The Food Production Environment and the Development of Antimicrobial Resistance in Human Pathogens of Animal Origin
Abstract
:1. Introduction
2. Mechanisms of Bacterial Antibiotic Resistance
3. Use of Antimicrobials in Food Production Environment
3.1. Historical Perspective
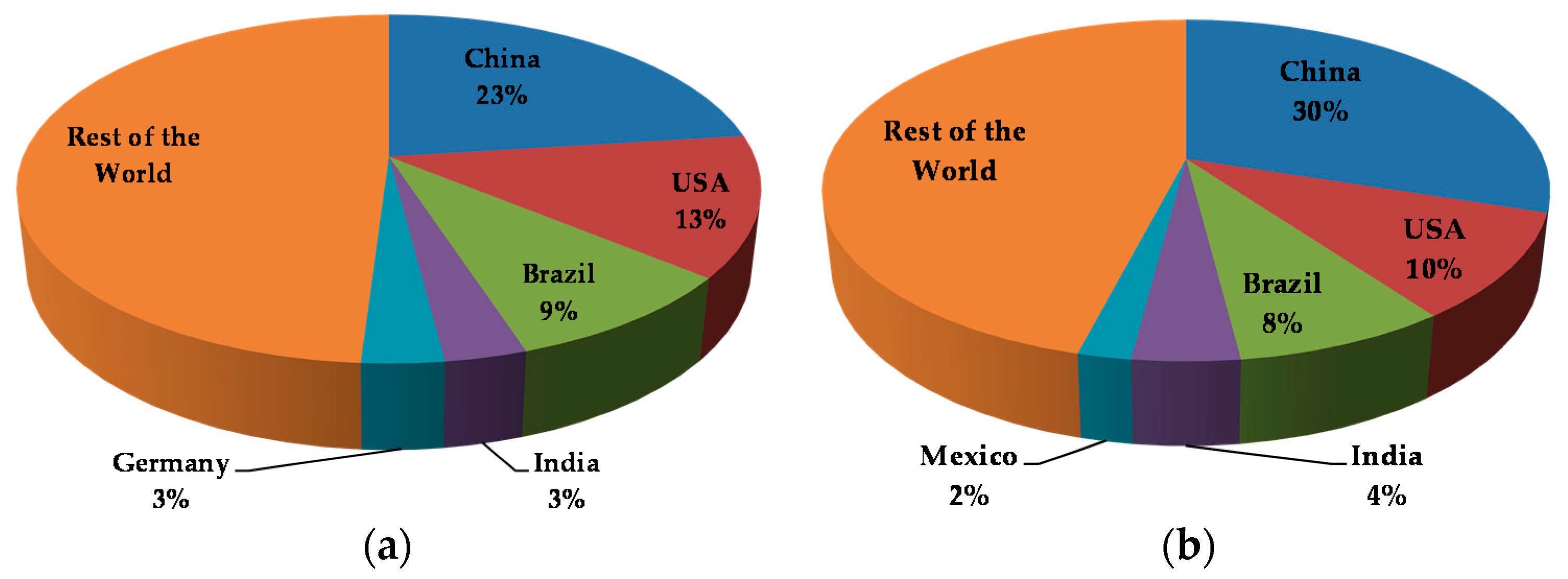
3.2. Current Trends in the Use of Antimicrobials in Food Production Environments
4. Antimicrobial Use and the Development of Resistance in Food Production Environments
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tampa, M.; Sarbu, I.; Matei, C.; Benea, V.; Georgescu, S. Brief History of Syphilis. J. Med. Life 2014, 7, 4–10. [Google Scholar] [PubMed]
- Aminov, R.I.; Otto, M.; Sommer, A. A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 1, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Farber, L. Antibiotics in Food Preservation. Annu. Rev. Microbiol. 1959, 13, 125–140. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. MMBR 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Frankel, R.B.; Kalmijn, A.J.; Amann, R.; Ludwig, W.; Petersen, N.; Arakaki, A.; Matsunaga, T.; Bleil, U.; Kirschvink, J.L.; Sievert, S.M.; et al. Sampling the Antibiotic Resistome. Science 2006, 311, 374–378. [Google Scholar]
- Čižman, M. The use and resistance to antibiotics in the community. Int. J. Antimicrob. Agents 2003, 21, 297–307. [Google Scholar] [CrossRef]
- Tenover, F.C. Mechanisms of antimicrobial resistance in bacteria. Am. J. Infect. Control 2006, 34, S3–S10. [Google Scholar] [CrossRef] [PubMed]
- Starrels, J.L.; Barg, F.K.; Metlay, J.P. Populations at Risk Patterns and Determinants of Inappropriate Antibiotic Use in Injection Drug Users. J. Gen. Intern. Med. 2009, 24, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013; p. 11.
