Insight into the Genome of Staphylococcus xylosus, a Ubiquitous Species Well Adapted to Meat Products
Abstract
:1. Occurrence of Staphylococcus xylosus in Meat Products
2. Adaptation to Substrates
2.1. Carbon Substrates
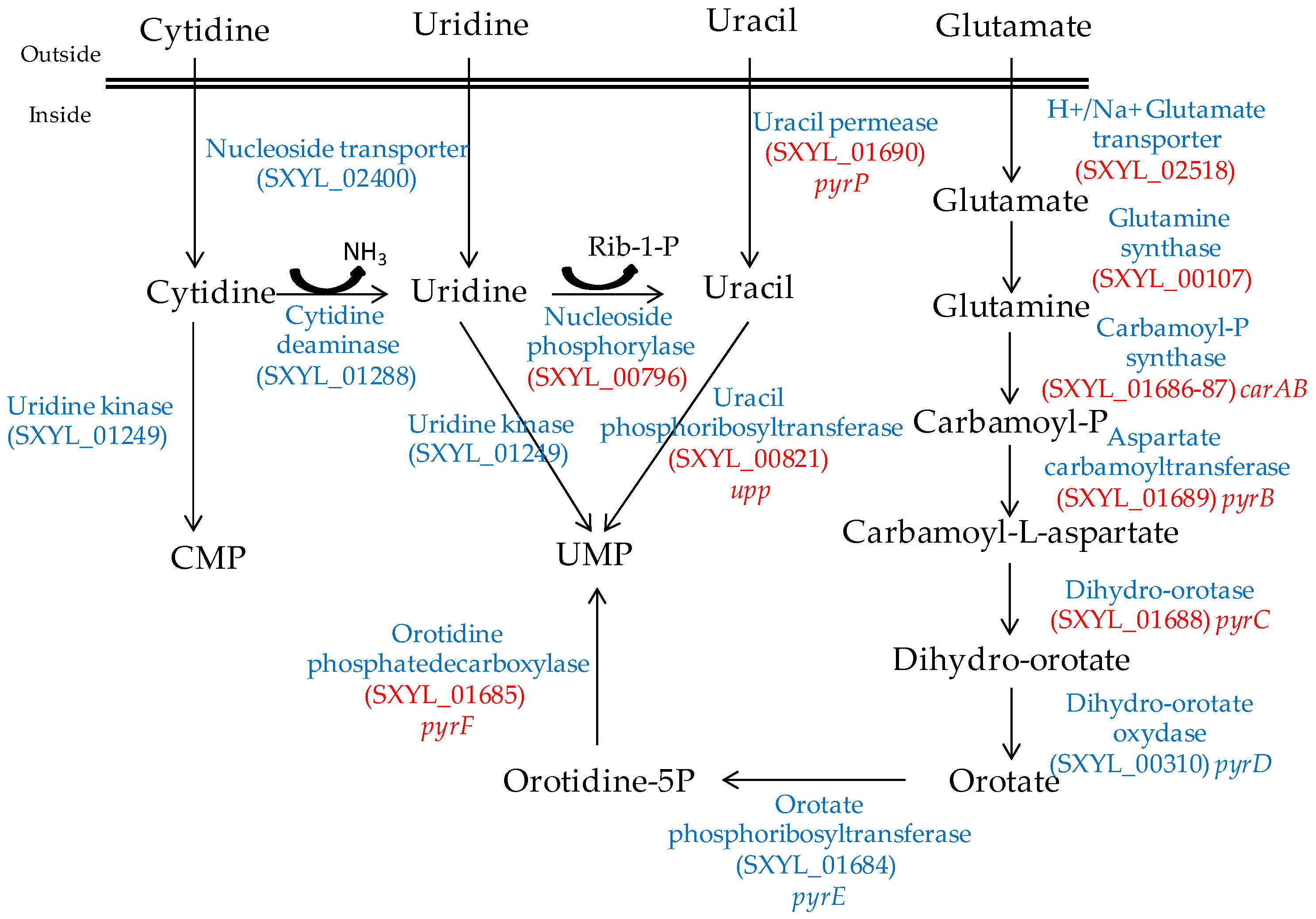
2.2. Nitrogen Substrates, Peptides, Amino Acids
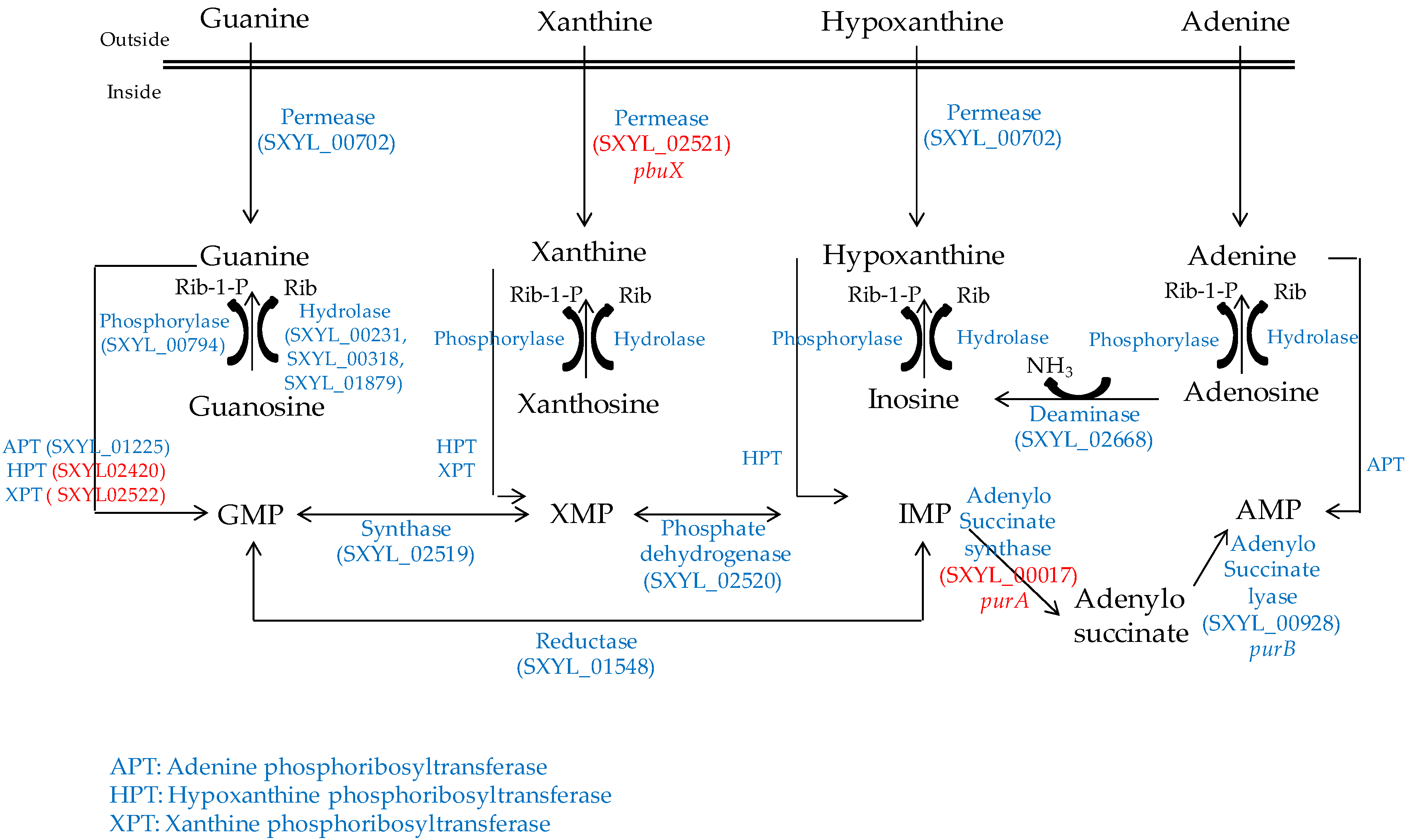
2.3. Nucleosides
2.4. Iron Uptake
3. Adaptation to Stressful Manufacturing Processes
3.1. Osmotic Stress
3.2. Oxidative, Nitrosative Stress
3.3. Acid Stress
4. Functional Properties
4.1. Colour Development
4.2. Flavour Development
4.2.1. Pyruvate Catabolism
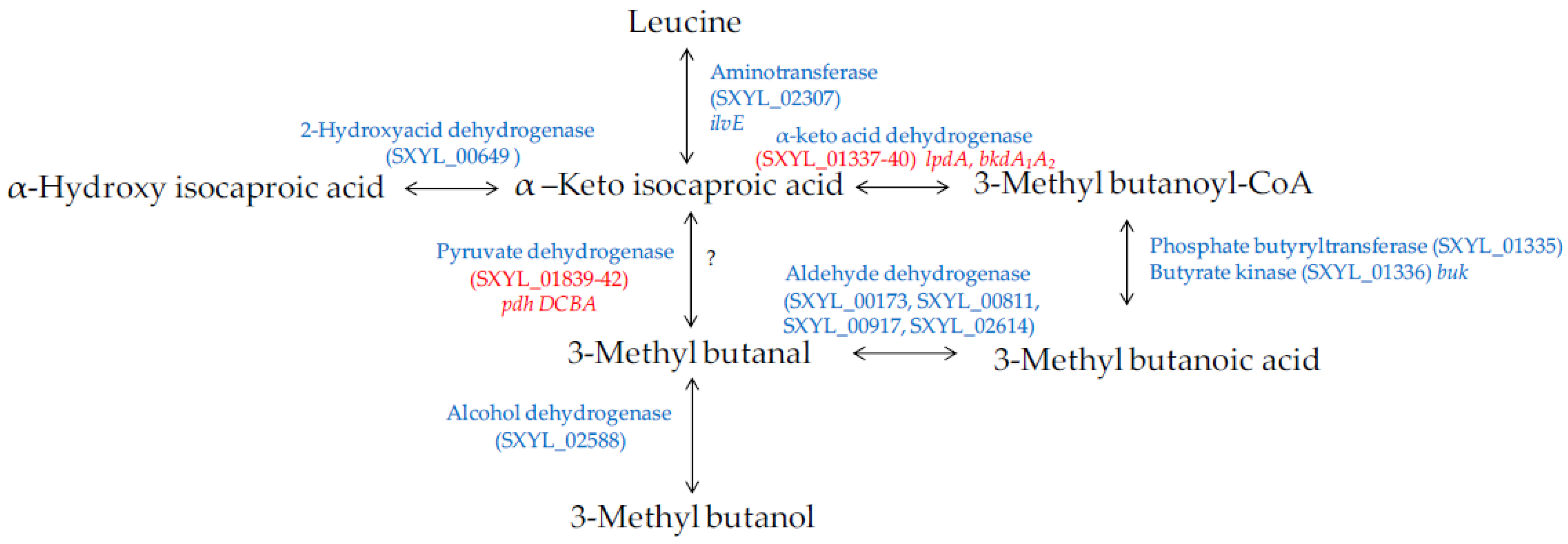
4.2.2. Amino Acid Catabolism
4.2.3. Lipolysis and Fatty Acid Oxidation
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Kloos, W.E.; Zimmerman, R.J.; Smith, R.F. Preliminary studies on the characterization and distribution of Staphylococcus and Micrococcus species on animal skin. Appl. Environ. Microbiol. 1976, 31, 53–59. [Google Scholar] [PubMed]
- Nagase, N.; Sasaki, A.; Yamashita, K.; Shimizu, A.; Wakita, Y.; Kitai, S.; Kawano, J. Isolation and species distribution of staphylococci from animal and human skin. J. Vet. Med. Sci. 2002, 64, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Rantsiou, K.; Iacumin, L.; Urso, R.; Cantoni, C.; Comi, G. Study of the ecology of fresh sausages and characterization of populations of lactic acid bacteria by molecular methods. Appl. Environ. Microbiol. 2004, 70, 1883–1894. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Iacumin, L.; Cantoni, C.; Comi, G.; Cocolin, L. Ecology and characterization by molecular methods of Staphylococcus species isolated from fresh sausages. Int. J. Food Microbiol. 2005, 97, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Chaillou, S.; Chaulot-Talmon, A.; Caekebeke, H.; Cardinal, M.; Christieans, S.; Denis, C.; Desmonts, M.H.; Dousset, X.; Feurer, C.; Hamon, E.; et al. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J. 2015, 9, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Talon, R.; Leroy, S. Fermented meat products and the role of starter cultures. In Encyclopedia of Food Microbiology; Batt, C.A., Tortorello, M.L., Eds.; Elsevier; Academic Press: London, UK, 2014; Volume 1, pp. 870–874. [Google Scholar]
- Ratsimba, A.; Leroy, S.; Chacornac, J.P.; Rakoto, D.; Arnaud, E.; Jeannoda, V.; Talon, R. Staphylococcal ecosystem of kitoza, a traditional malagasy meat product. Int. J. Food Microbiol. 2017, 246, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Manzano, M.; Aggio, D.; Cantoni, C.; Comi, G. A novel polymerase chain reaction (PCR)—Denaturing gradient gel electrophoresis (DGGE) for the identification of micrococcaceae strains involved in meat fermentations. Its application to naturally fermented italian sausages. Meat Sci. 2001, 58, 59–64. [Google Scholar] [CrossRef]
- Coppola, S.; Mauriello, G.; Aponte, M.; Moschetti, G.; Villani, F. Microbial succession during ripening of naples-type salami, a southern italian fermented sausage. Meat Sci. 2000, 56, 321–329. [Google Scholar] [CrossRef]
- Rossi, F.; Tofalo, R.; Torriani, S.; Suzzi, G. Identification by 16S-23S rDNA intergenic region amplification, genotypic and phenotypic clustering of Staphylococcus xylosus strains from dry sausages. J. Appl. Microbiol. 2001, 90, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Drosinos, E.H.; Mataragas, M.; Xiraphi, N.; Moschonas, G.; Gaitis, F.; Metaxopoulos, J. Characterization of the microbial flora from a traditional greek fermented sausage. Meat Sci. 2005, 69, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Garcίa-Varona, M.; Santos, E.M.; Jaime, I.; Rovira, J. Characterisation of micrococcaceae isolated from different varieties of chorizo. Int. J. Food Microbiol. 2000, 54, 189–195. [Google Scholar] [CrossRef]
- Greppi, A.; Ferrocino, I.; La Storia, A.; Rantsiou, K.; Ercolini, D.; Cocolin, L. Monitoring of the microbiota of fermented sausages by culture independent rRNA-based approaches. Int. J. Food Microbiol. 2015, 212, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Aymerich, T.; Martίn, B.; Garriga, M.; Hugas, M. Microbial quality and direct PCR identification of lactic acid bacteria and nonpathogenic staphylococci from artisanal low-acid sausages. Appl. Environ. Microbiol. 2003, 69, 4583–4594. [Google Scholar] [CrossRef] [PubMed]
