Metal Binding Properties of the N-Terminus of the Functional Amyloid Orb2
Abstract
:1. Introduction
2. Results
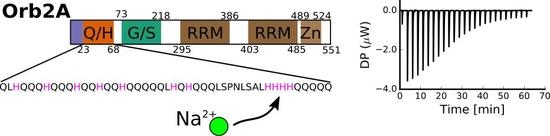
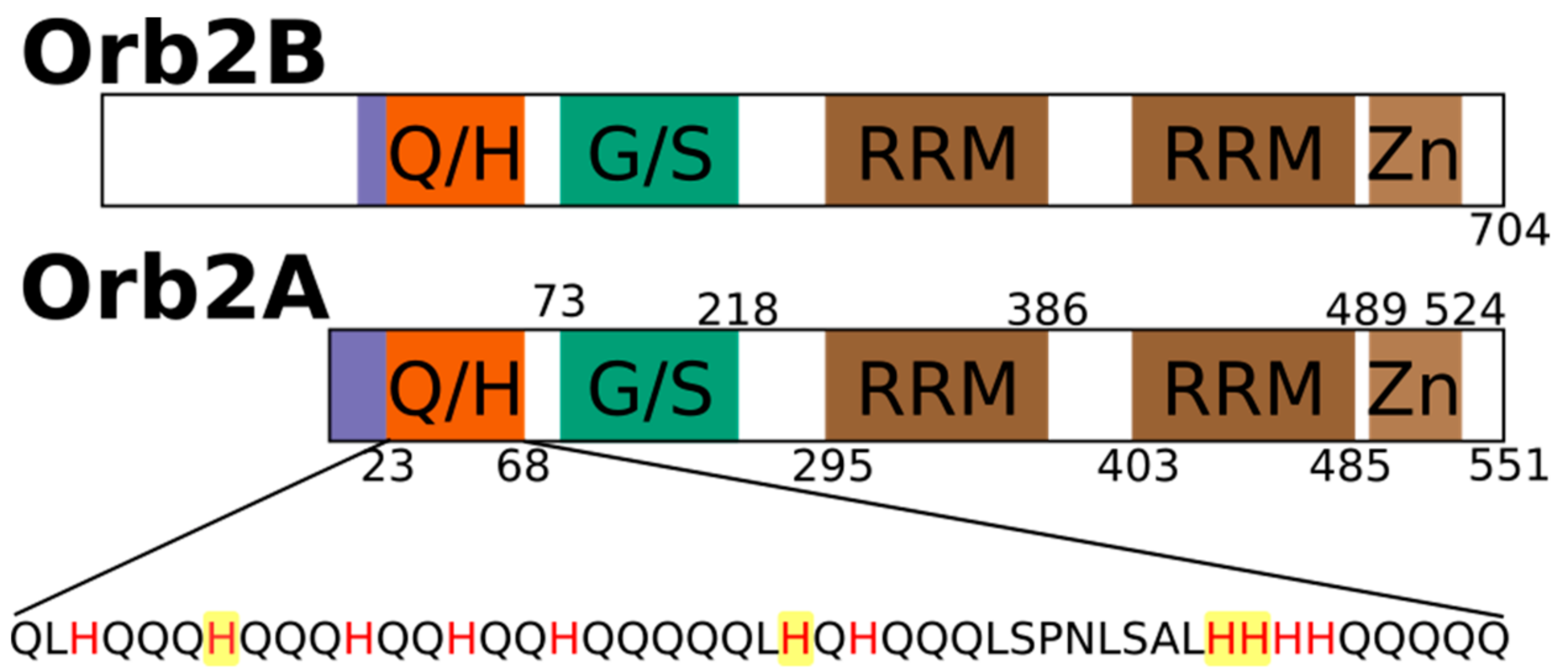
2.1. The N-Terminus of Orb2A Binds