supra 40VP | 1 |
3. The Problems
3.1. Technical Problems
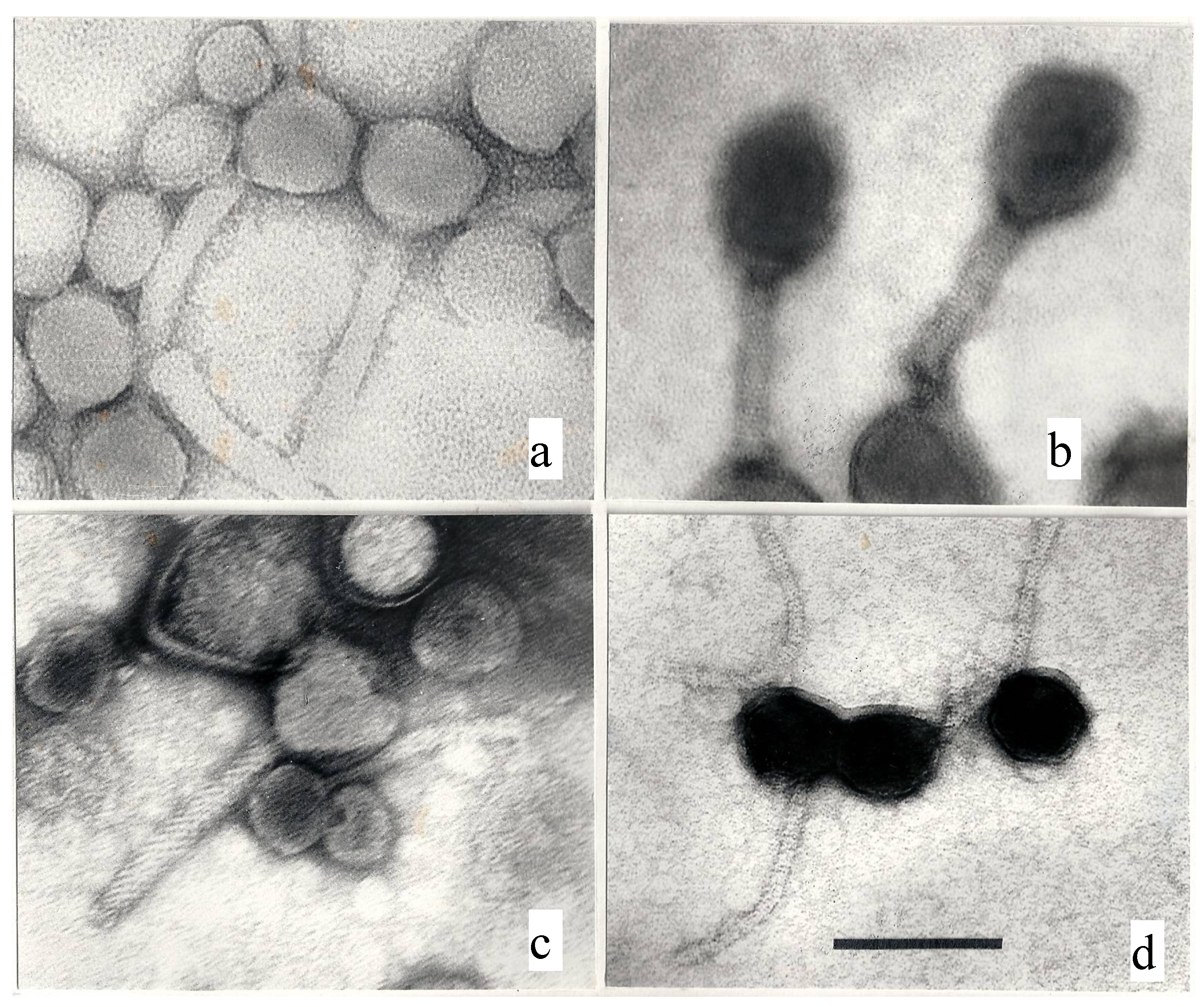
3.2. Human Responsibility
| Technical | Investigator-related | Journal |
|---|---|---|
| Image too small | Insufficient information on methods | Image too small |
| Magnification too low | Positive staining | Incompetent referees |
| Magnification not checked | Dirty samples | - |
| No contrast | Incomplete virus descriptions | - |
| Unsharp | No comparisons | - |
| No details visible | Misclassification | - |
4. Reasons for Declining Quality
5. Conclusions
| Countermeasures | Effectiveness | Comments |
|---|---|---|
| Courses in EM | Little | Too many different instruments in use |
| Reference laboratories | Impractical | Legal and monetary problems |
| Atlas of bacteriophages | Desirable | Copyright problems |
| Standards for pictures | Easy | Must be adopted by journals |
| Contrast improvement | Easy or not | Manual EMs: use graded filters; Digital EMs: use Photoshop or the like |
| More severe reviews | Great | Must be required by journals |
| Signed reviews | Great? | Probably unpopular |
| Blacklisting poor referees | Dubious | An internal matter for individual journals |
Acknowledgments
Conflicts of Interest
References
- Ackermann, H.-W.; Prangishvili, D. Prokaryote viruses studied by electron microscopy. Arch. Virol. 2012, 157, 1843–1849. [Google Scholar] [CrossRef]
- Hall, C.E. Electron densitometry of stained virus particles. J. Biophys. Biochem. Cytol. 1955, 1, 1–11. [Google Scholar] [CrossRef]
- Brenner, S.; Horne, R.W. A negative staining method for high resolution electron microscopy of viruses. Biochim. Biophys. Acta 1959, 34, 103–110. [Google Scholar] [CrossRef]
- Brenner, S.; Streisinger, G.; Horne, R.W.; Champe, S.P.; Barnett, L.; Benzer, S.; Rees, M.W. Structural components of bacteriophage. J. Mol. Biol. 1959, 1, 281–292. [Google Scholar] [CrossRef]
- Huxley, H.E.; Zubay, G. Preferential staining of nucleic acid-containing structures for electron microscopy. J. Biophys. Biochem. Cytol. 1961, 11, 273–296. [Google Scholar] [CrossRef]
- Ultrastructure of Animal Viruses and Bacteriophages: An Atlas; Dalton, A.J.; Haguenau, F. (Eds.) Academic Press: New York, NY, USA, 1973; p. 413.
- The Atlas of Insect and Plant Viruses: Including Mycoplasmaviruses and Viroids; Maramorosch, K. (Ed.) Academic Press: New York, NY, USA, 1977; p. 478.
- Palmer, E.; Martin, M.L. An Atlas of Mammalian Viruses; CRC Press: Boca Raton, FL, USA, 1982; p. 163. [Google Scholar]