- Leroy, S.; Giammarinaro, P.; Chacornac, J.P.; Lebert, I.; Talon, R. Biodiversity of indigenous staphylococci of naturally fermented dry sausages and manufacturing environments of small-scale processing units. Food Microbiol. 2010, 27, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Połka, J.; Rebecchi, A.; Pisacane, V.; Morelli, L.; Puglisi, E. Bacterial diversity in typical italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiol. 2015, 46, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Aquilanti, L.; Garofalo, C.; Osimani, A.; Clementi, F. Ecology of lactic acid bacteria and coagulase-negative cocci in fermented dry sausages manufactured in italy and other mediterranean countries: An overview. Int. Food Res. J. 2016, 23, 429–445. [Google Scholar]
- Rebecchi, A.; Pisacane, V.; Miragoli, F.; Połka, J.; Falasconi, I.; Morelli, L.; Puglisi, E. High-throughput assessment of bacterial ecology in hog, cow and ovine casings used in sausages production. Int. J. Food Microbiol. 2015, 212, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Rodrίguez, M.; Núñez, F.; Córdoba, J.J.; Sanabria, C.; Bermúdez, E.; Asensio, M.A. Characterization of Staphylococcus spp. and Micrococcus spp. isolated from iberian ham throughout the ripening process. Int. J. Food Microbiol. 1994, 24, 329–335. [Google Scholar] [CrossRef]
- Vilar, I.; García Fontán, M.C.; Prieto, B.; Tornadijo, M.E.; Carballo, J. A survey on the microbiological changes during the manufacture of dry-cured lacon, a spanish traditional meat product. J. Appl. Microbiol. 2000, 89, 1018–1026. [Google Scholar] [CrossRef] [PubMed]
- Cordero, M.R.; Zumalacárregui, J.M. Characterization of micrococcaceae isolated from salt used for spanish dry-cured ham. Lett. Appl. Microbiol. 2000, 31, 303–306. [Google Scholar] [CrossRef] [PubMed]
- Blaiotta, G.; Pennacchia, C.; Villani, F.; Ricciardi, A.; Tofalo, R.; Parente, E. Diversity and dynamics of communities of coagulase-negative staphylococci in traditional fermented sausages. J. Appl. Microbiol. 2004, 97, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Corbière Morot-Bizot, S.; Leroy, S.; Talon, R. Monitoring of staphylococcal starters in two french processing plants manufacturing dry fermented sausages. J. Appl. Microbiol. 2007, 102, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.J.; El Haddad, L.; Tremblay, D.M.; Plante, P.L.; Wasserscheid, J.; Dumaresq, J.; Dewar, K.; Corbeil, J.; Moineau, S. First complete genome sequence of Staphylococcus xylosus, a meat starter culture and a host to propagate Staphylococcus aureus phages. Genome Announc. 2014, 2, e00671-14. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.P.; Jiang, J.; Tun, H.M.; Mauroo, N.F.; Yuen, C.S.; Leung, F.C. Complete genome sequence of Staphylococcus xylosus HKUOPL8, a potential opportunistic pathogen of mammals. Genome Announc. 2014, 2, e00653-14. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Liu, L.; Liu, S.; Yang, D.; Liu, Y.; Yang, S.; Jia, A.; Qin, N. Genome of Staphylococcus xylosus and comparison with S. aureus and S. epidermidis. J. Genet. Genom. 2014, 41, 413–416. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Arora, A.; Sathyabama, S.; Mubin, N.; Verma, S.; Mayilraj, S.; Agrewala, J.N. Genome sequencing, assembly, annotation and analysis of Staphylococcus xylosus strain DMB3-Bh1 reveals genes responsible for pathogenicity. Gut Pathog. 2016, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Toldrá, F.; Reig, M. The biochemistry of meat and fat. In Handbook of Fermented Meat and Poultry, 2nd ed.; Toldrá, F., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2015; pp. 49–54. [Google Scholar]
- Götz, F.; Bannerman, T.; Schleifer, K.-H. The genera Staphylococcus and Macrococcus. Prokaryotes 2006, 4, 5–75. [Google Scholar] [CrossRef]
- Brückner, R.; Rosenstein, R. Carbohydrate catabolism: Pathways and regulation—Chapter 34. In Gram-Positive Pathogens, 2nd ed.; Fischetti, V., Novick, R., Ferretti, J., Portnoy, D., Rood, J., Eds.; American Society of Microbiology Press: Washington, DC, USA, 2006; pp. 427–433. [Google Scholar]
