Surface-Enhanced Raman Scattering Spectroscopy and Microfluidics: Towards Ultrasensitive Label-Free Sensing
Abstract
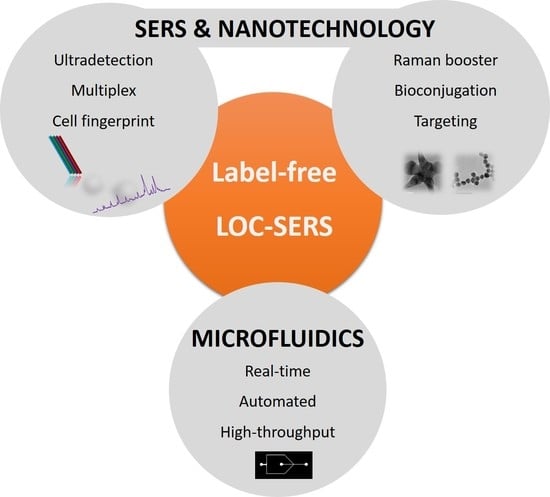
:1. Introduction
2. Considerations
3. SERS and Microfluidics for Label-Free Detection
3.1. Dispersed Nanoparticles and Continuous Flow
3.2. Immobilized Nanoparticles (Continuous/Static Flow)
3.3. Segmented Flow
4. Bottlenecks and Future Outlook
Funding
Conflicts of Interest
References
- Raman, C.V.; Krishnan, K.S. A New Type of Secondary Radiation. Nature 1928, 121, 501–502. [Google Scholar] [CrossRef]
- Fleischmann, M.; Hendra, P.J.; McQuillan, A.J. Raman spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 1974, 26, 163–166. [Google Scholar] [CrossRef]
- Li, Z.; Deen, M.; Kumar, S.; Selvaganapathy, P. Raman Spectroscopy for In-Line Water Quality Monitoring—Instrumentation and Potential. Sensors 2014, 14, 17275–17303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.-S.; Church, J.S. Raman spectroscopy in the analysis of food and pharmaceutical nanomaterials. J. Food Drug Anal. 2014, 22, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Premasiri, W.R.; Chen, Y.; Fore, J.; Brodeur, A.; Ziegler, L.D. SERS Biomedical Applications: Diagnostics, Forensics, and Metabolomics. In Frontiers and Advances in Molecular Spectroscopy; Elsevier: New York, NY, USA, 2018; pp. 327–367. ISBN 9780128112205. [Google Scholar]
- Prochazka, M. Medical Applications of SERS; Springer: Cham, Switzerland, 2016; pp. 149–211. [Google Scholar]
- Abalde-Cela, S.; Aldeanueva-Potel, P.; Mateo-Mateo, C.; Rodríguez-Lorenzo, L.; Alvarez-Puebla, R.A.; Liz-Marzán, L.M. Surface-enhanced Raman scattering biomedical applications of plasmonic colloidal particles. J. R. Soc. Interface 2010, 7. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, F.; Leona, M. Surface-enhanced Raman spectroscopy in art and archaeology. J. Raman Spectrosc. 2016, 47, 67–77. [Google Scholar] [CrossRef]
- Hakonen, A.; Andersson, P.O.; Stenbæk Schmidt, M.; Rindzevicius, T.; Käll, M. Explosive and chemical threat detection by surface-enhanced Raman scattering: A review. Anal. Chim. Acta 2015, 893, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Aroca, R. Surface Enhanced Vibrational Spectroscopy; Wiley: Hoboken, NJ, USA, 2006; ISBN 9780471607311. [Google Scholar]
- Jeanmaire, D.L.; Van Duyne, R.P. Surface Raman spectroelectrochemistry. J. Electroanal. Chem. Interfacial Electrochem. 1977, 84, 1–20. [Google Scholar] [CrossRef]
- Albrecht, M.G.; Creighton, J.A. Anomalously intense Raman spectra of pyridine at a silver electrode. J. Am. Chem. Soc. 1977, 99, 5215–5217. [Google Scholar] [CrossRef]
- Moskovits, M. Surface-enhanced spectroscopy. Rev. Mod. Phys. 1985, 57, 783–826. [Google Scholar] [CrossRef]
- Kneipp, K.; Wang, Y.; Kneipp, H.; Perelman, L.T.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Single molecule detection using surface-enhanced raman scattering (SERS). Phys. Rev. Lett. 1997, 78, 1667–1670. [Google Scholar] [CrossRef]