Transition Metals in the Absence of a Polyhistidine-Tag
2.2. Isothermal Titration Calorimetry to Characterize Metal Binding Affinity of Orb2A87
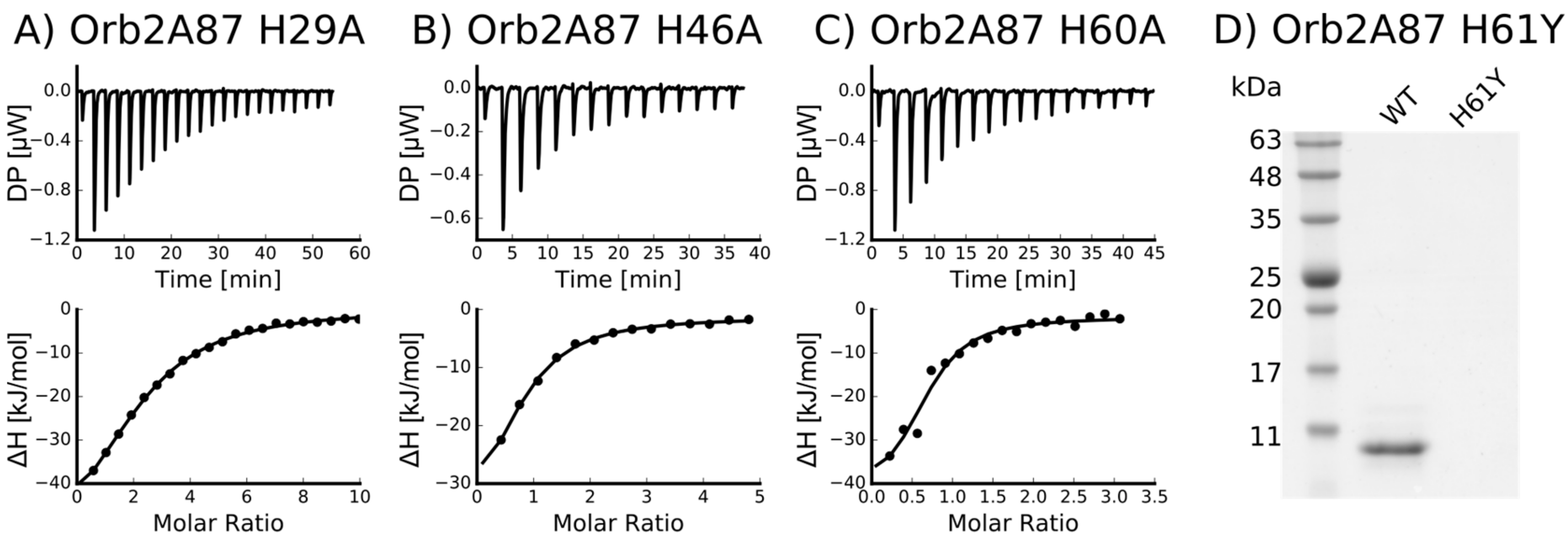
2.3. Several Histidines Are Important for