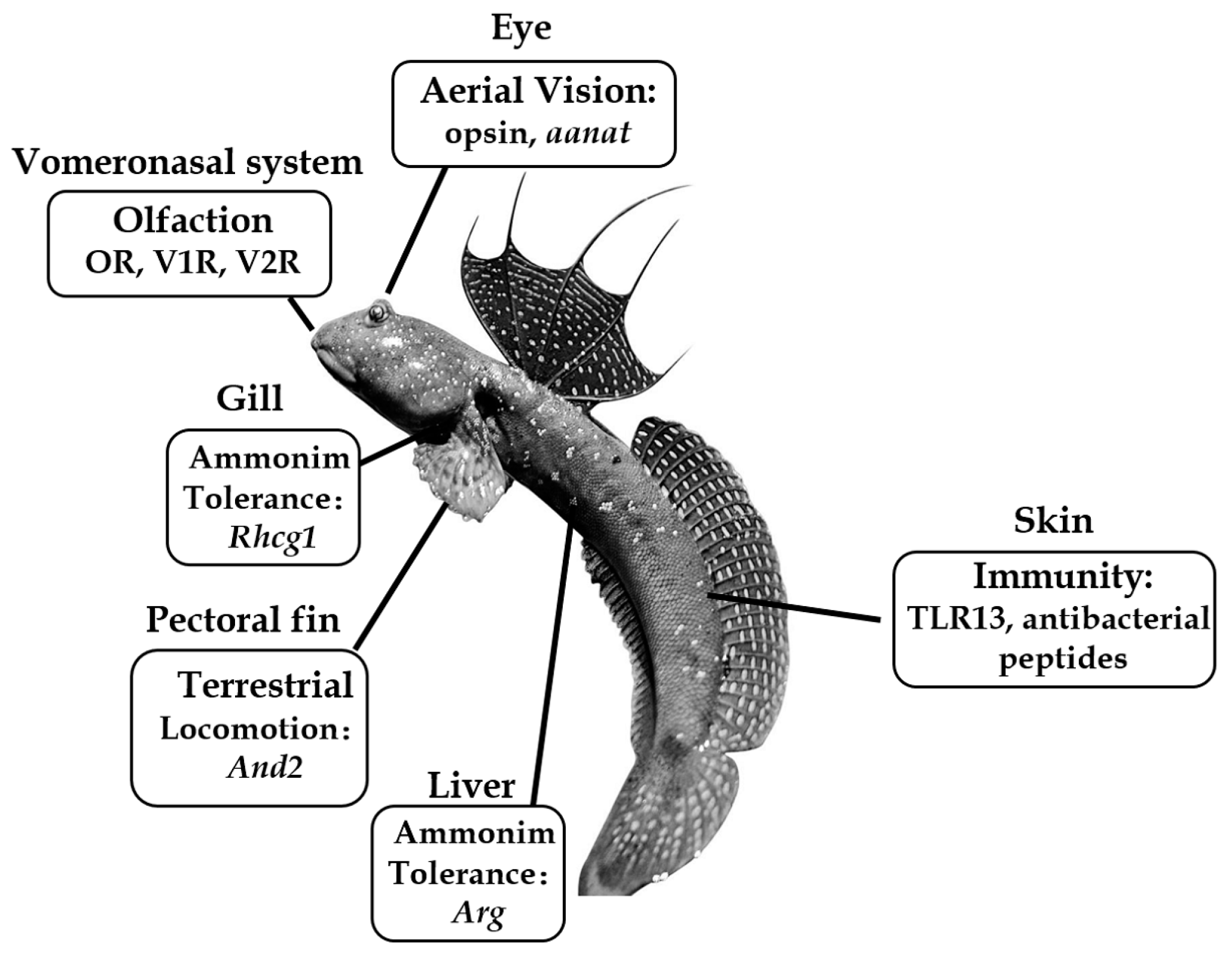
Mudskippers and Their Genetic Adaptations to an Amphibious Lifestyle
Abstract
:Simple Summary
Abstract
1. Introduction
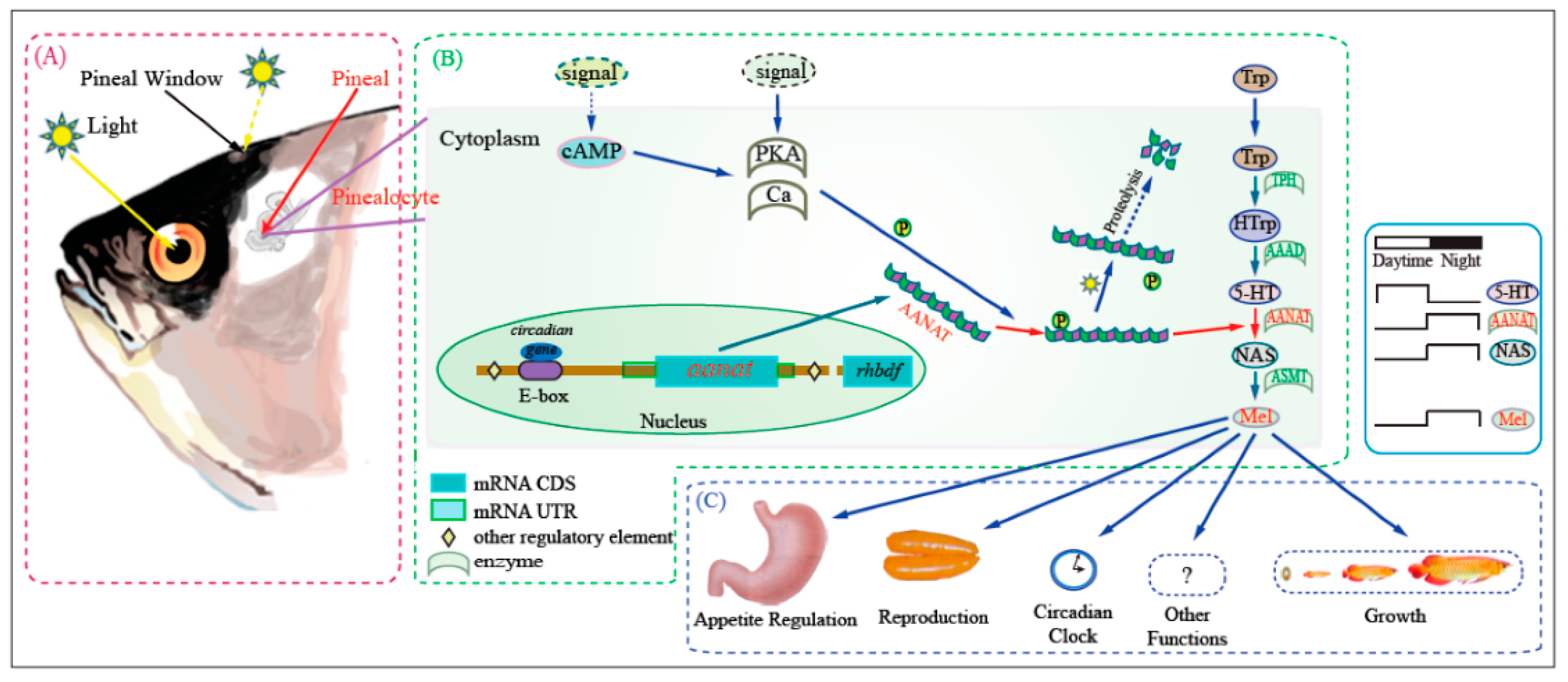
2. Modification of Aerial Vision
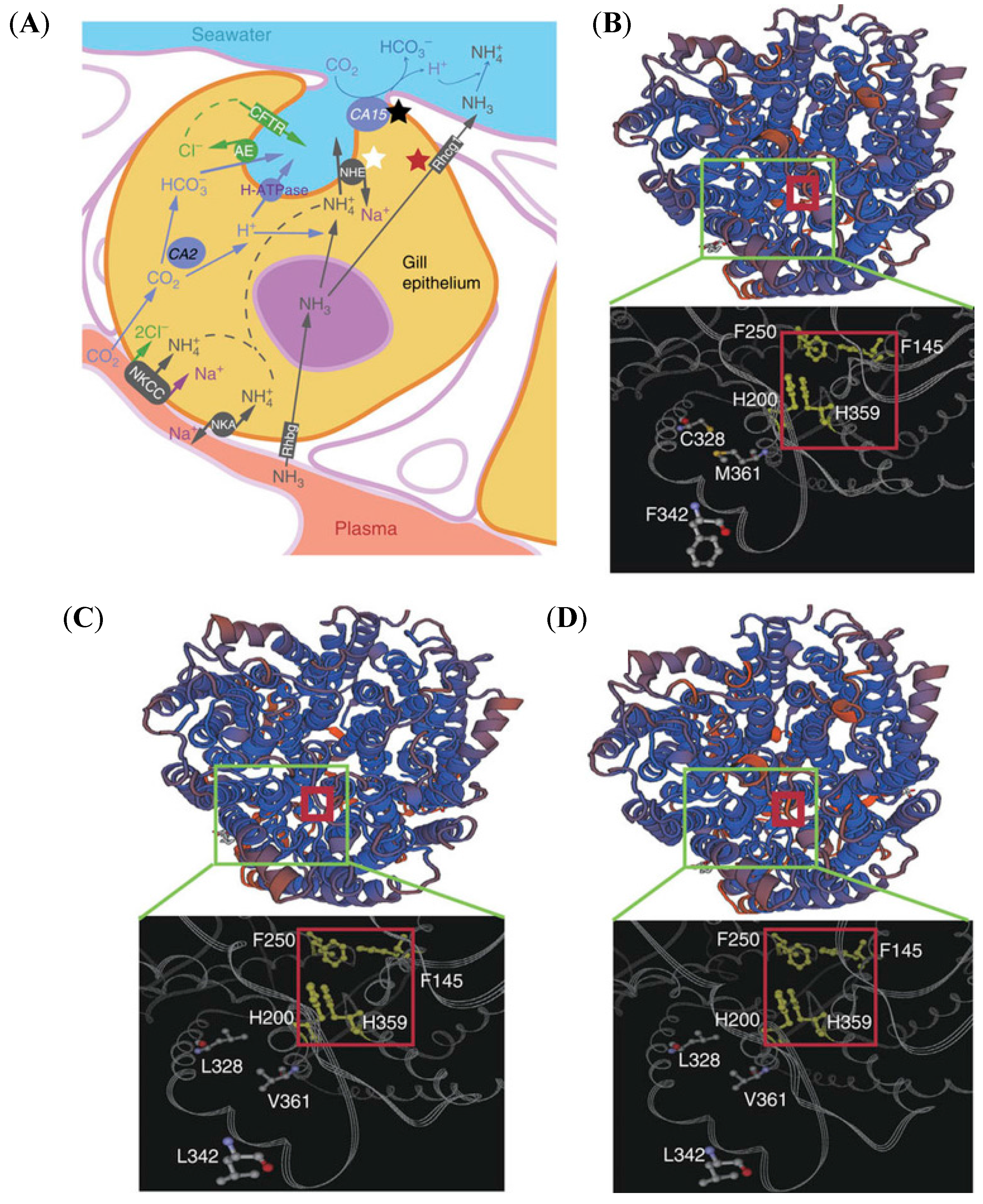
3. Higher Ammonia Tolerance
3.1. Ammonia Excretion Pathway
3.2. Amino Acid Metabolism for Ammonia Tolerance
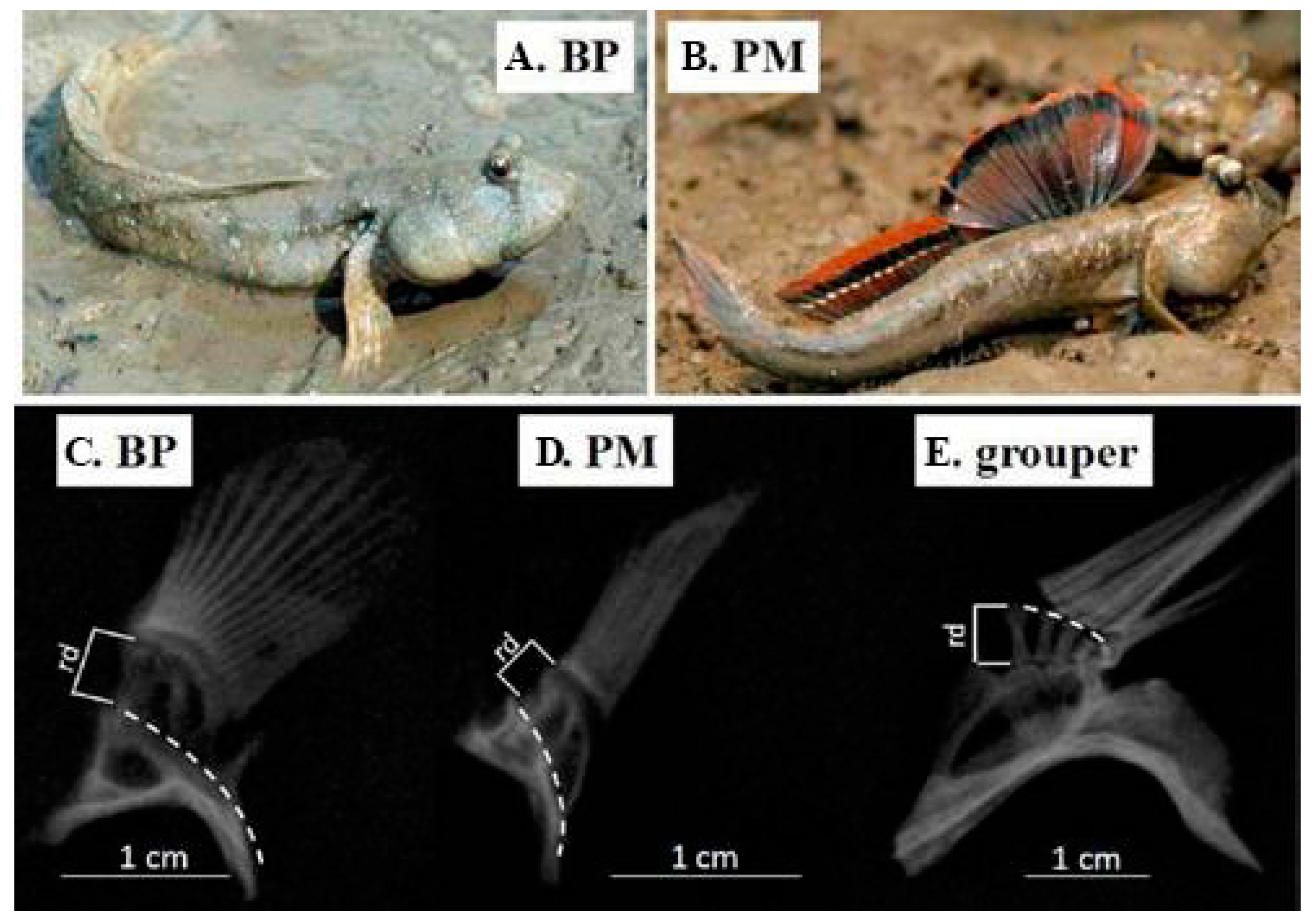
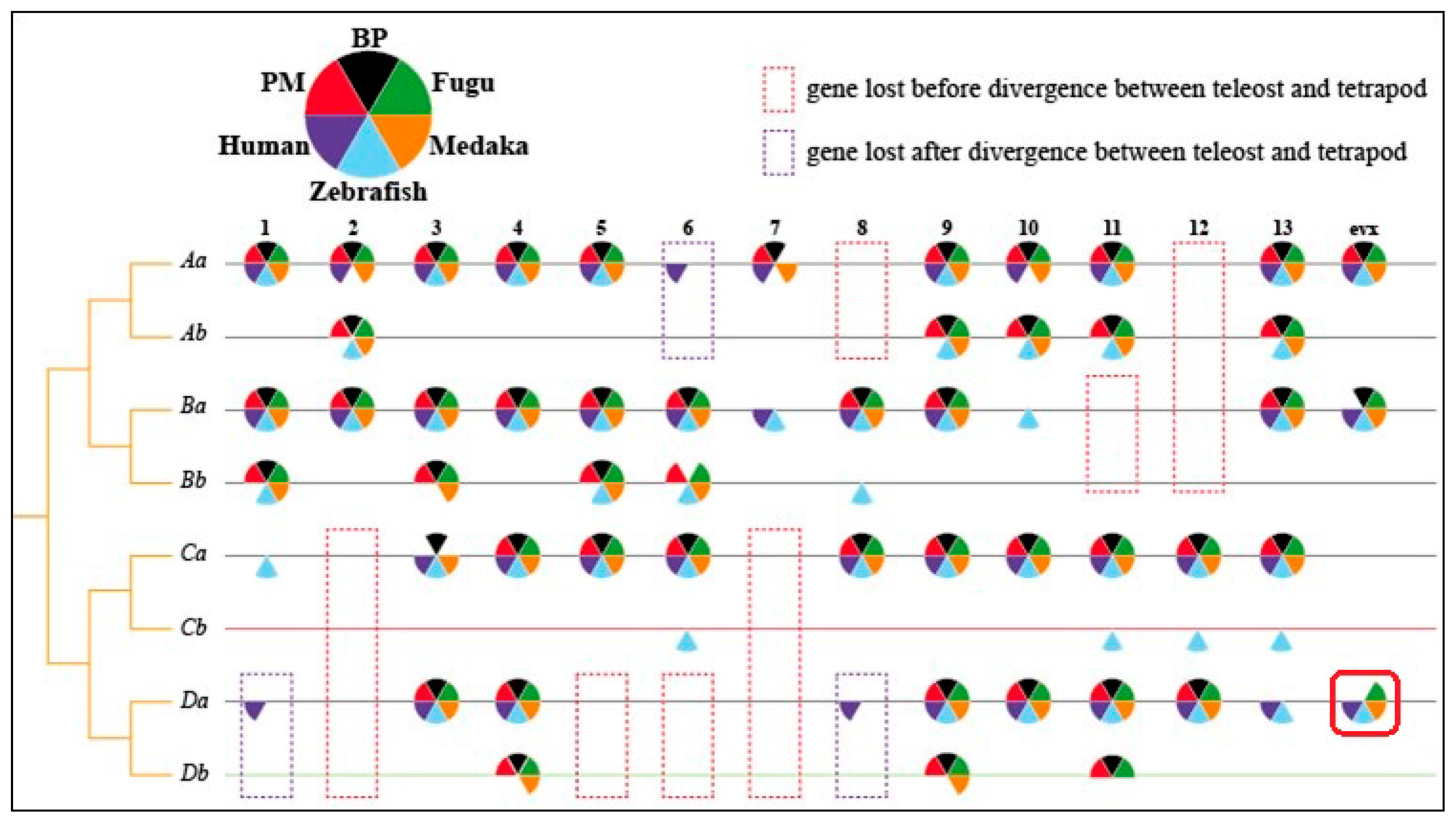
4. Terrestrial Locomotion
5. Immunological Difference
6. Olfaction Modification
7. Air Exposure
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gordon, M.S.; Boëtius, I.; Evans, D.H.; McCarthy, R.; Oglesby, L.C. Aspects of the physiology of terrestrial life in amphibious fishes. I. The mudskipper, Periophthalmus sobrinus. J. Exp. Biol. 1969, 50, 141–149. [Google Scholar]
- Ishimatsu, A.; Gonzales, T.T. Mudskippers: Front runners in the modern invasion of land. In The Biology of Gobies; Patzner, R.A., Van Tassell, J.L., Kovačić, M., Kapoor, B.G., Eds.; Science Publishers: Enfield, NH, USA, 2011; pp. 609–638. [Google Scholar]
- Kutschera, U.; Elliott, J.M. Do mudskippers and lungfishes elucidate the early evolution of four-limbed vertebrates? Evol. Educ. Outreach 2013, 6, 1–8. [Google Scholar] [CrossRef]
- Shi, Q.; Fan, M.; Zhang, Y. Economically Important Fish in China; Press of Central China University of Science and Technology: Wuhan, China, 2014; pp. 391–393. [Google Scholar]