- Hakonen, A.; Svedendahl, M.; Ogier, R.; Yang, Z.-J.; Lodewijks, K.; Verre, R.; Shegai, T.; Andersson, P.O.; Käll, M. Dimer-on-mirror SERS substrates with attogram sensitivity fabricated by colloidal lithography. Nanoscale 2015, 7, 9405–9410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardinal, M.F.; Vander Ende, E.; Hackler, R.A.; McAnally, M.O.; Stair, P.C.; Schatz, G.C.; Van Duyne, R.P. Expanding applications of SERS through versatile nanomaterials engineering. Chem. Soc. Rev. 2017, 46, 3886–3903. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhao, X.; Yi, Z.; Glushenkov, A.M.; Kong, L. Plasmonic substrates for surface enhanced Raman scattering. Anal. Chim. Acta 2017, 984, 19–41. [Google Scholar] [CrossRef] [PubMed]
- Abalde-Cela, S.; Carregal-Romero, S.; Coelho, J.P.; Guerrero-Martínez, A. Recent progress on colloidal metal nanoparticles as signal enhancers in nanosensing. Adv. Colloid Interface Sci. 2016, 233, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, M.; Mullen, E.R.; Korkmaz, A.; Wachsmann-Hogiu, S. Fundamentals and applications of SERS-based bioanalytical sensing. Nanophotonics 2017, 6, 831–852. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Korivi, N.S. Microfluidics: Technologies and applications. In Nanolithography; Elsevier: New York, NY, USA, 2014; pp. 424–443. ISBN 9780857095008. [Google Scholar]
- Best, R.J.; Lyczakowski, J.J.; Abalde-Cela, S.; Yu, Z.; Abell, C.; Smith, A.G. Label-Free Analysis and Sorting of Microalgae and Cyanobacteria in Microdroplets by Intrinsic Chlorophyll Fluorescence for the Identification of Fast Growing Strains. Anal. Chem. 2016, 88, 10445–10451. [Google Scholar] [CrossRef] [PubMed]
- Abalde-Cela, S.; Taladriz-Blanco, P.; De Oliveira, M.G.; Abell, C. Droplet microfluidics for the highly controlled synthesis of branched gold nanoparticles. Sci. Rep. 2018, 8, 2440. [Google Scholar] [CrossRef] [PubMed]
- Luka, G.; Ahmadi, A.; Najjaran, H.; Alocilja, E.; DeRosa, M.; Wolthers, K.; Malki, A.; Aziz, H.; Althani, A.; Hoorfar, M. Microfluidics Integrated Biosensors: A Leading Technology towards Lab-on-a-Chip and Sensing Applications. Sensors 2015, 15, 30011–30031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, J.W.; Nieuwoudt, M.K.; Vargas, M.J.T.; Bodley, O.L.C.; Yohendiran, T.S.; Oosterbeek, R.N.; Williams, D.E.; Cather Simpson, M. Raman on a disc: High-quality Raman spectroscopy in an open channel on a centrifugal microfluidic disc. Analyst 2017, 142, 1682–1688. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, S.; Zheng, J.; He, L. Surface-enhanced Raman spectroscopy (SERS) combined techniques for high-performance detection and characterization. TrAC Trends Anal. Chem. 2017, 90, 1–13. [Google Scholar] [CrossRef]
- White, I.M.; Yazdi, S.H.; Yu, W.W. Optofluidic SERS: Synergizing photonics and microfluidics for chemical and biological analysis. Microfluid. Nanofluid. 2012, 13, 205–216. [Google Scholar] [CrossRef]
- Oh, Y.-J.; Jeong, K.-H. Optofluidic SERS chip with plasmonic nanoprobes self-aligned along microfluidic channels. Lab Chip 2014, 14, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Choo, J. Recent advances in surface-enhanced Raman scattering detection technology for microfluidic chips. Electrophoresis 2008, 29, 1815–1828. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Choi, N.; Chang, S.-I.; Lee, E.K.; Choo, J. Real-time analysis of diaquat dibromide monohydrate in water with a SERS-based integrated microdroplet sensor. Nanoscale 2014, 6, 8781–8786. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, K.R.; Henkel, T.; Popp, J. Quantitative Online Detection of Low-Concentrated Drugs via a SERS Microfluidic System. ChemPhysChem 2007, 8, 2665–2670. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lee, S.; Seong, G.H.; Choo, J.; Lee, E.K.; Gweon, D.-G.; Lee, S. Quantitative Analysis of Methyl Parathion Pesticides in a Polydimethylsiloxane Microfluidic Channel Using Confocal Surface-Enhanced Raman Spectroscopy. Appl. Spectrosc. 2006, 60, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Yazdi, S.H.; White, I.M. Multiplexed detection of aquaculture fungicides using a pump-free optofluidic SERS microsystem. Analyst 2013, 138, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.-Y.; Hossain, M.K.; Lee, J.-H.; Han, J.; Lee, H.J.; Kim, K.-J.; Kim, J.-H.; Lee, K.-B.; Choi, J.-W. Selective isolation and noninvasive analysis of circulating cancer stem cells through Raman imaging. Biosens. Bioelectron. 2018, 102, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lee, Y.H.; Phang, I.Y.; Lee, C.K.; Ling, X.Y. Hierarchical 3D SERS Substrates Fabricated by Integrating Photolithographic Microstructures and Self-Assembly of Silver Nanoparticles. Small 2014, 10, 2703–2711. [Google Scholar] [CrossRef] [PubMed]
- Mikac, L.; Ivanda, M.; Gotić, M.; Mihelj, T.; Horvat, L. Synthesis and characterization of silver colloidal nanoparticles with different coatings for SERS application. J. Nanopart. Res. 2014, 16, 2748. [Google Scholar] [CrossRef]
- Willets, K.A. Super-resolution imaging of SERS hot spots. Chem. Soc. Rev. 2014, 43, 3854–3864. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Graña, S.; Fernández-López, C.; Polavarapu, L.; Salmon, J.-B.; Leng, J.; Pastoriza-Santos, I.; Pérez-Juste, J. Gold Nanooctahedra with Tunable Size and Microfluidic-Induced 3D Assembly for Highly Uniform SERS-Active Supercrystals. Chem. Mater. 2015, 27, 8310–8317. [Google Scholar] [CrossRef]
- Abalde-Cela, S.; Hermida-Ramón, J.M.; Contreras-Carballada, P.; De Cola, L.; Guerrero-Martínez, A.; Alvarez-Puebla, R.A.; Liz-Marzán, L.M. SERS chiral recognition and quantification of enantiomers through cyclodextrin supramolecular complexation. ChemPhysChem 2011, 12, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Lane, L.A.; Qian, X.; Nie, S. SERS Nanoparticles in Medicine: From Label-Free Detection to Spectroscopic Tagging. Chem. Rev. 2015, 115, 10489–10529. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Cáceres, R.; Abalde-Cela, S.; Guardia-Girós, P.; Fernández-Barbero, A.; Pérez-Juste, J.; Alvarez-Puebla, R.A.; Liz-Marzán, L.M. Multifunctional microgel magnetic/optical traps for SERS ultradetection. Langmuir 2011, 27, 4520–4525. [Google Scholar] [CrossRef] [PubMed]
- Abalde-Cela, S.; Auguie, B.; Fischlechner, M.; Huck, W.T.S.; Alvarez-Puebla, R.A.; Liz-Marzan, L.M.; Abell, C. Microdroplet fabrication of silver-agarose nanocomposite beads for SERS optical accumulation. Soft Matter 2011, 7, 1321–1325. [Google Scholar] [CrossRef]
- Spuch-Calvar, M.; Rodríguez-Lorenzo, L.; Morales, M.P.; Álvarez-Puebla, R.A.; Liz-Marzán, L.M. Bifunctional Nanocomposites with Long-Term Stability as SERS Optical Accumulators for Ultrasensitive Analysis. J. Phys. Chem. C 2009, 113, 3373–3377. [Google Scholar] [CrossRef]