- ECDC/EMEA. Joint Technical Report. The Bacterial Challenge: Time to React; European Centre for Disease Control: Stockholm, Sweden, September 2009.
- United States Food and Drug Administration. Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals; United States Food and Drug Administration: Silver Spring, MD, USA, September 2014. [Google Scholar]
- Wegener, H.C. Antibiotics in animal feed and their role in resistance development. Curr. Opin. Microbiol. 2016, 6, 439–445. [Google Scholar] [CrossRef]
- Heuer, O.E.; Kruse, H.; Grave, K.; Collignon, P.; Karunasagar, I.; Angulo, F.J. Human Health Consequences of Use of Antimicrobial Agents in Aquaculture. Clin. Infect. Dis. 2009, 49, 1248–1253. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, M.K.; Nosanchuk, J.D.; Einstein, A. Use of antibiotics as feed additives: A burning question. Front. Microbiol. 2014, 5, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Varela, M. Molecular mechanisms of bacterial resistance to antimicrobial agents. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education; Méndez-Vilas, A., Ed.; Formatex Research Center, Inc.: Badajoz, Spain, 2013; pp. 522–534. [Google Scholar]
- De Pascale, G.; Wright, G.D. Antibiotic resistance by enzyme inactivation: From mechanisms to solutions. Chembiochem Eur. J. Chem. Biol. 2010, 11, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.; Bush, K. β-Lactam resistance in the 21st century. In Frontiers in Antimicrobial Resistance; American Society for Microbiology: Washington, DC, USA, 2005. [Google Scholar]
- Michael, G.B.; Freitag, C.; Wendlandt, S.; Eidam, C.; Feßler, A.T.; Lopes, G.V.; Kadlec, K.; Schwarz, S. Emerging issues in antimicrobial resistance of bacteria from food-producing animals. Future Microbiol. 2015, 10, 427–443. [Google Scholar] [CrossRef] [PubMed]
- Smet, A.; Martel, A.; Persoons, D.; Dewulf, J.; Heyndrickx, M.; Herman, L.; Haesebrouck, F.; Butaye, P. Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: Molecular aspects, mobility and impact on public health. FEMS Microbiol. Rev. 2010, 34, 295–316. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, R. Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2002, 34, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Pyörälä, S.; Baptiste, K.E.; Catry, B.; van Duijkeren, E.; Greko, C.; Moreno, M.A.; Pomba, M.C.M.F.; Rantala, M.; Ružauskas, M.; Sanders, P.; et al. Macrolides and lincosamides in cattle and pigs: Use and development of antimicrobial resistance. Vet. J. Lond. Engl. 1997 2014, 200, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside modifying enzymes. Drug Resist. Updat. Rev. Comment. Antimicrob. Anticancer Chemother. 2010, 13, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Lambert, P.A. Bacterial resistance to antibiotics: Modified target sites. Adv. Drug Deliv. Rev. 2005, 57, 1471–1485. [Google Scholar] [CrossRef] [PubMed]
- Lambert, T. Antibiotics that affect the ribosome. Rev. Sci. Tech. Int. Off. Epizoot. 2012, 31, 57–64. [Google Scholar] [CrossRef]
- Healy, V.L.; Lessard, I.A.; Roper, D.I.; Knox, J.R.; Walsh, C.T. Vancomycin resistance in enterococci: Reprogramming of the D-ala-D-Ala ligases in bacterial peptidoglycan biosynthesis. Chem. Biol. 2000, 7, R109–R119. [Google Scholar] [CrossRef]
- Collin, F.; Karkare, S.; Maxwell, A. Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Appl. Microbiol. Biotechnol. 2011, 92, 479–497. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I. Bacterial RNA polymerase: A promising target for the discovery of new antimicrobial agents. Curr. Opin. Investig. Drugs Lond. Engl. 2000 2007, 8, 600–607. [Google Scholar]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: quinolone resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12–31. [Google Scholar] [CrossRef] [PubMed]