- You, X.; Bian, C.; Zan, Q.; Xu, X.; Liu, X.; Chen, J.; Wang, J.; Qiu, Y.; Li, W.; Zhang, X.; et al. Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nat. Commun. 2014, 5, 5594. [Google Scholar] [CrossRef] [PubMed]
- Sayer, M.D.J. Adaptations of amphibious fish for surviving life out of water. Fish Fish. 2005, 6, 186–211. [Google Scholar] [CrossRef]
- Chew, S.F.; Hiong, K.C.; Lam, S.P.; Ong, S.W.; Wee, W.L.; Wong, W.P.; Ip, Y.K. Functional roles of Na+/K+-ATPase in active ammonia excretion and seawater acclimation in the giant mudskipper, Periophthalmodon schlosseri. Front. Physiol. 2014, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.W.; Chew, S.F.; Lim, C.B.; Kuah, S.S.L.; Kok, W.K.; Ip, Y.K. The mudskippers Periophthalmodon schlosseri and Boleophthalmus boddaerti can tolerate environmental NH3 concentrations of 446 and 36 µM, respectively. Fish Physiol. Biochem. 1998, 19, 59–69. [Google Scholar] [CrossRef]
- Randall, D.J.; Ip, Y.K.; Chew, S.F.; Wilson, J.M. Air breathing and ammonia excretion in the giant mudskipper, Periophthalmodon schlosseri. Physiol. Biochem. Zool. 2004, 77, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.M.; You, X.X.; Ruan, Z.Q.; Hong, W.S.; Chen, S.X.; Bian, C.; Shi, Q. A genomic study on the arm-like pectoral fins in amphibious mudskippers. Ann. Mar. Biol. Res. 2016, 3, 1013. [Google Scholar]
- Tytler, P.; Vaughan, T. Thermal ecology of the mudskippers, Periophthalmus koelreuteri (Pallas) and Boleophthalmus boddarti (Pallas) of Kuwait Bay. J. Fish Biol. 1983, 23, 327–337. [Google Scholar] [CrossRef]