- Kasera, S.; Herrmann, L.O.; del Barrio, J.; Baumberg, J.J.; Scherman, O.A. Quantitative multiplexing with nano-self-assemblies in SERS. Sci. Rep. 2015, 4, 6785. [Google Scholar] [CrossRef] [PubMed]
- Fraire, J.C.; Sueldo Ocello, V.N.; Allende, L.G.; Veglia, A.V.; Coronado, E.A. Toward the Design of Highly Stable Small Colloidal SERS Substrates with Supramolecular Host–Guest Interactions for Ultrasensitive Detection. J. Phys. Chem. C 2015, 119, 8876–8888. [Google Scholar] [CrossRef]
- Zhang, J.; Coulston, R.J.; Jones, S.T.; Geng, J.; Scherman, O.A.; Abell, C. One-Step Fabrication of Supramolecular Microcapsules from Microfluidic Droplets. Science 2012, 335, 690–694. [Google Scholar] [CrossRef] [PubMed]
- Guerrini, L.; Graham, D. Molecularly-mediated assemblies of plasmonic nanoparticles for Surface-Enhanced Raman Spectroscopy applications. Chem. Soc. Rev. 2012, 41, 7085–7107. [Google Scholar] [CrossRef] [PubMed]
- Ngo, H.T.; Wang, H.-N.; Fales, A.M.; Vo-Dinh, T. Label-free DNA Biosensor Based on SERS Molecular Sentinel on Nanowave Chip. Anal. Chem. 2013, 85, 6378–6383. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Lorenzo, L.; Krpetic, Z.; Barbosa, S.; Alvarez-Puebla, R.A.; Liz-Marzán, L.M.; Prior, I.A.; Brust, M. Intracellular mapping with SERS-encoded gold nanostars. Integr. Biol. 2011, 3, 922–926. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Juste, J.; Pastoriza-Santos, I.; Liz-Marzán, L.M.; Biomagune, C.; Fern Andez-L Opez, C.; Mateo-Mateo, C.; On, R.; Alvarez-Puebla, A.; Erez-Juste, J.P.; Liz-Marz, L.M. Highly Controlled Silica Coating of PEG-Capped Metal Nanoparticles and Preparation of SERS-Encoded Particles. Langmuir 2009, 25, 13894–13899. [Google Scholar]
- Song, J.; Zhou, J.; Duan, H. Self-Assembled Plasmonic Vesicles of SERS-Encoded Amphiphilic Gold Nanoparticles for Cancer Cell Targeting and Traceable Intracellular Drug Delivery. J. Am. Chem. Soc. 2012, 134, 13458–13469. [Google Scholar] [CrossRef] [PubMed]
- Fabris, L. Gold-based SERS tags for biomedical imaging. J. Opt. 2015, 17, 114002. [Google Scholar] [CrossRef]
- Fabris, L. SERS Tags: The Next Promising Tool for Personalized Cancer Detection? ChemNanoMat 2016, 2, 249–258. [Google Scholar] [CrossRef]
- Wu, L.; Wang, Z.; Zong, S.; Cui, Y. Rapid and reproducible analysis of thiocyanate in real human serum and saliva using a droplet SERS-microfluidic chip. Biosens. Bioelectron. 2014, 62, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-Y.; Chang, C.-L.; Wang, Y.-N.; Fu, L.-M. Microfluidic Mixing: A Review. Int. J. Mol. Sci. 2011, 12, 3263–3287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malekzad, H.; Sahandi Zangabad, P.; Mohammadi, H.; Sadroddini, M.; Jafari, Z.; Mahlooji, N.; Abbaspour, S.; Gholami, S.; Ghanbarpour Houshangi, M.; Pashazadeh, R.; et al. Noble metal nanostructures in optical biosensors: Basics, and their introduction to anti-doping detection. TrAC Trends Anal. Chem. 2018, 100, 116–135. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Zhou, D.; Chen, T.; Streets, A.M.; Huang, Y. Label-Free Digital Quantification of Lipid Droplets in Single Cells by Stimulated Raman Microscopy on a Microfluidic Platform. Anal. Chem. 2016, 88, 4931–4939. [Google Scholar] [CrossRef] [PubMed]