- Fabrega, A.; Sánchez-Céspedes, J.; Soto, S.; Vila, J. Quinolone resistance in the food chain. Int. J. Antimicrob. Agents 2008, 31, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C. Tetracycline resistance determinants: Mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 1996, 19, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Schnappinger, D.; Hillen, W. Tetracyclines: Antibiotic action, uptake, and resistance mechanisms. Arch. Microbiol. 1996, 165, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Doan, T.-L.; Fung, H.B.; Mehta, D.; Riska, P.F. Tigecycline: A glycylcycline antimicrobial agent. Clin. Ther. 2006, 28, 1079–1106. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Cai, Y.; Liu, X.; Bai, N.; Liang, B.; Wang, R. The emergence of clinical resistance to tigecycline. Int. J. Antimicrob. Agents 2013, 41, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Bacterial resistance to antibiotics as a function of outer membrane permeability. J. Antimicrob. Chemother. 1988, 22, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 2009, 1794, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Preventing drug access to targets: Cell surface permeability barriers and active efflux in bacteria. Semin. Cell Dev. Biol. 2001, 12, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [PubMed]
- Floyd, J.; Kumar, S.; Mukherjee, M.; He, G.; Varela, M. A review of the molecular mechanisms of drug efflux in pathogenic bacteria: A structure-function perspective. In Recent Research Developments in Membrane Biology; Research Signpost Inc.: Kerala, India, 2013; pp. 15–66. [Google Scholar]
- Kumar, S.; Varela, M.F. Biochemistry of bacterial multidrug efflux pumps. Int. J. Mol. Sci. 2012, 13, 4484–4495. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B. Active efflux mechanisms for antimicrobial resistance. Antimicrob. Agents Chemother. 1992, 36, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B. Active efflux, a common mechanism for biocide and antibiotic resistance. Symp. Ser. Soc. Appl. Microbiol. 2002, 65S–71S. [Google Scholar] [CrossRef]
- Saier, M.H.; Paulsen, I.T.; Sliwinski, M.K.; Pao, S.S.; Skurray, R.A.; Nikaido, H. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 1998, 12, 265–274. [Google Scholar]
- Davidson, A.L.; Maloney, P.C. ABC transporters: How small machines do a big job. Trends Microbiol. 2007, 15, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Krämer, R. Functional principles of solute transport systems: Concepts and perspectives. Biochim. Biophys. Acta 1994, 1185, 1–34. [Google Scholar] [CrossRef]
- Poolman, B.; Konings, W.N. Secondary solute transport in bacteria. Biochim. Biophys. Acta 1993, 1183, 5–39. [Google Scholar] [CrossRef]
- Ruggerone, P.; Murakami, S.; Pos, K.M.; Vargiu, A.V. RND efflux pumps: Structural information translated into function and inhibition mechanisms. Curr. Top. Med. Chem. 2013, 13, 3079–3100. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.J.; Saier, M.H. SMR-type multidrug resistance pumps. Curr. Opin. Drug Discov. Devel. 2001, 4, 237–245. [Google Scholar] [PubMed]
- Kumar, S.; Floyd, J.T.; He, G.; Varela, M.F. Bacterial Antimicrobial Efflux Pumps of the MFS and Mate Transporter Families: A Review; Research Signpost Inc.: Kerala, India, 2013; pp. 1–21. [Google Scholar]
- Kuroda, T.; Tsuchiya, T. Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta 2009, 1794, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Ranaweera, I.; Shrestha, U.; Ranjana, K.C.; Kakarla, P.; Willmon, T.M.; Hernandez, A.J.; Mukherjee, M.M.; Barr, S.R.; Varela, M.F. Structural comparison of bacterial multidrug efflux pumps of the major facilitator superfamily. Trends Cell Mol. Biol. 2015, 10, 131–140. [Google Scholar] [PubMed]
