Interaction Analysis of Longevity Interventions Using Survival Curves
Abstract
:1. Introduction
2. Results
2.1. Composition Principles
2.2. Competing Risks CP
2.3. Generalized Multiplicative CP
2.4. Generalized Scaling CP
2.5. Data Set
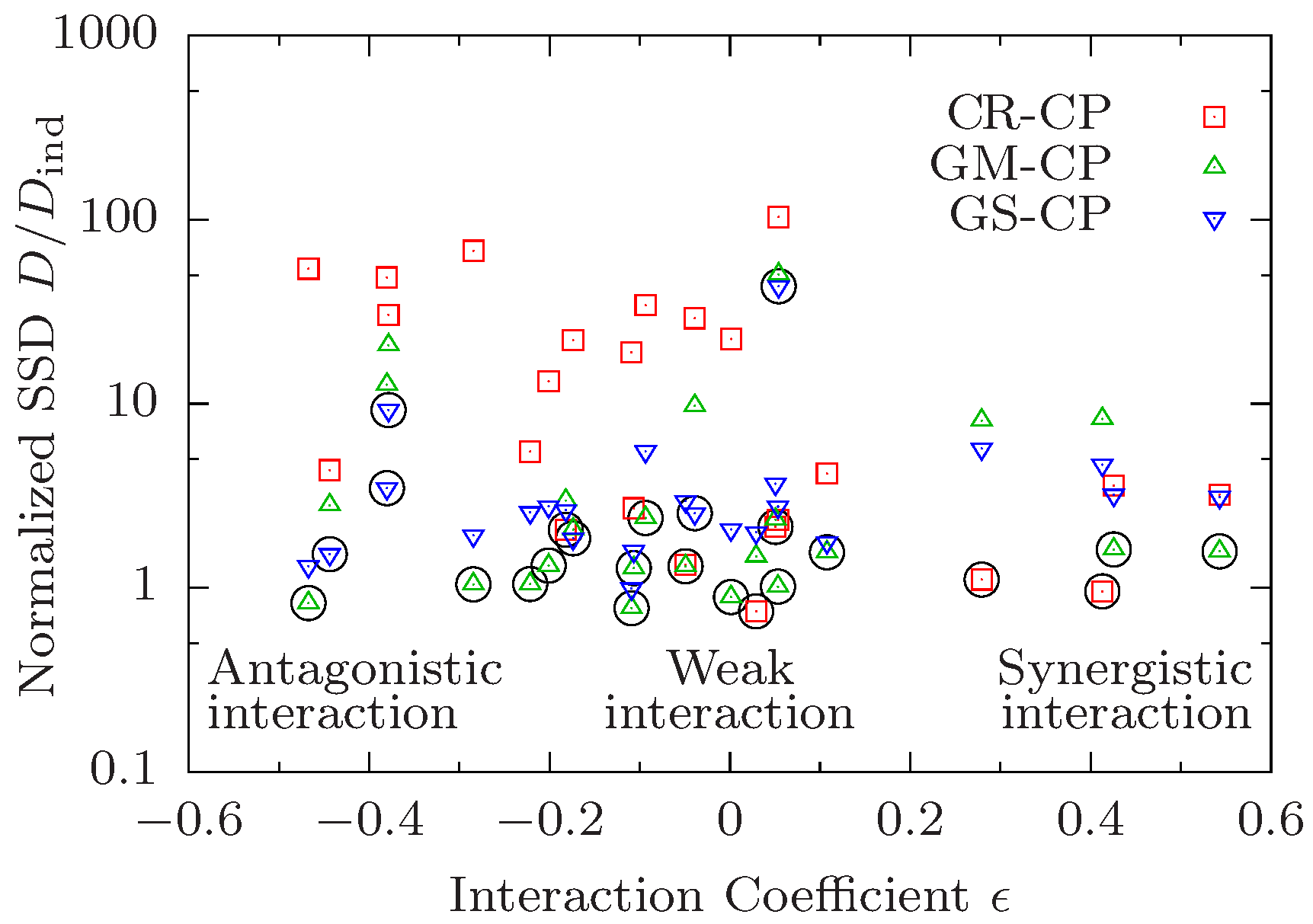
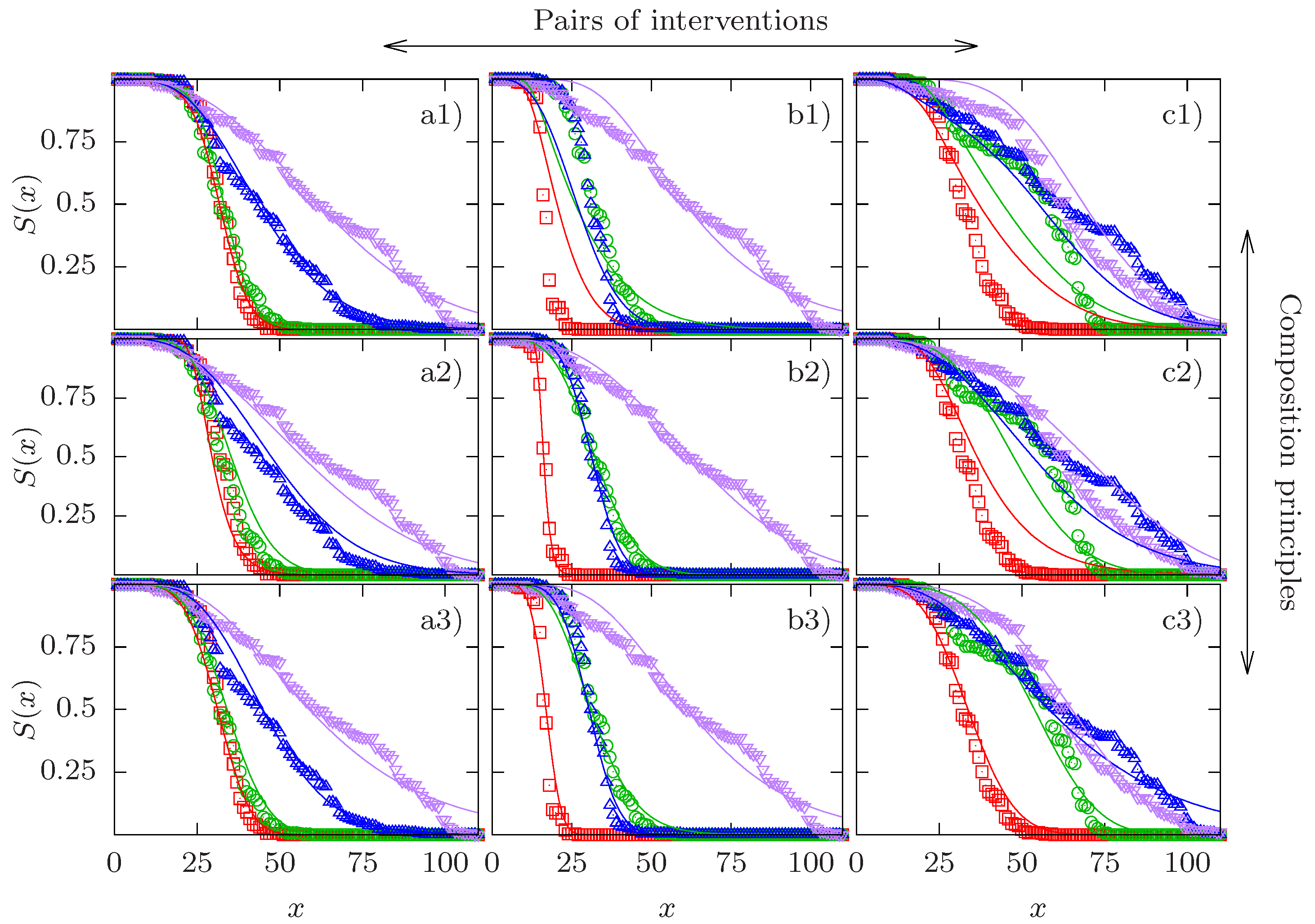
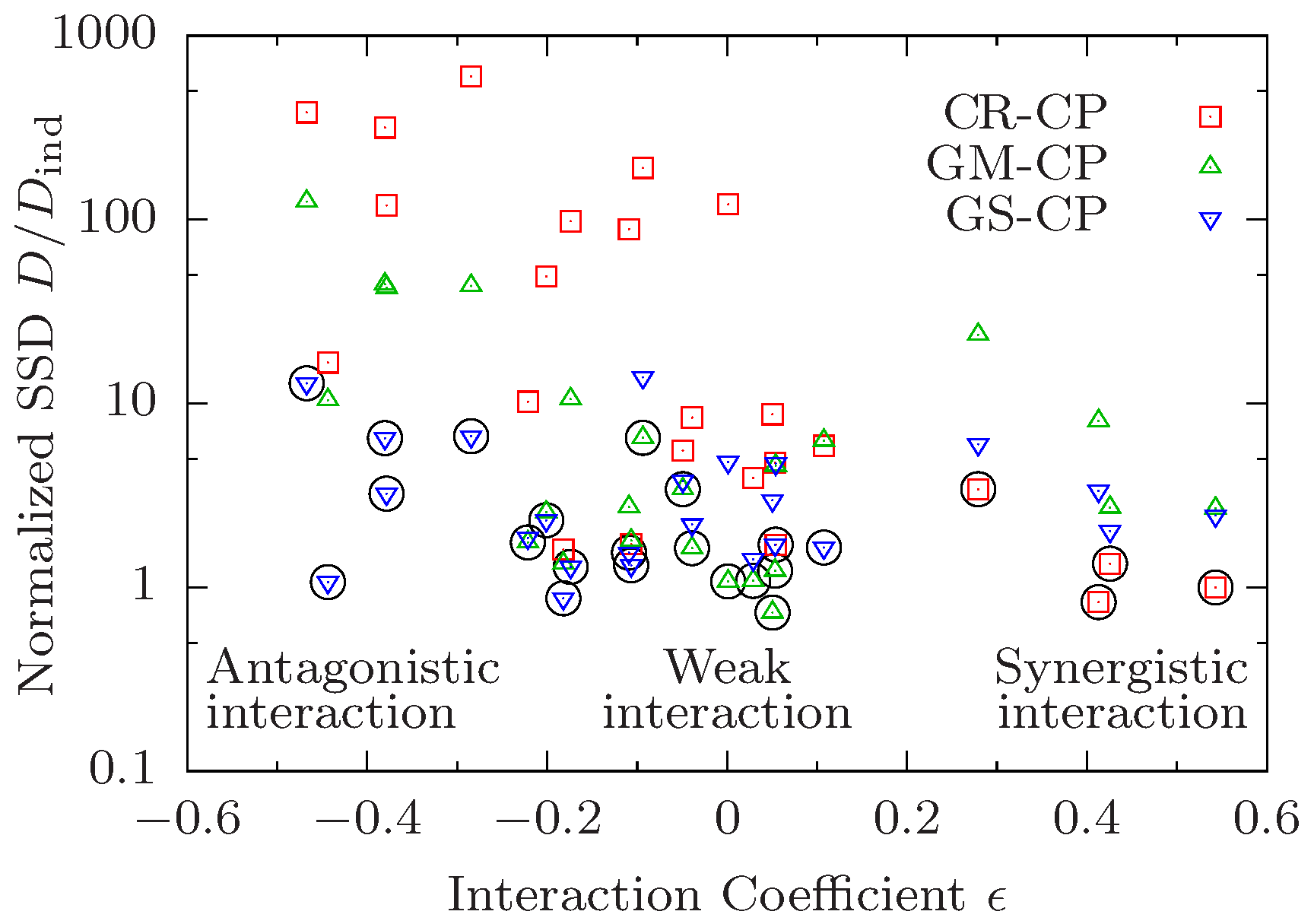
2.6. Test of Composition Principles
- A direct fitting algorithm constrained to satisfy the CR-CP (3) will in most cases fail to converge to a valid survival curve. This reflects the restrictive conditions on the individual curves imposed by this CP. To overcome this difficulty, we further constrained the fitting procedure by demanding that the four survival curves in the quadruple take the specific formwhere the are again represented by three-parameter functions (15). This enforces the validity of the CR-CP (3) but also implies that the curves have to be ordered according to for all x.
- For the GM-CP, the survival curves , and are represented by three survival functions of the form (15), and the fourth curve is constructed according to (11) using the numerical computation of inverse functions. The nine parameters entering the three functions are then adjusted to optimize the fit to the data quadruple.
3. Discussion
4. Materials and Methods
- The algorithm is initialized with a population of n quadruples of survival functions. Initial parameter values are .
- Next, m offspring are created that descend from randomly chosen parents. The parameters of the children are equal to the parents’ parameters multiplied with a factor , where X is uniform random variable on and is the mutation strength.