- Li, J.; You, X.; Bian, C.; Yu, H.; Coon, S.L.; Shi, Q. Molecular evolution of aralkylamine N-acetyltransferase in fish: A genomic survey. Int. J. Mol. Sci. 2016, 17, 51. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhang, J.; Kang, B. Structure and function of corneal surface of mudskipper fishes. Fish Physiol. Biochem. 2016, 1, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Van Norren, D.; Schellekens, P. Blue light hazard in rat. Vis. Res. 1990, 30, 1517–1520. [Google Scholar] [CrossRef]
- Shi, Y.; Yokoyama, S. Molecular analysis of the evolutionary significance of ultraviolet vision in vertebrates. Proc. Natl. Acad. Sci. USA 2003, 100, 8308–8313. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Bian, C.; Chen, J.; Sun, Y.; Shi, Q. Recent advances in genomic study on amphibious mudskippers. Mar. Fish. 2015, 37, 479–484. (In Chinese) [Google Scholar]
- Zilberman-Peled, B.; Ron, B.; Gross, A.; Finberg, J.P.M.; Gothilf, Y. A possible new role for fish retinal serotonin N-acetyltransferase-1 (AANAT1): Dopamine metabolism. Brain Res. 2006, 1073, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Feldkaemper, M.; Schaeffel, F. An updated view on the role of dopamine in myopia. Exp. Eye Res. 2013, 114, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Weihrauch, D.; Wilkie, M.P.; Walsh, P.J. Ammonia and urea transporters in gills of fish and aquatic crustaceans. J. Exp. Biol. 2009, 212, 1716–1730. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Chen, J.; Bian, C.; Yi, Y.; Ruan, Z.; Li, J.; Zhang, X.; Yu, H.; Xu, J.; Shi, Q. Transcriptomic evidence of adaptive tolerance to high environmental ammonia in mudskippers. Genomics 2018, in press. [Google Scholar]