- Smith, K.P.; Kumar, S.; Varela, M.F. Identification, cloning, and functional characterization of EmrD-3, a putative multidrug efflux pump of the major facilitator superfamily from Vibrio cholerae O395. Arch. Microbiol. 2009, 191, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.L.; He, G.-X.; Kakarla, P.; Ranjana, K.C.; Kumar, S.; Lakra, W.S.; Mukherjee, M.M.; Ranaweera, I.; Shrestha, U.; Tran, T.; et al. Multidrug efflux pumps from Enterobacteriaceae, Vibrio cholerae and Staphylococcus aureus bacterial food pathogens. Int. J. Environ. Res. Public Health 2015, 12, 1487–1547. [Google Scholar] [CrossRef] [PubMed]
- Floyd, J.L.; Smith, K.P.; Kumar, S.H.; Floyd, J.T.; Varela, M.F. LmrS is a multidrug efflux pump of the major facilitator superfamily from Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 5406–5412. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, U.; Ranaweera, I.; Kumar, S.; Ranjana, K.; Kakarla, P.; lakhra, W.; He, G.; Andersen, J.; Varela, M. Multidrug resistance efflux pumps of Salmonella enterica. In Salmonella: Prevalence, Risk Factors and Treatment Options; Hackett, C.B., Ed.; Nova Science Publishers Inc.: New York, NY, USA, 2015; pp. 1–30. [Google Scholar]
- Kumar, S.; Mukherjee, M.M.; Varela, M.F. Modulation of Bacterial Multidrug Resistance Efflux Pumps of the Major Facilitator Superfamily. Int. J. Bacteriol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; He, G.; Kakarla, P.; Shrestha, U.; Ranjana, K.C.; Ranaweera, I.; Willmon, T.M.; Barr, S.R.; Hernandez, A.J.; Varela, M.F. Bacterial Multidrug Efflux Pumps of the Major Facilitator Superfamily as Targets for Modulation. Infect. Disord. Drug Targets 2016, 16, 28–43. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A. Classics in infectious diseases: On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae by Alexander Fleming, Reprinted from the British Journal of Experimental Pathology 10:226–236, 1929. Rev. Infect. Dis. 1980, 2, 129–139. [Google Scholar] [PubMed]
- Jesman, C.; Młudzik, A.; Cybulska, M. History of antibiotics and sulphonamides discoveries. Pol. Merkur. Lek. Organ Pol. Tow. Lek. 2011, 30, 320–322. [Google Scholar]
- Schatz, A.; Bugie, E.; Waksman, S. Streptomycin, a substance exhibiting antibiotic activity against Gram-positive and Gram-negative bacteria. Proc. Soc. Exptl. Biol. Med. 1944, 55, 66–69. [Google Scholar] [CrossRef]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef] [PubMed]
- Jukes, T.H.; Williams, W.L. Nutritional effects of antibiotics. Pharmacol. Rev. 1953, 5, 381–420. [Google Scholar] [PubMed]
- Wahlstrom, R.C.; Terrill, S.W.; Johnson, B.C. Effect of antibacterial agents on growth of baby pigs fed a “synthetic” diet. Proc. Soc. Exp. Biol. Med. Soc. Exp. Biol. Med. N. Y. N 1950, 75, 710–711. [Google Scholar] [CrossRef]
- Prescott, J.; Baggot, J. Antimicrobial Therapy in Veterinary Medicine, 2nd ed.; Iowa State University Press: Iowa City, IA, USA, 1993. [Google Scholar]
- Butaye, P.; Devriese, L.A.; Haesebrouck, F. Antimicrobial Growth Promoters Used in Animal Feed: Effects of Less Well Known Antibiotics on Gram-Positive Bacteria. Clin. Microbiol. Rev. 2003, 16, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance Review. In Antimicrobials in Agriculture and the Environment: Reducing Unnecessary Use and Wastage. The Review on Antimicrobial Resistance; Antimicrobial Resistance Review (AMR): London, UK, 2015.
- Barton, M.D. Impact of antibiotic use in the Swine industry. Curr. Opin. Microbiol. 2014, 19, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Center for Disease Dynamics, Economics & Policy. The State of the World’s Antibiotics 2015; Center for Disease Dynamics, Economics & Policy: Washington, DC, USA, 2015. [Google Scholar]
- NIAA. Antibiotic Use in Food Animals: A Dialogue for a Common Purpose. In Proceedings of the Symposium by National Institute for Animal Agriculture and Conducted, Chicago, IL, USA, 26–27 October 2011.