- Out of the total population of the individuals, the n with lowest SSD survive. These individuals make up the next generation.
- Mutation strength u is decreased by a constant factor, and the algorithm continues with the second step.
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CP | Composition principle |
| CR-CP | Competing risks CP |
| GM-CP | Generalized multiplicative CP |
| GS-CP | Generalized scaling CP |
| SSD | Sum of squares of mean square deviations |
Appendix A. Interaction for Median and Most Likely LifeSpan
Appendix A.1. CR-CP Applied to Weibull Survival Curves
Appendix A.2. GS-CP Applied to Gompertz Survival Curves
Appendix B. Alternative Fits Using a Logistic Model
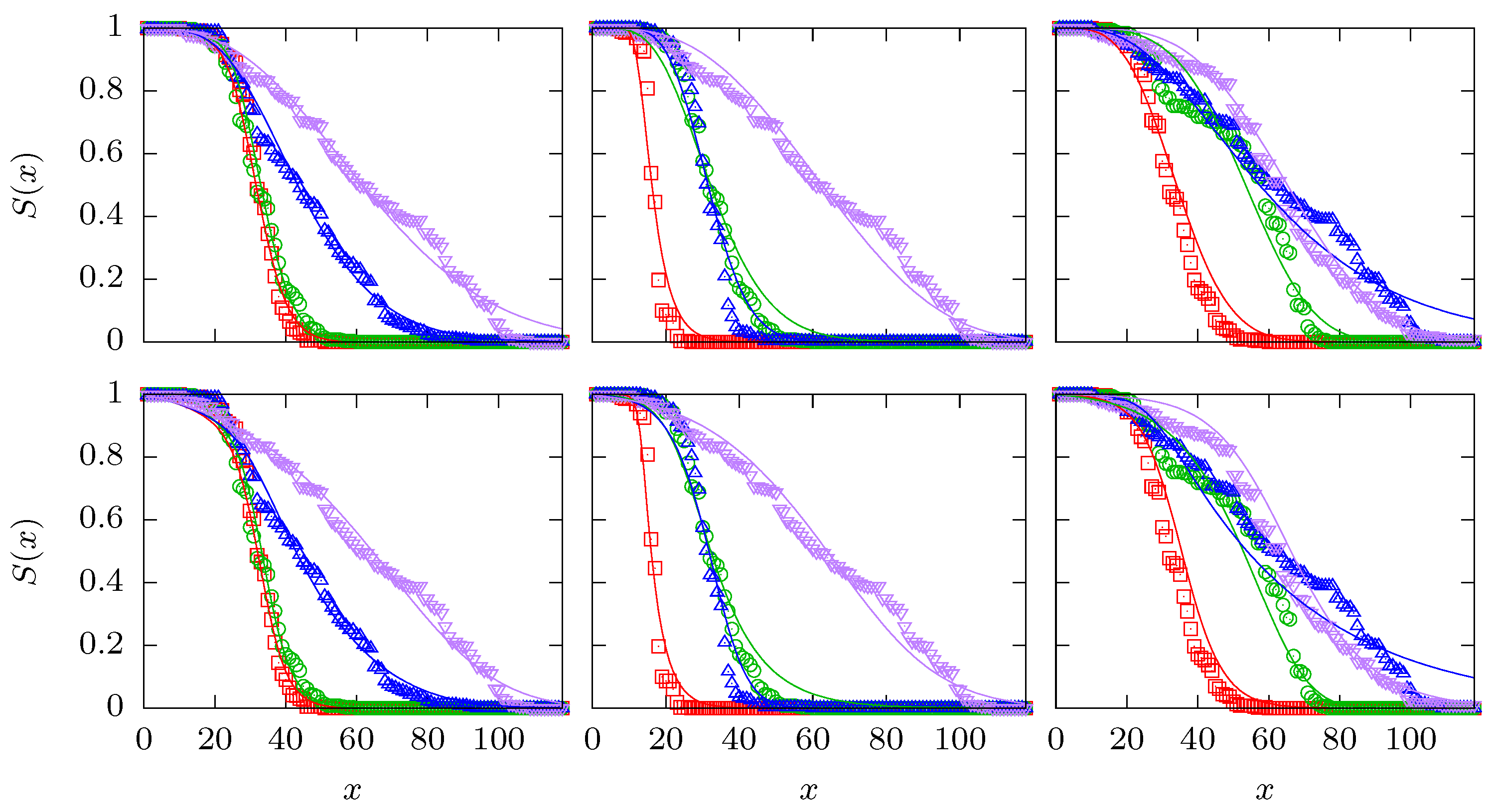
Appendix C. Interaction Analysis for a Single Set of Survival Curves
References
- Guarente, L.; Kenyon, C. Genetic pathways that regulate ageing in model organisms. Nature 2000, 408, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Partridge, L.; Gems, D. Mechanisms of ageing: Public or private? Nat. Rev. Genet. 2002, 3, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Antebi, A. Genetics of Aging in Caenorhabditis elegans. PLoS Genet. 2007, 3, e129. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, C.J. The genetics of ageing. Nature 2010, 464, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Gems, D.; Partridge, L. Genetics of longevity in model organisms: Debates and paradigm shifts. Annu. Rev. Physiol. 2013, 75, 621–644. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Partridge, L. Promoting health and longevity through diet: From model organisms to humans. Cell 2015, 161, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Brunet, A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 2009, 8, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Hekimi, S.; Benard, C.; Branicky, R.; Burgess, J.; Hihi, A.K.; Rea, S. Why only time will tell. Mech. Ageing Dev. 2001, 122, 571–594. [Google Scholar] [CrossRef]
- Gems, D.; Pletcher, S.; Partridge, L. Interpreting interactions between treatments that slow aging. Aging Cell 2002, 1, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yen, K.; Mobbs, C.V. Evidence for only two independent pathways for decreasing senescence in Caenorhabditis elegans. AGE 2010, 32, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Cordell, H.J. Epistasis: What it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 2002, 11, 2463–2468. [Google Scholar] [CrossRef] [PubMed]
- Phillips, P.C. Epistasis—The essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 2008, 9, 855–867. [Google Scholar] [CrossRef] [PubMed]