- Leung, S.-A.; Winkle, R.F.; Wootton, R.C.R.; DeMello, A.J. A method for rapid reaction optimisation in continuous-flow microfluidic reactors using online Raman spectroscopic detection. Analyst 2005, 130, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Lin, Y.; Gu, B.; Panwar, N.; Tjin, S.C.; Song, J.; Qu, J.; Yong, K.-T. Optical trapping-assisted SERS platform for chemical and biosensing applications: Design perspectives. Coord. Chem. Rev. 2017, 339, 138–152. [Google Scholar] [CrossRef]
- Pu, H.; Xiao, W.; Sun, D.-W. SERS-microfluidic systems: A potential platform for rapid analysis of food contaminants. Trends Food Sci. Technol. 2017, 70, 114–126. [Google Scholar] [CrossRef]
- Hassoun, M.; Rüger, J.; Kirchberger-Tolstik, T.; Schie, I.W.; Henkel, T.; Weber, K.; Cialla-May, D.; Krafft, C.; Popp, J. A droplet-based microfluidic chip as a platform for leukemia cell lysate identification using surface-enhanced Raman scattering. Anal. Bioanal. Chem. 2018, 410, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Mühlig, A.; Bocklitz, T.; Labugger, I.; Dees, S.; Henk, S.; Richter, E.; Andres, S.; Merker, M.; Stöckel, S.; Weber, K.; et al. LOC-SERS: A promising closed system for the identification of mycobacteria. Anal. Chem. 2016, 88, 7998–8004. [Google Scholar] [CrossRef] [PubMed]
- Abalde-Cela, S.; Abell, C.; Alvarez-Puebla, R.A.; Liz-Marzán, L.M. Real time dual-channel multiplex SERS ultradetection. J. Phys. Chem. Lett. 2014, 5, 73–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parisi, J.; Su, L.; Lei, Y. In situ synthesis of silver nanoparticle decorated vertical nanowalls in a microfluidic device for ultrasensitive in-channel SERS sensing. Lab Chip 2013, 13, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- Tycova, A.; Prikryl, J.; Foret, F. Recent strategies toward microfluidic-based surface-enhanced Raman spectroscopy. Electrophoresis 2017, 38, 1977–1987. [Google Scholar] [CrossRef] [PubMed]
- Fei, J.; Wu, L.; Zhang, Y.; Zong, S.; Wang, Z.; Cui, Y. Pharmacokinetics-on-a-Chip Using Label-Free SERS Technique for Programmable Dual-Drug Analysis. ACS Sens. 2017, 2, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Ko, J.; Cha, K.; Ho Jeon, J.; Rhie, G.; Choi, J.; deMello, A.J.; Choo, J. Fast and sensitive detection of an anthrax biomarker using SERS-based solenoid microfluidic sensor. Biosens. Bioelectron. 2015, 72, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Mungroo, N.A.; Oliveira, G.; Neethirajan, S. SERS based point-of-care detection of food-borne pathogens. Microchim. Acta 2016, 183, 697–707. [Google Scholar] [CrossRef]
- Yap, L.W.; Chen, H.; Gao, Y.; Petkovic, K.; Liang, Y.; Si, K.J.; Wang, H.; Tang, Z.; Zhu, Y.; Cheng, W. Bifunctional plasmonic-magnetic particles for an enhanced microfluidic SERS immunoassay. Nanoscale 2017, 9, 7822–7829. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yi, Y.; Zhou, C.; Ying, G.; Zhou, X.; Fu, C.; Zhu, Y.; Shen, Y. SERS detection of microRNA biomarkers for cancer diagnosis using gold-coated paramagnetic nanoparticles to capture SERS-active gold nanoparticles. RSC Adv. 2017, 7, 52782–52793. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Fu, C.; Yi, Y.; Zhou, X.; Zhou, C.; Ying, G.; Shen, Y.; Zhu, Y. A magnetic-based SERS approach for highly sensitive and reproducible detection of cancer-related serum microRNAs. Anal. Methods 2018, 10, 624–633. [Google Scholar] [CrossRef]