- Fiegler, H.; Bassias, J.; Jankovic, I.; Brückner, R. Identification of a gene in Staphylococcus xylosus encoding a novel glucose uptake protein. J. Bacteriol. 1999, 181, 4929–4936. [Google Scholar] [PubMed]
- Jankovic, I.; Brückner, R. Carbon catabolite repression by the catabolite control protein CcpA in Staphylococcus xylosus. J. Mol. Microbiol. Biotechnol. 2002, 4, 309–314. [Google Scholar] [PubMed]
- Christiansen, I.; Hengstenberg, W. Staphylococcal phosphoenolpyruvate-dependent phosphotransferase system—Two highly similar glucose permeases in Staphylococcus carnosus with different glucoside specificity: Protein engineering in vivo? Microbiology 1999, 145, 2881–2889. [Google Scholar] [CrossRef] [PubMed]
- Arkouledos, J.S.; Nychas, G.J. Comparative studies of the growth of Staphylococcus carnosus with or without glucose. Lett. Appl. Microbiol. 1995, 20, 19–24. [Google Scholar]
- Ferreira, M.T.; Manso, A.S.; Gaspar, P.; Pinho, M.G.; Neves, A.R. Effect of oxygen on glucose metabolism: Utilization of lactate in Staphylococcus aureus as revealed by in vivo NMR studies. PLoS ONE 2013, 8, e58277. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Mainar, M.; Matheuse, F.; De Vuyst, L.; Leroy, F. Effects of glucose and oxygen on arginine metabolism by coagulase-negative staphylococci. Food Microbiol. 2017, 65, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Seidl, K.; Muller, S.; Francois, P.; Kriebitzsch, C.; Schrenzel, J.; Engelmann, S.; Bischoff, M.; Berger-Bachi, B. Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 2009, 9, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vermassen, A.; Dordet-Frisoni, E.; de La Foye, A.; Micheau, P.; Laroute, V.; Leroy, S.; Talon, R. Adaptation of Staphylococcus xylosus to nutrients and osmotic stress in a salted meat model. Front. Microbiol. 2016, 7, 87. [Google Scholar] [CrossRef] [PubMed]
- Brückner, R.; Wagner, E.; Götz, F. Characterization of a sucrase gene from Staphylococcus xylosus. J. Bacteriol. 1993, 175, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Jankovic, I.; Brückner, R. Carbon catabolite repression of sucrose utilization in Staphylococcus xylosus: Catabolite control protein CcpA ensures glucose preference and autoregulatory limitation of sucrose utilization. J. Mol. Microbiol. Biotechnol. 2007, 12, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Gering, M.; Brückner, R. Transcriptional regulation of the sucrase gene of Staphylococcus xylosus by the repressor ScrR. J. Bacteriol. 1996, 178, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Poolman, B.; Knol, J.; van der Does, C.; Henderson, P.J.; Liang, W.J.; Leblanc, G.; Pourcher, T.; Mus-Veteau, I. Cation and sugar selectivity determinants in a novel family of transport proteins. Mol. Microbiol. 1996, 19, 911–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassias, J.; Brückner, R. Regulation of lactose utilization genes in Staphylococcus xylosus. J. Bacteriol. 1998, 180, 2273–2279. [Google Scholar] [PubMed]
- Hughes, M.C.; Kerry, J.P.; Arendt, E.K.; Kenneally, P.M.; McSweeney, P.L.; O’Neill, E.E. Characterization of proteolysis during the ripening of semi-dry fermented sausages. Meat Sci. 2002, 62, 205–216. [Google Scholar] [CrossRef]
- Miralles, M.C.; Flores, J.; Perez-Martinez, G. Biochemical tests for the selection of Staphylococcus strains as potential meat starter cultures. Food Microbiol. 1996, 13, 227–236. [Google Scholar] [CrossRef]
- Aro Aro, J.M.; Nyam-Osor, P.; Tsuji, K.; Shimada, K.I.; Fukushima, M.; Sekikawa, M. The effect of starter cultures on proteolytic changes and amino acid content in fermented sausages. Food Chem. 2010, 119, 279–285. [Google Scholar] [CrossRef]
- Monnet, V. Bacterial oligopeptide-binding proteins. Cell. Mol. Life Sci. 2003, 60, 2100–2114. [Google Scholar] [CrossRef] [PubMed]
- Fiegler, H.; Brückner, R. Identification of the serine acetyltransferase gene of Staphylococcus xylosus. FEMS Microbiol. Lett. 1997, 148, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Majerczyk, C.D.; Dunman, P.M.; Luong, T.T.; Lee, C.Y.; Sadykov, M.R.; Somerville, G.A.; Bodi, K.; Sonenshein, A.L. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 2010, 192, 2861–2877. [Google Scholar] [CrossRef] [PubMed]