- World Health Organization. Critically Important Antimicrobials for Human Medicine 3rd Revision 2011; WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR); World Health Organization: Geneva, Switzerland, 2012. [Google Scholar]
- Hao, H.; Cheng, G.; Iqbal, Z.; Ai, X.; Hussain, H.I.; Huang, L.; Dai, M.; Wang, Y.; Liu, Z.; Yuan, Z. Benefits and risks of antimicrobial use in food-producing animals. Front. Microbiol. 2014, 5, 288. [Google Scholar] [CrossRef] [PubMed]
- Jensen, B. The impact of feed additives on the microbial ecology of the gut in young pigs. J. Anim. Feed Sci. 1998, 7, 45–64. [Google Scholar]
- Stutz, M.W.; Lawton, G.C. Effects of diet and antimicrobials on growth, feed efficiency, intestinal Clostridium perfringens, and ileal weight of broiler chicks. Poult. Sci. 1984, 63, 2036–2042. [Google Scholar] [CrossRef] [PubMed]
- Wegener, H. Use of Antimicrobial Growth Promoters in Food Animals and Enterococcus faecium Resistance to Therapeutic Antimicrobial Drugs in Europe. Emerg. Infect. Dis. 1999, 5, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Wegener, H.C. Historical yearly usage of glycopeptides for animals and humans: The American-European paradox revisited. Antimicrob. Agents Chemother. 1998, 42, 3049. [Google Scholar] [PubMed]
- McEwen, S.A.; Fedorka-Cray, P.J. Antimicrobial use and resistance in animals. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2002, 34, S93–S106. [Google Scholar] [CrossRef] [PubMed]
- Kyriakis, S.C.; Tsinas, A.; Lekkas, S.; Sarris, K.; Bourtzi-Hatzopoulou, E. Clinical evaluation of in-feed zinc bacitracin for the control of porcine intestinal adenomatosis in growing/fattening pigs. Vet. Rec. 1996, 138, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Manning, J.G.; Hargis, B.M.; Hinton, A.; Corrier, D.E.; DeLoach, J.R.; Creger, C.R. Effect of selected antibiotics and anticoccidials on Salmonella enteritidis cecal colonization and organ invasion in Leghorn chicks. Avian Dis. 1994, 38, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.; Wang, W.; Regev-Yochay, G.; Lipsitch, M.; Hanage, W.P. Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl. 2015, 8, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Chantziaras, I.; Boyen, F.; Callens, B.; Dewulf, J. Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: A report on seven countries. J. Antimicrob. Chemother. 2014, 69, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Wegener, H.C.; Collignon, P. Resistance in bacteria of the food chain: Epidemiology and control strategies. Expert Rev. Anti-Infect. Ther. 2008, 6, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Witte, W. Medical consequences of antibiotic use in agriculture. Science 1998, 279, 996–997. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, X.; Li, W.; Lu, X.; Liu, B.; Wang, J. The residues and environmental risks of multiple veterinary antibiotics in animal faeces. Environ. Monit. Assess. 2013, 185, 2211–2220. [Google Scholar] [CrossRef] [PubMed]
- Finley, R.L.; Collignon, P.; Larsson, D.G.J.; McEwen, S.A.; Li, X.-Z.; Gaze, W.H.; Reid-Smith, R.; Timinouni, M.; Graham, D.W.; Topp, E. The Scourge of Antibiotic Resistance: The Important Role of the Environment. Clin. Infect. Dis. 2013, 57, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Cabello, F.C.; Godfrey, H.P.; Tomova, A.; Ivanova, L.; Dölz, H.; Millanao, A.; Buschmann, A.H. Antimicrobial use in aquaculture re-examined: Its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 2013, 15, 1917–1942. [Google Scholar] [CrossRef] [PubMed]
- Starr, M.P.; Reynolds, D.M. Streptomycin resistance of coliform bacteria from turkeys fed streptomycin. Am. J. Public Health Nations Health 1951, 41, 1375–1380. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.E.; Besser, J.M.; Hedberg, C.W.; Leano, F.T.; Bender, J.B.; Wicklund, J.H.; Johnson, B.P.; Moore, K.A.; Osterholm, M.T. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992–1998. Investigation Team. N. Engl. J. Med. 1999, 340, 1525–1532. [Google Scholar] [CrossRef] [PubMed]
- Endtz, H.P.; Ruijs, G.J.; van Klingeren, B.; Jansen, W.H.; van der Reyden, T.; Mouton, R.P. Quinolone resistance in campylobacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. J. Antimicrob. Chemother. 1991, 27, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Dunlop, R.H.; McEwen, S.A.; Meek, A.H.; Clarke, R.C.; Black, W.D.; Friendship, R.M. Associations among antimicrobial drug treatments and antimicrobial resistance of fecal Escherichia coli of swine on 34 farrow-to-finish farms in Ontario, Canada. Prev. Vet. Med. 1998, 34, 283–305. [Google Scholar] [CrossRef]