- Wang, Z.; Zong, S.; Wang, Z.; Wu, L.; Chen, P.; Yun, B.; Cui, Y. Microfluidic chip based micro RNA detection through the combination of fluorescence and surface enhanced Raman scattering techniques. Nanotechnology 2017, 28, 105501. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Jiang, Y.; Zheng, X.; Jia, S.; Zhu, Z.; Ren, B.; Ma, H. Facile fabrication of microfluidic surface-enhanced Raman scattering devices via lift-up lithography. R. Soc. Open Sci. 2018, 5, 172034. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Lee, K.; Kim, K.H.; Oh, K.W.; Choo, J. SERS-based immunoassay using a gold array-embedded gradient microfluidic chip. Lab Chip 2012, 12, 3720–3727. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Kang, T.; Cho, H.; Choi, Y.; Lee, L.P. Additional amplifications of SERSvia an optofluidic CD-based platform. Lab Chip 2009, 9, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Madiyar, F.; Yu, C.; Li, J. Detection of extremely low concentration waterborne pathogen using a multiplexing self-referencing SERS microfluidic biosensor. J. Biol. Eng. 2017, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhao, F.; Zeng, J.; Qi, J.; Lu, J.; Shih, W.-C. Microfluidic surface-enhanced Raman scattering sensor with monolithically integrated nanoporous gold disk arrays for rapid and label-free biomolecular detection. J. Biomed. Opt. 2014, 19, 111611. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-Y.; Choi, C.J.; Cunningham, B.T. Plasmonic Nanogap-Enhanced Raman Scattering Using a Resonant Nanodome Array. Small 2012, 8, 2878–2885. [Google Scholar] [CrossRef] [PubMed]
- Novara, C.; Lamberti, A.; Chiadò, A.; Virga, A.; Rivolo, P.; Geobaldo, F.; Giorgis, F. Surface-enhanced Raman spectroscopy on porous silicon membranes decorated with Ag nanoparticles integrated in elastomeric microfluidic chips. RSC Adv. 2016, 6, 21865–21870. [Google Scholar] [CrossRef]
- Kalantarifard, A.; Saateh, A.; Elbuken, C. Label-Free Sensing in Microdroplet-Based Microfluidic Systems. Chemosensors 2018, 6, 23. [Google Scholar] [CrossRef]
- Salmon, A.R.; Esteban, R.; Taylor, R.W.; Hugall, J.T.; Smith, C.A.; Whyte, G.; Scherman, O.A.; Aizpurua, J.; Abell, C.; Baumberg, J.J. Monitoring Early-Stage Nanoparticle Assembly in Microdroplets by Optical Spectroscopy and SERS. Small 2016, 12, 1788–1796. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Choi, N.; Chang, S.-I.; Kang, S.H.; Song, J.M.; Cho, S.I.; Lim, D.W.; Choo, J. Highly sensitive trace analysis of paraquat using a surface-enhanced Raman scattering microdroplet sensor. Anal. Chim. Acta 2010, 681, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Jiang, W.; Wang, H.; Yang, X.; Zhang, S.; Yuan, Y.; Wu, T.; Du, Y. A Surface Enhanced Raman Scattering (SERS) microdroplet detector for trace levels of crystal violet. Microchim. Acta 2013, 180, 997–1004. [Google Scholar] [CrossRef]
- Wang, G.; Lim, C.; Chen, L.; Chon, H.; Choo, J.; Hong, J.; deMello, A.J. Surface-enhanced Raman scattering in nanoliter droplets: Towards high-sensitivity detection of mercury (II) ions. Anal. Bioanal. Chem. 2009, 394, 1827–1832. [Google Scholar] [CrossRef] [PubMed]