- Nuxoll, A.S.; Halouska, S.M.; Sadykov, M.R.; Hanke, M.L.; Bayles, K.W.; Kielian, T.; Powers, R.; Fey, P.D. CcpA regulates arginine biosynthesis in Staphylococcus aureus through repression of proline catabolism. PLoS Pathog. 2012, 8, e1003033. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Mainar, M.; Weckx, S.; Leroy, F. Coagulase-negative staphylococci favor conversion of arginine into ornithine despite a widespread genetic potential for nitric oxide synthase activity. Appl. Environ. Microbiol. 2014, 80, 7741–7751. [Google Scholar] [CrossRef] [PubMed]
- Janssens, M.; Van der Mijnsbrugge, A.; Sanchez Mainar, M.; Balzarini, T.; De Vuyst, L.; Leroy, F. The use of nucleosides and arginine as alternative energy sources by coagulase-negative staphylococci in view of meat fermentation. Food Microbiol. 2014, 39, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Vermassen, A.; de la Foye, A.; Loux, V.; Talon, R.; Leroy, S. Transcriptomic analysis of Staphylococcus xylosus in the presence of nitrate and nitrite in meat reveals its response to nitrosative stress. Front. Microbiol. 2014, 5, 691. [Google Scholar] [CrossRef] [PubMed]
- Batlle, N.; Aristoy, M.C.; Toldrá, F. ATP metabolites during aging of exudative and nonexudative pork meats. J. Food Sci. 2001, 66, 68–71. [Google Scholar] [CrossRef]
- Switzer, R.L.; Zalkin, H.; Saxild, H.H. Purine, pyrimidine, and pyridine nucleotide metabolism. In Bacillus Subtilis and Its Closest Relatives; Sonenshein, A., Hoch, J.A., Losick, R., Eds.; American Society of Microbiology Press: Washington, DC, USA, 2002. [Google Scholar]
- Chaillou, S.; Champomier-Vergès, M.C.; Cornet, M.; Crutz-Le Coq, A.M.; Dudez, A.M.; Martin, V.; Beaufils, S.; Darbon-Rongère, E.; Bossy, R.; Loux, V.; et al. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 2005, 23, 1527–1533. [Google Scholar] [CrossRef] [PubMed]
- Rimaux, T.; Vrancken, G.; Vuylsteke, B.; De Vuyst, L.; Leroy, F. The pentose moiety of adenosine and inosine is an important energy source for the fermented-meat starter culture Lactobacillus sakei CTC 494. Appl. Environ. Microbiol. 2011, 77, 6539–6550. [Google Scholar] [CrossRef] [PubMed]
- Batlle, N.; Aristoy, M.C.; Toldrá, F. Early postmortem detection of exudative pork meat based on nucleotide content. J. Food Sci. 2000, 65, 413–416. [Google Scholar] [CrossRef]
- Vermassen, A.; Talon, R.; Leroy, S. Ferritin, an iron source in meat for Staphylococcus xylosus? Int. J. Food Microbiol. 2016, 225, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Skaar, E.P.; Schneewind, O. Iron-regulated surface determinants (isd) of Staphylococcus aureus: Stealing iron from heme. Microbes Infect. 2004, 6, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, J.R.; Heinrichs, D.E. The iron-regulated staphylococcal lipoproteins. Front. Cell. Infect. Microbiol. 2012, 2, 41. [Google Scholar] [CrossRef] [PubMed]
- Hammer, N.D.; Skaar, E.P. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 2011, 65, 129–147. [Google Scholar] [CrossRef] [PubMed]
- Rosenstein, R.; Futter-Bryniok, D.; Götz, F. The choline-converting pathway in Staphylococcus xylosus C2a: Genetic and physiological characterization. J. Bacteriol. 1999, 181, 2273–2278. [Google Scholar] [PubMed]
- Hammes, W.P. Metabolism of nitrate in fermented meats: The characteristic feature of a specific group of fermented foods. Food Microbiol. 2012, 29, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Gaupp, R.; Ledala, N.; Somerville, G.A. Staphylococcal response to oxidative stress. Front. Cell. Infect. Microbiol. 2012, 2, 33. [Google Scholar] [CrossRef] [PubMed]
- Barrière, C.; Brückner, R.; Talon, R. Characterization of the single superoxide dismutase of Staphylococcus xylosus. Appl. Environ. Microbiol. 2001, 67, 4096–4104. [Google Scholar] [CrossRef] [PubMed]