- De Visser, J.A.G.M.; Cooper, T.F.; Elena, S.F. The causes of epistasis. Proc. R. Soc. B: Biol. Sci. 2011, 278, 3617–3624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szendro, I.G.; Schenk, M.F.; Franke, J.; Krug, J.; de Visser, J.A.G.M. Quantitative analyses of empirical fitness landscapes. J. Stat. Mech. Theory Exp. 2013. [Google Scholar] [CrossRef]
- Weinreich, D.M.; Lan, Y.; Wylie, C.S.; Heckendorn, R.B. Should evolutionary geneticists worry about higher-order epistasis? Curr. Opin. Genet. Dev. 2013, 23, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Mani, R.; St Onge, R.P.; Hartman, J.L., 4th; Giaever, G.; Roth, F.P. Defining genetic interaction. Proc. Natl. Acad. Sci. USA 2008, 105, 3461–3466. [Google Scholar] [CrossRef] [PubMed]
- Mair, W.; Goymer, P.; Pletcher, S.D.; Partridge, L. Demography of Dietary Restriction and Death in Drosophila. Science 2003, 301, 1731–1733. [Google Scholar] [CrossRef] [PubMed]
- Baudisch, A. The pace and shape of ageing. Meth. Ecol. Evol. 2011, 2, 375–382. [Google Scholar] [CrossRef]
- Jones, O.R.; Scheuerlein, A.; Salguero-Gómez, R.; Camarda, C.G.; Schaible, R.; Casper, B.B.; Dahlgren, J.P.; Ehrlén, J.; García, M.B.; Menges, E.S.; et al. Diversity of ageing across the tree of life. Nature 2014, 505, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Atlan, H.; Miquel, J.; Helmle, L.C.; Dolkas, C.B. Thermodynamics of aging in Drosophila melanogaster. Mech. Ageing Dev. 1976, 5, 371–387. [Google Scholar] [CrossRef]
- Wilson, D.L. The analysis of survival (mortality) data: Fitting Gompertz, Weibull and logistic functions. Mech. Ageing Devel. 1994, 74, 15–33. [Google Scholar] [CrossRef]
- Vanfleteren, J.R.; De Vreese, A.; Braeckmann, B.P. Two-parameter logistic and Weibull equations provide better fits to survival data from isogenic populations of Caenorhabditis elegans in axenic culture than does the Gompertz model. J. Gerontol. 1998, 53A, B393–B403. [Google Scholar] [CrossRef]
- Pletcher, S.D. Model fitting and hypothesis testing for age-specific mortality data. J. Evol. Biol. 1999, 12, 430–439. [Google Scholar] [CrossRef]
- Pletcher, S.D.; Khazaeli, A.A.; Curtsinger, J.W. Why do life spans differ? Partitioning mean longevity differences in terms of age-specific mortality parameters. J. Gerontol. 2000, 55, B381–B389. [Google Scholar] [CrossRef]
- Johnson, T.E.; Wu, D.; Tedesco, P.; Dames, S.; Vaupel, J.W. Age-Specific Demographic Profiles of Longevity Mutants in Caenorhabditis elegans Show Segmental Effects. J. Gerontol. 2001, 56, B331–B339. [Google Scholar] [CrossRef]