- Meier, T.-A.; Beulig, R.J.; Klinge, E.; Fuss, M.; Ohla, S.; Belder, D. On-chip monitoring of chemical syntheses in microdroplets via surface-enhanced Raman spectroscopy. Chem. Commun. 2015, 51, 8588–8591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strelau, K.K.; Kretschmer, R.; Möller, R.; Fritzsche, W.; Popp, J. SERS as tool for the analysis of DNA-chips in a microfluidic platform. Anal. Bioanal. Chem. 2010, 396, 1381–1384. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Shi, G.; Wang, H.; Zhang, Q.; Li, Y. In Situ Functionalization of Stable 3D Nest-Like Networks in Confined Channels for Microfluidic Enrichment and Detection. Adv. Funct. Mater. 2014, 24, 1017–1026. [Google Scholar] [CrossRef]
- Guo, Y.; Oo, M.K.; Reddy, K.; Fan, X. Ultrasensitive Optofluidic Surface-Enhanced Raman Scattering Detection with Flow-Through Multihole Capillaries. ACS Nano 2012, 6, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Knauer, M.; Ivleva, N.P.; Niessner, R.; Haisch, C. A flow-through microarray cell for the online SERS detection of antibody-captured E. coli bacteria. Anal. Bioanal. Chem. 2012, 402, 2663–2667. [Google Scholar] [CrossRef] [PubMed]




| SERS Substrate | Device Type | Strategy | Target | LOD | Refs. |
|---|---|---|---|---|---|
| Ag NPs | PDMS | Dispersed NPs continuous flow | Carboxyfluorescein-labelled DNA | 0.5 µM | [85] |
| Fe3O4@Au NPs@Ag | PDMS | Dispersed NPs continuous flow | IgG | 1 pg mL−1 | [68] |
| Au@M NPs | PDMS | Dispersed NPs continuous flow | miRNA | 100 fM | [69] |
| Ag NPs | PDMS | Dispersed NPs continuous flow | Foodborne bacteria (8 species) | 7 CFU | [67] |
| Ag NPs | PDMS | Dispersed NPs continuous flow | Dyes | 0.9 nM | [62] |
| Ag NPs | PDMS | Immobilized NPs continuous flow | miRNA | 10−8 M | [71] |
| Nanoporous Au disks | PDMS | Immobilized NPs continuous flow | R6G DA Urea | 5.16 nM 32.4 nM 0.67 mM | [76] |
| Ag Zn(OH)F@ZnS | Capillary | Immobilized NPs continuous flow | Bovine serum albumin (BSA) | 1 pM | [86] |
| Au NPs | Glass | Immobilized NPs continuous flow | R6G | 0.5 pM | [87] |
| Ag NPs | PDMS | Immobilized NPs static condition | E. coli | 4.5 × 103 cells/mL | [88] |
| Au NPs | PDMS | Immobilized NPs static condition | E.coli O157:H7 | 1 CFU/mL | [75] |
| Ag NPs | PDMS | Microdroplets | Mycobacteria species | 93.8% | [61] |
| Ag NPs | PDMS | Microdroplets | 2-aminothiazole | [84] | |
| Au@Ag NRs | PDMS | Microdroplets | Thiocyanate | 1 µM | [53] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kant, K.; Abalde-Cela, S. Surface-Enhanced Raman Scattering Spectroscopy and Microfluidics: Towards Ultrasensitive Label-Free Sensing. Biosensors 2018, 8, 62. https://doi.org/10.3390/bios8030062
Kant K, Abalde-Cela S. Surface-Enhanced Raman Scattering Spectroscopy and Microfluidics: Towards Ultrasensitive Label-Free Sensing. Biosensors. 2018; 8(3):62. https://doi.org/10.3390/bios8030062
Chicago/Turabian StyleKant, Krishna, and Sara Abalde-Cela. 2018. "Surface-Enhanced Raman Scattering Spectroscopy and Microfluidics: Towards Ultrasensitive Label-Free Sensing" Biosensors 8, no. 3: 62. https://doi.org/10.3390/bios8030062