- Wilson, J.M.; Randall, D.J.; Donowitz, M.; Vogl, A.W.; Ip, A.K. Immunolocalization of ion-transport proteins to branchial epithelium mitochondria-rich cells in the mudskipper (Periophthalmodon schlosseri). J. Exp. Biol. 2000, 203, 2297–2310. [Google Scholar] [PubMed]
- Ip, Y.K.; Lim, C.K.; Lee, S.L.M.; Wong, W.P.; Chew, S.F. Postprandial increases in nitrogenous excretion and urea synthesis in the giant mudskipper Periophthalmodon schlosseri. J. Exp. Biol. 2004, 207, 3015–3023. [Google Scholar] [CrossRef] [PubMed]
- Ip, Y.K.; Lim, C.B.; Chew, S.F.; Wilson, J.M.; Randall, D.J. Partial amino acid catabolism leading to the formation of alanine in Periophthalmodon schlosseri (mudskipper), a strategy that facilitates the use of amino acids as an energy source during locomotor activity on land. J. Exp. Biol. 2001, 204, 1615–1624. [Google Scholar] [PubMed]
- Iwata, K.; Deguichi, M. Metabolic fate and distribution of 15N-ammonia in an ammonotelic amphibious fish, Periophthalmus modestus, following immersion in 15N-ammonium sulfate, a long term experiment. Zool. Sci. 1995, 12, 175–184. [Google Scholar] [CrossRef]
- Swanson, B.O.; Gibb, A.C. Kinematics of aquatic and terrestrial escape responses in mudskippers. J. Exp. Biol. 2004, 207, 4037–4044. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.M.; Gibb, A.C. Mudskipper pectoral fin kinematics in aquatic and terrestrial environments. J. Exp. Biol. 2009, 212, 2279–2286. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, M.; Liu, B.; Jiang, T.; Zhang, S.; Yang, J. Experimental study on morphology and kinematics of mudskipper in amphibious environments. In Proceedings of the IEEE International Conference on Robotics and Biomimetics (ROBIO), Shenzhen, China, 12–14 December 2013; pp. 1095–1100. [Google Scholar]
- Soshnikova, N. Hox genes regulation in vertebrate. Dev. Dyn. 2014, 243, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Zakany, J.; Denis, D. The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev. 2007, 17, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Amemiya, C.T.; Powers, T.P.; Prohaska, S.J.; Grimwood, J.; Schmutz, J.; Dickson, M.; Miyake, T.; Schoenborn, M.A.; Myers, R.M.; Ruddle, F.H.; et al. Complete HOX cluster characterization of the coelacanth provides further evidence for slow evolution of its genome. Proc. Natl. Acad. Sci. USA 2010, 107, 3622–3627. [Google Scholar] [CrossRef] [PubMed]
- Grandel, H.; Lun, K.; Rauch, G.J.; Rhinn, M.; Piotrowsi, T.; Houart, C.; Sordino, P.; Küchler, A.M.; Schulte-Merker, S.; Geisler, R.; et al. Retinoic acid signaling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development 2002, 129, 2851–2865. [Google Scholar] [PubMed]