- Tikhonenko, A.S. Ultrastructure of Bacterial Viruses; Plenum Press: New York, NY, USA, 1970; p. 294. [Google Scholar]
- Ackermann, H.-W. Declining Electron Microscopy. Available online: http://www.mansfield.ohio-state.edu/~sabedon/bgnws021.htm#editorial (accessed on 10 December 2013).
- Ackermann, H-W. A Bacteriophage Bibliography for 1965–2012. Available online: http://www.phage.ulaval.ca/fileadmin/phages/documents/Bibliographie/Bibliography_2012-04-11.pdf (accessed on 30 September 2013).
- Tiekotter, K.L.; Ackermann, H.-W. High-quality virus images obtained by TEM and CCD technology. J. Virol. Methods 2009, 159, 87–92. [Google Scholar] [CrossRef]
- De Carlo, S.; Harris, J.R. Negative staining and cryo-negative stainng of macromolecules and viruses for TEM. Micron 2011, 42, 117–131. [Google Scholar] [CrossRef]
- Ackermann, H.-W.; Tiekotter, K.L. Murphy's law—If anything can go wrong, it will: Problems in phage electron microscopy. Bacteriophage 2012, 2, 122–129. [Google Scholar] [CrossRef]
- Luftig, R.B. An accurate measurement of the catalase crystal period and its use as an internal marker for electron microscopy. J. Ultrastruct. Res. 1967, 20, 91–102. [Google Scholar] [CrossRef]
- Bohannon, J. Who is afraid of peer review? Science 2013, 342, 60–65. [Google Scholar] [CrossRef]
- Krzywy, T.; Slopek, S. Morfologia i Ultrastruktura Bakteriofagów Shigella i Klebsiella(Morphology and Ultrastructure of Shigella and Klebsiella Phages); Polish Medical Publishers: Warsaw, Poland, 1974; p. 166. [Google Scholar]
- Adobe Photoshop CS3. Available online: http://www.adobe.com/support/downloads/detail.jsp?ftpID=3793 (accessed on 18 October 2013).
- Paint.NET. Available online: http://www.getpaint.net/ (accessed on 18 October 2013).
- GIMP (GNU Image Manipulation Program). Available online: http://www.gimp.org/ (accessed on 18 October 2013).
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ackermann, H.-W. Sad State of Phage Electron Microscopy. Please Shoot the Messenger. Microorganisms 2014, 2, 1-10. https://doi.org/10.3390/microorganisms2010001
Ackermann H-W. Sad State of Phage Electron Microscopy. Please Shoot the Messenger. Microorganisms. 2014; 2(1):1-10. https://doi.org/10.3390/microorganisms2010001
Chicago/Turabian StyleAckermann, Hans-W. 2014. "Sad State of Phage Electron Microscopy. Please Shoot the Messenger" Microorganisms 2, no. 1: 1-10. https://doi.org/10.3390/microorganisms2010001