- Rosenstein, R.; Nerz, C.; Biswas, L.; Resch, A.; Raddatz, G.; Schuster, S.C.; Götz, F. Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl. Environ. Microbiol. 2009, 75, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Blaiotta, G.; Fusco, V.; Ercolini, D.; Pepe, O.; Coppola, S. Diversity of Staphylococcus species strains based on partial kat (catalase) gene sequences and design of a PCR-restriction fragment length polymorphism assay for identification and differentiation of coagulase-positive species (S. aureus, S. delphini, S. hyicus, S. intermedius, S. pseudintermedius, and S. schleiferi subsp. coagulans). J. Clin. Microbiol. 2010, 48, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Barrière, C.; Brückner, R.; Centeno, D.; Talon, R. Characterisation of the katA gene encoding a catalase and evidence for at least a second catalase activity in Staphylococcus xylosus, bacteria used in food fermentation. FEMS Microbiol. Lett. 2002, 216, 277–283. [Google Scholar] [CrossRef]
- Tavares, A.F.; Nobre, L.S.; Melo, A.M.; Saraiva, L.M. A novel nitroreductase of Staphylococcus aureus with S-nitrosoglutathione reductase activity. J. Bacteriol. 2009, 191, 3403–3406. [Google Scholar] [CrossRef] [PubMed]
- Ras, G.; Zuliani, V.; Derkx, P.; Seibert, T.M.; Leroy, S.; Talon, R. Evidence for nitric oxide synthase activity in Staphylococcus xylosus mediating nitrosoheme formation. Front. Microbiol. 2017, 8, 598. [Google Scholar] [CrossRef] [PubMed]
- Van Sorge, N.M.; Beasley, F.C.; Gusarov, I.; Gonzalez, D.J.; von Köckritz-Blickwede, M.; Anik, S.; Borkowski, A.W.; Dorrestein, P.C.; Nudler, E.; Nizet, V. Methicillin-resistant Staphylococcus aureus bacterial nitric-oxide synthase affects antibiotic sensitivity and skin abscess development. J. Biol. Chem. 2013, 288, 6417–6426. [Google Scholar] [CrossRef] [PubMed]
- Sapp, A.M.; Mogen, A.B.; Almand, E.A.; Rivera, F.E.; Shaw, L.N.; Richardson, A.R.; Rice, K.C. Contribution of the nos-pdt operon to virulence phenotypes in methicillin-sensitive Staphylococcus aureus. PLoS ONE 2014, 9, e108868. [Google Scholar] [CrossRef] [PubMed]
- Clauditz, A.; Resch, A.; Wieland, K.P.; Peschel, A.; Götz, F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 2006, 74, 4950–4953. [Google Scholar] [CrossRef] [PubMed]
- Uziel, O.; Borovok, I.; Schreiber, R.; Cohen, G.; Aharonowitz, Y. Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 2004, 186, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Moskovitz, J. Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: Expression of activity androles in tolerance of oxidative stress. Microbiology 2003, 149, 2739–2747. [Google Scholar] [CrossRef] [PubMed]
- Demeyer, D.; Raemaekers, M.; Rizzo, A.; Holck, A.; De Smedt, A.; Ten Brink, B.; Hagen, B.; Montel, C.; Zanardi, E.; Murbrekk, E.; et al. Control of bioflavour and safety in fermented sausages: First results of a european project. Food Res. Int. 2000, 33, 171–180. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C. Surviving the acid test: Responses of Gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 2003, 67, 429–453. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, F.C.; Baddiley, J. A continuum of anionic charge: Structures and functions of d-alanyl-teichoic acids in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 686–723. [Google Scholar] [CrossRef] [PubMed]
- Boyd, D.A.; Cvitkovitch, D.G.; Bleiweis, A.S.; Kiriukhin, M.Y.; Debabov, D.V.; Neuhaus, F.C.; Hamilton, I.R. Defects in d-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J. Bacteriol. 2000, 182, 6055–6065. [Google Scholar] [CrossRef] [PubMed]
- Bore, E.; Langsrud, S.; Langsrud, O.; Rode, T.M.; Holck, A. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 2007, 153, 2289–2303. [Google Scholar] [CrossRef] [PubMed]
- Leroy, S.; Vermassen, A.; Talon, R. Staphylococcus: Occurrence and properties. In Encyclopedia of Food and Health; Cabalerro, B., Finglas, P.M., Toldrá, F., Eds.; Elsevier Limited Publisher: Oxford, UK, 2016; Volume 5, pp. 104–145. [Google Scholar]
- Sánchez Mainar, M.; Stavropoulou, D.A.; Leroy, F. Exploring the metabolic heterogeneity of coagulase-negative staphylococci to improve the quality and safety of fermented meats: A review. Int. J. Food Microbiol. 2016, 247, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Cocconcelli, P.S.; Fontana, C. Bacteria. In Handbook of Fermented Meat and Poultry; Toldrá, F., Hui, H., Astiasarán, I., Sebranek, J.G., Talon, R., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2014; pp. 117–128. [Google Scholar]