- Lin, Q.; Fan, S.; Zhang, Y.; Xu, M.; Zhang, H.; Yang, Y.; Lee, A.P.; Woltering, J.M.; Ravi, V.; Gunter, H.M.; et al. The seahorse genome and the evolution of its specialized morphology. Nature 2016, 540, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Van de Peer, Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). Bioessays 2005, 27, 937–945. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Yan, Y.L.; Eberhart, J.K.; Herpin, A.; Wagner, T.U.; Schartl, M.; Postlethwait, J.H. miR-196 regulates axial patterning and pectoral appendage initiation. Dev. Biol. 2011, 357, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Gilland, E.; Bass, A.H.; Baker, R. Ancestry of motor innervation to pectoral fin and forelimb. Nat. Commun. 2010, 1, 49. [Google Scholar] [CrossRef] [PubMed]
- Sordino, P.; Duboule, D.; Kondo, T. Zebrafish Hoxa and Evx-2 genes: Cloning, developmental expression and implications for the functional evolution of posterior Hox genes. Mech. Dev. 1996, 59, 165–175. [Google Scholar] [CrossRef]
- Hérault, Y.; Hraba-Renevey, S.; van der Hoeven, F.; Duboule, D. Function of the Evx-2 gene in the morphogenesis of vertebrate limbs. EMBO J. 1996, 15, 6727–6738. [Google Scholar] [PubMed]
- McLean, D.L.; Fetcho, J.R. Using imaging and genetics in zebrafish to study developing spinal circuits in vivo. Dev. Neurobiol. 2008, 68, 817–834. [Google Scholar] [CrossRef] [PubMed]
- McLean, D.L.; Masino, M.A.; Koh, I.Y.Y.; Lindquist, W.B.; Fetcho, J.R. Continuous shifts in the active set of spinal interneurons during changes in locomotor speed. Nat. Neurosci. 2008, 11, 1419–1429. [Google Scholar] [CrossRef] [PubMed]