- Bager, F.; Madsen, M.; Christensen, J.; Aarestrup, F.M. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev. Vet. Med. 1997, 31, 95–112. [Google Scholar] [CrossRef]
- Levy, S.B.; FitzGerald, G.B.; Macone, A.B. Changes in intestinal flora of farm personnel after introduction of a tetracycline-supplemented feed on a farm. N. Engl. J. Med. 1976, 295, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.M.; Levy, S.B. Food Animals and Antimicrobials: Impacts on Human Health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Van Cleef, B.A.G.L.; van Benthem, B.H.B.; Verkade, E.J.M.; van Rijen, M.M.L.; Kluytmans-van den Bergh, M.F.Q.; Graveland, H.; Bosch, T.; Verstappen, K.M.H.W.; Wagenaar, J.A.; Bos, M.E.H.; et al. Livestock-associated MRSA in household members of pig farmers: Transmission and dynamics of carriage, a prospective cohort study. PLoS ONE 2015, 10, e0127190. [Google Scholar] [CrossRef] [PubMed]
- Price, L.B.; Graham, J.P.; Lackey, L.G.; Roess, A.; Vailes, R.; Silbergeld, E. Elevated risk of carrying gentamicin-resistant Escherichia coli among U.S. poultry workers. Environ. Health Perspect. 2007, 115, 1738–1742. [Google Scholar] [CrossRef] [PubMed]
- Klare, I.; Badstübner, D.; Konstabel, C.; Böhme, G.; Claus, H.; Witte, W. Decreased incidence of VanA-type vancomycin-resistant enterococci isolated from poultry meat and from fecal samples of humans in the community after discontinuation of avoparcin usage in animal husbandry. Microb. Drug Resist. Larchmt. N 1999, 5, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Seyfarth, A.M.; Emborg, H.D.; Pedersen, K.; Hendriksen, R.S.; Bager, F. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 2001, 45, 2054–2059. [Google Scholar] [CrossRef] [PubMed]
- Inglis, G.D.; McAllister, T.A.; Busz, H.W.; Yanke, L.J.; Morck, D.W.; Olson, M.E.; Read, R.R. Effects of subtherapeutic administration of antimicrobial agents to beef cattle on the prevalence of antimicrobial resistance in Campylobacter jejuni and Campylobacter hyointestinalis. Appl. Environ. Microbiol. 2005, 71, 3872–3881. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Critically Important Antimicrobials for Human Medicine, 2nd ed.; Department of Food Safety and Zoonoses, World Health Organization: Geneva, Switzerland, 2009. [Google Scholar]
- Gupta, A.; Nelson, J.M.; Barrett, T.J.; Tauxe, R.V.; Rossiter, S.P.; Friedman, C.R.; Joyce, K.W.; Smith, K.E.; Jones, T.F.; Hawkins, M.A.; et al. Antimicrobial resistance among Campylobacter strains, United States, 1997–2001. Emerg. Infect. Dis. 2004, 10, 1102–1109. [Google Scholar] [CrossRef] [PubMed]
- Engberg, J.; Aarestrup, F.M.; Taylor, D.E.; Gerner-Smidt, P.; Nachamkin, I. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: Resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 2001, 7, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.M.; Chiller, T.M.; Powers, J.H.; Angulo, F.J. Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: A public health success story. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2007, 44, 977–980. [Google Scholar] [CrossRef] [PubMed]
- United States Food and Drug Administration. National Antimicrobial Resistance Monitoring System—Enteric Bacteria (NARMS); United States Food and Drug Administration: Silver Spring, MD, USA, 2014. [Google Scholar]
- Alexander, T.W.; Yanke, L.J.; Topp, E.; Olson, M.E.; Read, R.R.; Morck, D.W.; McAllister, T.A. Effect of subtherapeutic administration of antibiotics on the prevalence of antibiotic-resistant Escherichia coli bacteria in feedlot cattle. Appl. Environ. Microbiol. 2008, 74, 4405–4416. [Google Scholar] [CrossRef] [PubMed]
- Varga, C.; Rajić, A.; McFall, M.E.; Reid-Smith, R.J.; Deckert, A.E.; Checkley, S.L.; McEwen, S.A. Associations between reported on-farm antimicrobial use practices and observed antimicrobial resistance in generic fecal Escherichia coli isolated from Alberta finishing swine farms. Prev. Vet. Med. 2009, 88, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Varga, C.; Rajić, A.; McFall, M.E.; Reid-Smith, R.J.; McEwen, S.A. Associations among antimicrobial use and antimicrobial resistance of Salmonella spp. isolates from 60 Alberta finishing swine farms. Foodborne Pathog. Dis. 2009, 6, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Rajić, A.; McFall, M.E.; Deckert, A.E.; Reid-Smith, R.; Manninen, K.; Poppe, C.; Dewey, C.E.; McEwen, S.A. Antimicrobial resistance of Salmonella isolated from finishing swine and the environment of 60 Alberta swine farms. Vet. Microbiol. 