- Sánchez Mainar, M.; Leroy, F. Process-driven bacterial community dynamics are key to cured meat colour formation by coagulase-negative staphylococci via nitrate reductase or nitric oxide synthase activities. Int. J. Food Microbiol. 2015, 212, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Mauriello, G.; Casaburi, A.; Blaiotta, G.; Villani, F. Isolation and technological properties of coagulase negative staphylococci from fermented sausages of southern italy. Meat Sci. 2004, 67, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Pantel, I.; Lindgren, P.E.; Neubauer, H.; Götz, F. Identification and characterization of the Staphylococcus carnosus nitrate reductase operon. Mol. Gen. Genet. 1998, 259, 105–114. [Google Scholar] [PubMed]
- Neubauer, H.; Pantel, I.; Götz, F. Molecular characterization of the nitrite-reducing system of Staphylococcus carnosus. J. Bacteriol. 1999, 181, 1481–1488. [Google Scholar] [PubMed]
- Schlag, S.; Fuchs, S.; Nerz, C.; Gaupp, R.; Engelmann, S.; Liebeke, M.; Lalk, M.; Hecker, M.; Götz, F. Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J. Bacteriol. 2008, 190, 7847–7858. [Google Scholar] [CrossRef] [PubMed]
- Morita, H.; Sakata, R.; Nagata, Y. Nitric oxide complex of iron(ii) myoglobin converted from metmyoglobin by Staphylococcus xylosus. J. Food Sci. 1998, 63, 352–355. [Google Scholar] [CrossRef]
- Li, P.; Kong, B.; Chen, Q.; Zheng, D.; Liu, N. Formation and identification of nitrosylmyoglobin by Staphylococcus xylosus in raw meat batters: A potential solution for nitrite substitution in meat products. Meat Sci. 2013, 93, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Sondergaard, A.K.; Stahnke, L.H. Growth and aroma production by Staphylococcus xylosus, S. carnosus and S. equorum—A comparative study in model systems. Int. J. Food Microbiol. 2002, 75, 99–109. [Google Scholar] [CrossRef]
- Stahnke, L.H. Volatiles produced by Staphylococcus xylosus and Staphylococcus carnosus during growth in sausage minces—Part II. The influence of growth parameters. LWT Food Sci. Technol. 1999, 32, 365–371. [Google Scholar] [CrossRef]
- Talon, R.; Leroy-Setrin, S.; Fadda, S. Dry fermented sausages. In Handbook of Food and Beverage Fermentation Technology; Hui, Y.H., Meunier-Goddick, L., Solvejg Hansen, A., Josephsen, J., Nip, W.K., Stanfield, P.S., Toldrá, F., Eds.; Marcel Dekker, Inc.: New York, NY, USA, 2004; pp. 397–416. [Google Scholar]
- Ravyts, F.; Steen, L.; Goemaere, O.; Paelinck, H.; De Vuyst, L.; Leroy, F. The application of staphylococci with flavour-generating potential is affected by acidification in fermented dry sausages. Food Microbiol. 2010, 27, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Flores, M.; Olivares, A. Flavor. In Handbook of Fermented Meat and Poultry, 2nd ed.; Toldrá, F., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2015; pp. 217–226. [Google Scholar]
- Møller, J.K.S.; Hinrichsen, L.L.; Andersen, H.J. Formation of amino acid (l-leucine, l-phenylalanine) derived volatile flavour compounds by Moraxella phenylpyruvica and Staphylococcus xylosus in cured meat model systems. Int. J. Food Microbiol. 1998, 42, 101–117. [Google Scholar] [CrossRef]
- Ravyts, F.; Vrancken, G.; D’Hondt, K.; Vasilopoulos, C.; De Vuyst, L.; Leroy, F. Kinetics of growth and 3-methyl-1-butanol production by meat-borne, coagulase-negative staphylococci in view of sausage fermentation. Int. J. Food Microbiol. 2009, 134, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulou, D.A.; Borremans, W.; De Vuyst, L.; De Smet, S.; Leroy, F. Amino acid conversions by coagulase-negative staphylococci in a rich medium: Assessment of inter- and intraspecies heterogeneity. Int. J. Food Microbiol. 2015, 212, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Tjener, K.; Stahnke, L.H.; Andersen, L.; Martinussen, J. The pH-unrelated influence of salt, temperature and manganese on aroma formation by Staphylococcus xylosus and Staphylococcus carnosus in a fermented meat model system. Int. J. Food Microbiol. 2004, 97, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Olesen, P.T.; Meyer, A.S.; Stahnke, L.H. Generation of flavour compounds in fermented sausages-the influence of curing ingredients, Staphylococcus starter culture and ripening time. Meat Sci. 2004, 66, 675–687. [Google Scholar] [CrossRef]