- Farin, H.F.; Lüdtke, T.H.W.; Schmidt, M.K.; Placzko, S.; Schuster-Gossler, K.; Petry, M.; Christoffels, V.M.; Kispert, A. Tbx2 terminates shh/fgf signaling in the developing mouse limb bud by direct repression of gremlin1. PLoS Genet. 2013, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, M.; Krüger, A.; Ferstl, R.; Kaufmann, A.; Nees, G.; Sigmund, A.; Bathke, B.; Lauterbach, H.; Suter, M.; Dreher, S.; et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 2012, 337, 1111–1115. [Google Scholar] [CrossRef] [PubMed]
- Kuciel, M.; Lauriano, E.R.; Silvestri, G.; Żuwała, K.; Pergolizzi, S.; Zaccone, D. The structural organization and immunohistochemistry of G-protein alpha subunits in the olfactory system of the air-breathing mudskipper, Periophthalmus barbarus (Linnaeus, 1766) (Gobiidae, Oxudercinae). Acta Histochem. 2014, 116, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Boschat, C.; Pélofi, C.; Randin, O.; Roppolo, D.; Lüscher, C.; Broillet, M.C.; Rodriguez, I. Pheromone detection mediated by a V1r vomeronasal receptor. Nat. Neurosci. 2002, 5, 1261–1262. [Google Scholar] [CrossRef] [PubMed]
- Leinders-Zufall, T.; Brennan, P.; Widmayer, P.; Chandramani, S.P.; Maul-Pavicic, A.; Jäger, M.; Li, X.; Breer, H.; Zufall, F.; Boehm, T. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science 2004, 306, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Nishiyama, Y.; Ikeda, A.; Takahashi, H.; Hyodo, S.; Kagawa, N.; Sakamoto, H. Neurohypophysial hormones regulate amphibious behaviour in the mudskipper goby. PLoS ONE 2015, 10, e0134605. [Google Scholar] [CrossRef] [PubMed]