- Yen, K.; Steinsaltz, D.; Mobbs, C.V. Validated analysis of mortality rates demonstrates distinct genetic mechanisms that influence lifespan. Exp. Gerontol. 2008, 43, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Hughes, B.G.; Hekimi, S. Different Mechanisms of Longevity in Long-Lived Mouse and Caenorhabditis elegans Mutants Revealed by Statistical Analysis of Mortality Rates. Genetics 2016, 204, 905–920. [Google Scholar] [CrossRef] [PubMed]
- Stroustrup, N.; Anthony, W.E.; Nash, Z.M.; Gowda, V.; Gomez, A.; López-Moyado, I.F.; Apfeld, J.; Fontana, W. The temporal scaling of Caenorhabditis elegans ageing. Nature 2016, 530, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Witten, M. A return to Time, Cells, Systems and Aging III: Gompertzian models of biological aging and some possible roles for critical elements. Mech. Ageing Dev. 1985, 32, 141–177. [Google Scholar] [CrossRef]
- Gavrilov, L.A.; Gavrilova, N.S. The Reliability Theory of Aging and Longevity. J. Theor. Biol. 2001, 213, 527–545. [Google Scholar] [CrossRef] [PubMed]
- Kalbfleisch, J.D.; Prentice, R.L. The Statistical Analysis of Failure Time Data; Wiley: Hoboken, NJ, USA, 2002. [Google Scholar]
- Sailer, Z.R.; Harms, M.J. Detecting High-Order Epistasis in Nonlinear Genotype-Phenotype Maps. Genetics 2017, 205, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Stroustrup, N.; Ulmschneider, B.E.; Nash, Z.M.; López-Moyado, I.F.; Apfeld, J.; Fontana, W. The Caenorhabditis elegans lifespan machine. Nat. Meth. 2013, 10, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Spivey, E.C.; Finkelstein, I.J. From cradle to grave: High-throughput studies of aging in model organisms. Mol. BioSyst. 2014, 10, 1658–1667. [Google Scholar] [CrossRef] [PubMed]
| Intervention | Binary | Intervention | Binary |
|---|---|---|---|
| None/control | 0000 | Dietary Restriction (DR) | 0001 |
| 16 °C | 1000 | DR at 16 °C | 1001 |
| daf-2 | 0100 | daf-2 & DR | 0101 |
| daf-2 at 16 °C | 1100 | daf-2 & DR at 16 °C | 1101 |
| clk-1 | 0010 | clk-1 & DR | 0011 |
| clk-1 at 16 °C | 1010 | clk-1 & DR at 16 °C | 1011 |
| clk-1 & daf-2 | 0110 | clk-1& daf-2 & DR | 0111 |
| clk-1 & daf-2 at 16 °C | 1110 | clk-1 & daf-2 & DR at 16 °C | 1111 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowak, S.; Neidhart, J.; Szendro, I.G.; Rzezonka, J.; Marathe, R.; Krug, J. Interaction Analysis of Longevity Interventions Using Survival Curves. Biology 2018, 7, 6. https://doi.org/10.3390/biology7010006
Nowak S, Neidhart J, Szendro IG, Rzezonka J, Marathe R, Krug J. Interaction Analysis of Longevity Interventions Using Survival Curves. Biology. 2018; 7(1):6. https://doi.org/10.3390/biology7010006
Chicago/Turabian StyleNowak, Stefan, Johannes Neidhart, Ivan G. Szendro, Jonas Rzezonka, Rahul Marathe, and Joachim Krug. 2018. "Interaction Analysis of Longevity Interventions Using Survival Curves" Biology 7, no. 1: 6. https://doi.org/10.3390/biology7010006