- Beck, H.C.; Hansen, A.M.; Lauritsen, F.R. Catabolism of leucine to branched-chain fatty acids in Staphylococcus xylosus. J. Appl. Microbiol. 2004, 96, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Beck, H.C.; Hansen, A.M.; Lauritsen, F.R. Metabolite production and kinetics of branched-chain aldehyde oxidation in Staphylococcus xylosus. Enzym. Microb. Technol. 2002, 31, 94–101. [Google Scholar] [CrossRef]
- Yvon, M.; Rijnen, L. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 2001, 11, 185–201. [Google Scholar] [CrossRef]
- Talon, R.; Leroy-Sétrin, S.; Fadda, S. Bacterial starters involved in the quality of fermented meat products—Chapter 10. In Research Advances in Quality of Meat and Meat Products; Toldrá, F., Ed.; Research Signpost: Trivandrum, Kerala, India, 2002; pp. 175–191. [Google Scholar]
- Rosenstein, R.; Götz, F. Staphylococcal lipases: Biochemical and molecular characterization. Biochimie 2000, 82, 1005–1014. [Google Scholar] [CrossRef]
- Mosbah, H.; Sayari, A.; Mejdoub, H.; Dhouib, H.; Gargouri, Y. Biochemical and molecular characterization of Staphylococcus xylosus lipase. Biochim. Biophys. Acta 2005, 1723, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Bouaziz, A.; Horchani, H.; Ben Salem, N.; Gargouri, Y.; Sayari, A. Expression, purification of a novel alkaline Staphylococcus xylosus lipase acting at high temperature. Biochem. Eng. J. 2011, 54, 93–102. [Google Scholar] [CrossRef]
- Bertoldo, J.B.; Razzera, G.; Vernal, J.; Brod, F.C.; Arisi, A.C.; Terenzi, H. Structural stability of Staphylococcus xylosus lipase is modulated by Zn(2+) ions. Biochim. Biophys. Acta 2011, 1814, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- Brod, F.C.; Pelisser, M.R.; Bertoldo, J.B.; Vernal, J.; Bloch, C., Jr.; Terenzi, H.; Arisi, A.C. Heterologous expression and purification of a heat-tolerant Staphylococcus xylosus lipase. Mol. Biotechnol. 2010, 44, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Iacumin, L.; Cocolin, L.; Cantoni, C.; Comi, G. Preliminary analysis of the lipase gene (gehM) expression of Staphylococcus xylosus in vitro and during fermentation of naturally fermented sausages (in situ). J. Food Prot. 2007, 70, 2665–2669. [Google Scholar] [CrossRef] [PubMed]
- Talon, R.; Walter, D.; Montel, M.C. Growth and effect of staphylococci and lactic acid bacteria on unsaturated free fatty acids. Meat Sci. 2000, 54, 41–47. [Google Scholar] [CrossRef]
- Barrière, C.; Centeno, D.; Lebert, A.; Leroy-Sétrin, S.; Berdague, J.L.; Talon, R. Roles of superoxide dismutase and catalase of Staphylococcus xylosus in the inhibition of linoleic acid oxidation. FEMS Microbiol. Lett. 2001, 201, 181–185. [Google Scholar] [CrossRef]
- Talon, R.; Leroy, S. Diversity and safety hasards of bacteria involved in meat fermentations. Meat Sci. 2011, 89, 303–309. [Google Scholar] [CrossRef] [PubMed]


| Phosphotransferase System (PTS) | PTS-Independent | ||||
|---|---|---|---|---|---|
| Glucose | Glucose | ||||
| SXYL_00369 | ptsG | EIICBA | SXYL_00698 | gdh | Glucose 1-dehydrogenase |
| SXYL_00253 | EIIBC | SXYL_00699 | glcU | Glucose uptake protein | |
| SXYL_01421 | crr | EIIA | SXYL_01308 | glkA | Glucokinase |
| Sucrose | Lactate | ||||
| SXYL_00555 | scrA | EIIBC | SXYL_00250 | Lactate permease | |
| SXYL_00886 | scrR | Repressor | SXYL_00577 | Lactate permease | |
| SXYL_00887-88 | scrBK | Hydrolase, Fructokinase | SXYL_00170 | lactate-quinone oxidoreductase | |
| others | Lactose | ||||
| SXYL_00060, SXYL_00626 | Arbutin, EIIBC | SXYL_00082 | lacR | Transcription activator | |
| SXYL_00528 | Beta-glucoside, EIIABC | SXYL_00083-84 | lacPH | Permease, Beta-galactosidase | |
| SXYL_00257-60 | Cellobiose, EIIC, EIIBA | SXYL_00671 | galR | Transcriptional regulator | |