- Hamasaki, S.; Mukuda, T.; Kaidoh, T.; Yoshida, M.; Uematsu, K. Impact of dehydration on the forebrain preoptic recess walls in the mudskipper, Periophthalmus modestus: A possible locus for the center of thirst. J. Comp. Physiol. B 2016, 186, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; You, X.; Bian, C.; Chen, S.; Lv, Z.; Qiu, L.; Shi, Q. High-throughput identification of antimicrobial peptides from amphibious mudskippers. Mar. Drugs 2017, 15, 364. [Google Scholar] [CrossRef] [PubMed]

| Tissue | BP | PM | ||
|---|---|---|---|---|
| Up-Regulated | Down-Regulated | Up-Regulated | Down-Regulated | |
| Brain | 117 | 177 | 519 | 409 |
| Gill | 192 | 1435 | 682 | 730 |
| Liver | 1353 | 638 | 553 | 553 |
| Muscle | 580 | 1448 | 429 | 124 |
| Skin | 175 | 272 | 487 | 1510 |
| Total | 2207 | 3444 | 2305 | 2917 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, X.; Sun, M.; Li, J.; Bian, C.; Chen, J.; Yi, Y.; Yu, H.; Shi, Q. Mudskippers and Their Genetic Adaptations to an Amphibious Lifestyle. Animals 2018, 8, 24. https://doi.org/10.3390/ani8020024
You X, Sun M, Li J, Bian C, Chen J, Yi Y, Yu H, Shi Q. Mudskippers and Their Genetic Adaptations to an Amphibious Lifestyle. Animals. 2018; 8(2):24. https://doi.org/10.3390/ani8020024
Chicago/Turabian StyleYou, Xinxin, Min Sun, Jia Li, Chao Bian, Jieming Chen, Yunhai Yi, Hui Yu, and Qiong Shi. 2018. "Mudskippers and Their Genetic Adaptations to an Amphibious Lifestyle" Animals 8, no. 2: 24. https://doi.org/10.3390/ani8020024