2004, 104, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Burow, E.; Simoneit, C.; Tenhagen, B.-A.; Käsbohrer, A. Oral antimicrobials increase antimicrobial resistance in porcine E. coli—A systematic review. Prev. Vet. Med. 2014, 113, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.C.; Wardyn, S.E. Human Infections with Staphylococcus aureus CC398. Curr. Environ. Health Rep. 2015, 2, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.S. Drug resistance in Salmonella typhimurium and its implications. Br. Med. J. 1968, 3, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B. The challenge of antibiotic resistance. Sci. Am. 1998, 278, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B.; FitzGerald, G.B.; Macone, A.B. Spread of antibiotic-resistant plasmids from chicken to chicken and from chicken to man. Nature 1976, 260, 40–42. [Google Scholar] [CrossRef] [PubMed]
- Frost, L.S.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Summers, A.O. Genetic linkage and horizontal gene transfer, the roots of the antibiotic multi-resistance problem. Anim. Biotechnol. 2006, 17, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Witte, W. Selective pressure by antibiotic use in livestock. Int. J. Antimicrob. Agents 2000, 16, S19–S24. [Google Scholar] [CrossRef]
- Giraffa, G. Functionality of enterococci in dairy products. Int. J. Food Microbiol. 2003, 88, 215–222. [Google Scholar] [CrossRef]
- Walther, C.; Rossano, A.; Thomann, A.; Perreten, V. Antibiotic resistance in Lactococcus species from bovine milk: Presence of a mutated multidrug transporter mdt(A) gene in susceptible Lactococcus garvieae strains. Vet. Microbiol. 2008, 131, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Danielsen, M.; Wind, A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 2003, 82, 1–11. [Google Scholar] [CrossRef]
- Bortolaia, V.; Espinosa-Gongora, C.; Guardabassi, L. Human health risks associated with antimicrobial-resistant enterococci and Staphylococcus aureus on poultry meat. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2016, 22, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Szmolka, A.; Nagy, B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front. Microbiol. 2013, 4, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Lin, J.; Pereira, S. Fluoroquinolone-resistant Campylobacter in animal reservoirs: Dynamics of development, resistance mechanisms and ecological fitness. Anim. Health Res. Rev. 2003, 4, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Karthik, K.; Tiwari, R.; Shabbir, M.Z.; Barbuddhe, S.; Malik, S.V.S.; Singh, R.K. Listeriosis in animals, its public health significance (food-borne zoonosis) and advances in diagnosis and control: A comprehensive review. Vet. Q. 2015, 35, 211–235. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vargas, F.M.; Abu-El-Haija, M.A.; Gómez-Duarte, O.G. Salmonella infections: An update on epidemiology, management, and prevention. Travel Med. Infect. Dis. 2011, 9, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, E.; Oprea, S.F.; Donabedian, S.M.; Perri, M.; Bozigar, P.; Bartlett, P.; Zervos, M.J. Epidemiology of antimicrobial resistance in enterococci of animal origin. J. Antimicrob. Chemother. 2005, 55, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Silbergeld, E.K.; Graham, J.; Price, L.B. Industrial food animal production, antimicrobial resistance, and human health. Annu. Rev. Public Health 2008, 29, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Levy, S. The Antibiotic Paradox. How the Misuse of Antibiotics Destroys Their Curative Powers, 2nd ed.; Perseus Publishing: Cambridge, MA, USA, 2002. [Google Scholar]
- EMA (European Medicines Agency) and EFSA (European Food Safety Authority). Joint Scientific Opinion on measures to reduce the need to use antimicrobial agents in animal husbandry in the European Union, and the resulting impacts on food safety (RONAFA). EFSA J. 2017, 15, 4666. [Google Scholar]
- Phillips, I.; Casewell, M.; Cox, T.; Groot, B.D.; Friis, C.; Jones, R.; Nightingale, C.; Preston, R.; Waddell, J. Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J. Antimicrob. Chemother. 2004, 53, 28–52. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, M.; Copeland, D. Is there human health harm following fluoroquinolone use in poultry? In Proceedings of the Fifty-Third Western Poultry Disease Conference, Sacramento, CA, USA, 6–9 March 2004; pp. 27–29.