| SXYL_02148-50 | fruAK | Fructose, EIIABC, catabolism, repression | SXYL_00672-74 | galKET | Galactokinase, epimerase, P-uridylyltransferase |
| SXYL_00277-78 | Fructose, regulation, EII | ||||
| SXYL_00773-76 | mtlD, A | Mannitol, EIIACB, catabolism, regulation | others | ||
| SXYL_02255 | Maltose, EIICB | SXYL_00122-26 | araRBDAT | Arabinose, transport, catabolism, regulation | |
| SXYL_01138 | N-acetylglucosamine, EIIBC | SXYL_01576-78, SXYL_01581 | glpDKF, P | Glycerol, transport, catabolism, regulation | |
| SXYL_02455-57 | Trehalose, regulation, catabolism, EIIBC | SXYL_00438-40 | gntRKP | Gluconate, transport, catabolism, regulation | |
| SXYL_00159, SXYL_02351 | Gluconate, transport | ||||
| SXYL_01518-22 | rbsBCADR | Ribose, transport, catabolism, regulation | |||
| SXYL_00132-35 | xylEBAR | Xylulose, transport, catabolism, regulation | |||
| Peptide transport | SXYL_00298-302, SXYL_01936-40 (oppAFDCB) |
| Peptidases | SXYL_00314 (sspA), SXYL_00502 (pcp), SXYL_00620, SXYL_00948 (map), SXYL_00957 (ampS), SXYL_01078, SXYL_01082, SXYL_01120 (pepA), SXYL_01136, SXYL_01247-48, SXYL_01324, SXYL_01348, SXYL_01489, SXYL_01511, SXYL_01806, SXYL_01931, SXYL_01980 (ampA), SXYL_02073, SXYL_02105 (pepT) |
| Arginine catabolism | SXYL_00252 (arcB), SXYL_02488 (arcC), SXYL_00769 (arg), SXYL_00290-97 (ureDGFECBA) |
| Glutamate catabolism | SXYL_02518 (gltT), SXYL_01964 (gluD1), SXYL_02326 (gluD2), SXYL_02459-61 (gltBCD), SXYL_00105-108 |
| Proline Uptake | SXYL_00427 (putP1), SXYL_00935 (putP2) |
|---|---|
| Serine/alanine/glycine uptake | SXYL_01171 (aapA), SXYL_00317 |
| Glycine betaine/carnitine/choline uptake and glycine betaine synthesis | SXYL_00488-91 (opuCABCD), SXYL_00486 (lcdH), SXYL_00223-26 (cudTCA, betA), SXYL_00743 (opuD2), SXYL_01535 (opuD1), SXYL_01095, SXYL_02127-28 |
| Na+/H+ antiporter | SXYL_01970-76 (mnhA1B1C1D1E1F1G1), SXYL_02220-26 (mnhG2 F2E2D2C2B2A2) |
| Detoxifying Enzymes | Protein Damage Repair | Iron Homeostasis | |||
|---|---|---|---|---|---|
| SXYL_01303 (sodA) | Superoxide dismutase [Mn/Fe] | SXYL_00374 | Thioredoxin | SXYL_00747-50 (sfaDCBA) | Siderophore biosynthesis Staphyloferrin A |
| SXYL_02505 (katA) | Catalase A | SXYL_01797 (trxA) | Thioredoxin | SXYL_00751-3 (htsABC) | Iron compound ABC transporter, Staphyloferrin A * |
| SXYL_01551 (katB) | Catalase B * | SXYL_02083 (trxB) | Thioredoxin reductase * | SXYL_02113-16 (sstDCBA) | Iron compound ABC transporter, Staphyloferrin B * |
| SXYL_02533 (katC) | Catalase C * | SXYL_00519 | Thioredoxin-like protein | SXYL_02201-3 (fhuGBC) | ABC-type cobalamin/Fe3+-siderophores transport system * |
| SXYL_01572 (bsaA) | Glutathione peroxidase | SXYL_01851 | Glutaredoxin | SXYL_02681 (fhuD2) | Iron(3+)-hydroxamate-binding protein * |
| SXYL_01153 (tpx) | Thiol peroxidase | SXYL_01517 (msrA1) | Peptide methionine sulphoxide reductase | SXYL_00667 (fhuD1) | Iron(3+)-hydroxamate-binding protein * |
| SXYL_02534-35 (ahpCF) | Alkyl hydroperoxide reductase * | SXYL_01516 | Regulator MsrR | SXYL_00944 (ftnA) | Ferritin * |
| SXYL_00973 (bcp) | Bacterioferritin comigratory protein Thioreductase peroxidase | SXYL_0019-21 (msrA2BA3) | Peptide methionine sulphoxide reductase | SXYL_00973 (bcp) | Bacterioferritin comigratory protein |
| SXYL_02021 | Nitroreductase | ||||
| SXYL_00229 | Nitroreductase | ||||
| SXYL_00410 | Nitroreductase | ||||
| SXYL_00895 | Nitroreductase * | ||||
| SXYL_00923 (nos) | Nitric oxide synthase | ||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leroy, S.; Vermassen, A.; Ras, G.; Talon, R. Insight into the Genome of Staphylococcus xylosus, a Ubiquitous Species Well Adapted to Meat Products. Microorganisms 2017, 5, 52. https://doi.org/10.3390/microorganisms5030052
Leroy S, Vermassen A, Ras G, Talon R. Insight into the Genome of Staphylococcus xylosus, a Ubiquitous Species Well Adapted to Meat Products. Microorganisms. 2017; 5(3):52. https://doi.org/10.3390/microorganisms5030052
Chicago/Turabian StyleLeroy, Sabine, Aurore Vermassen, Geoffrey Ras, and Régine Talon. 2017. "Insight into the Genome of Staphylococcus xylosus, a Ubiquitous Species Well Adapted to Meat Products" Microorganisms 5, no. 3: 52. https://doi.org/10.3390/microorganisms5030052