- World Health Organization. Who Global Principles for the Containment of Antimicrobial Resistance in Animals Intended for Food. Available online: http://www.foodsafetynews.com/WHO_CDS_CSR_APH_2000.4[1].pdf (accessed on 26 August 2016).
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- FAO. Joint FAO/WHO/OIE expert meeting on critically important antimicrobials. In Proceedings of the FAO/WHO/OIE Expert Meeting FAO Headquarters, Rome, Italy, 26–30 November 2007.
- Wierup, M. The Swedish experience of the 1986 year ban of antimicrobial growth promoters, with special reference to animal health, disease prevention, productivity, and usage of antimicrobials. Microb. Drug Resist. 2001, 7, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R. U.S.-EU Poultry Dispute on the Use of Pathogen Reduction Treatments (PRTs); Congressional Research Service: Washington, DC, USA, 2012. [Google Scholar]
- Johnson, R.; Becker, G.S. U.S.-Russia Meat and Poultry Trade Issues; Congressional Research Service: Washington, DC, USA, 2010. [Google Scholar]
- Maron, D.F.; Smith, T.J.; Nachman, K.E. Restrictions on antimicrobial use in food animal production: An international regulatory and economic survey. Glob. Health 2013, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tollefson, L.; Altekruse, S.F.; Potter, M.E. Therapeutic antibiotics in animal feeds and antibiotic resistance. Rev. Sci. Tech. Int. Off. Epizoot. 1997, 16, 709–715. [Google Scholar] [CrossRef]
| Antibiotic Group | Antibiotic(s) Used as Growth Promoters |
|---|---|
| Glycolipids | Bambermycin, avoparcin, ardacin |
| Streptogramins | Virginiamycin |
| Oligosaccharide | Avilamycin |
| Polypeptide | Bacitracin |
| Ionophore | Monensin, salinomycin |
| Macrolide | Tylosin, spiramycin, erythromycin |
| Tetracycline | Chlortetracycline, oxytetracycline |
| Quinoxalines | Carbadox, olaquidox |
| Elfamycin | Efrotomycin |
| Pleuromutilins | Tiamulin |
| β-Lactam | Penicillin |
| Antibiotic Class | Examples of Antibiotics Used in Animals | Importance in Human Medicine * |
|---|---|---|
| β-lactams | Penicillin, amoxicillin, ceftiofur | Critically important |
| Macrolides and lincosamides | Erythromycin, tylosin $, tilmicosin $, tulathromycin $, lincomycin $ | Critically important |
| Aminoglycosides | Gentamicin, neomycin | Critically important |
| Fluroquinolones | Ciprofloxacin, enrofloxacin, danofloxacin $ | Critically important |
| Tetracyclines | Tetracycline, oxytetracycline, chlortetracycline | Highly important |
| Sulfonamides | Several sulfonamides and sulfonamide derivatives | Important |
| Streptogramins | Virginiamycin $ | Highly important |
| Polypeptides | Bacitracin | Important |
| Phenicols | Florfenicol $ | Highly important |
| Pleuromultilin | Tiamulin $ | - |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lekshmi, M.; Ammini, P.; Kumar, S.; Varela, M.F. The Food Production Environment and the Development of Antimicrobial Resistance in Human Pathogens of Animal Origin. Microorganisms 2017, 5, 11. https://doi.org/10.3390/microorganisms5010011
Lekshmi M, Ammini P, Kumar S, Varela MF. The Food Production Environment and the Development of Antimicrobial Resistance in Human Pathogens of Animal Origin. Microorganisms. 2017; 5(1):11. https://doi.org/10.3390/microorganisms5010011
Chicago/Turabian StyleLekshmi, Manjusha, Parvathi Ammini, Sanath Kumar, and Manuel F. Varela. 2017. "The Food Production Environment and the Development of Antimicrobial Resistance in Human Pathogens of Animal Origin" Microorganisms 5, no. 1: 11. https://doi.org/10.3390/microorganisms5010011