the Metal Binding Affinity of Orb2A87
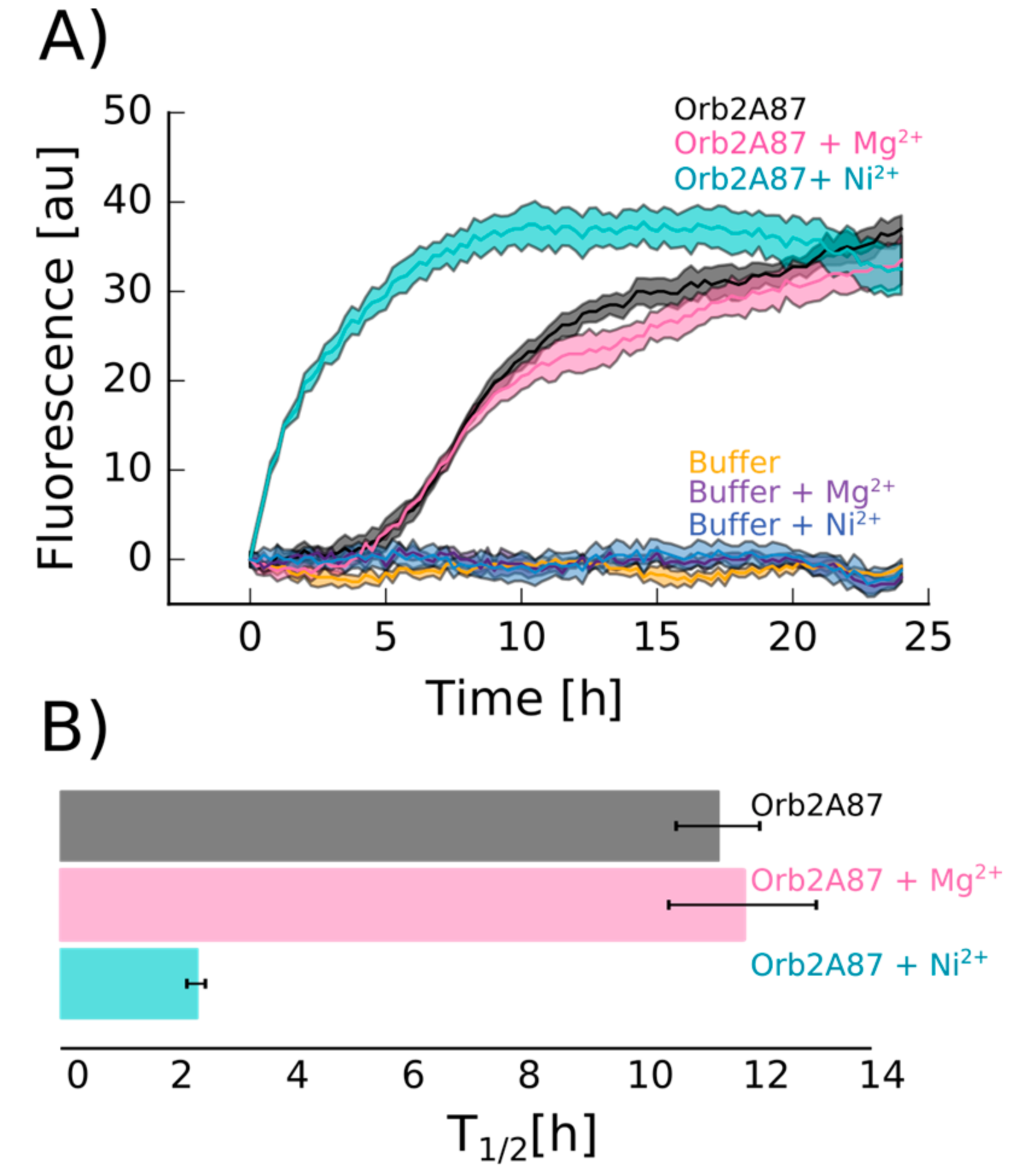
2.4. Orb2A–Metal Interaction Affects the Aggregation of Orb2A87
3. Discussion
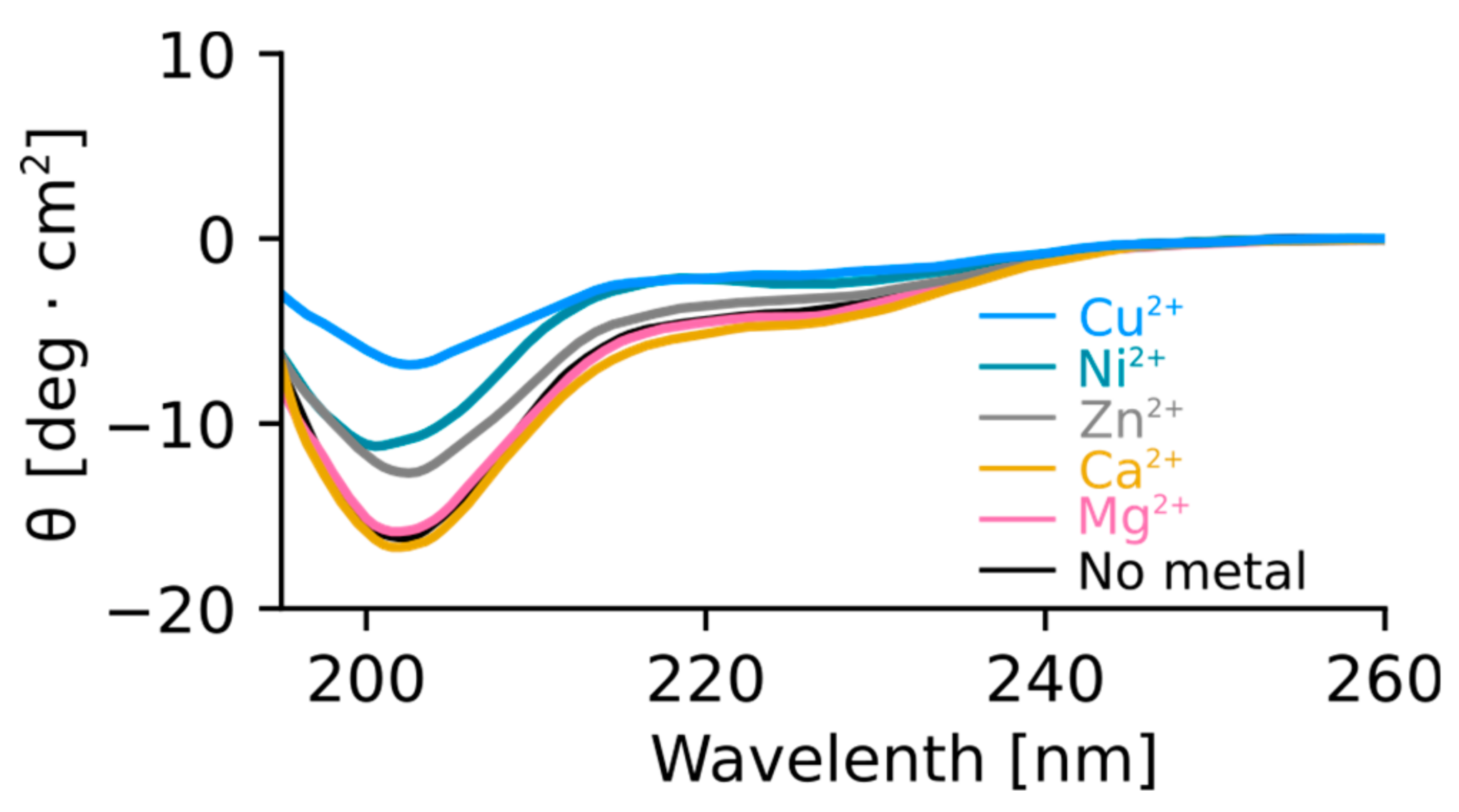
The N-Terminus of Orb2A Has an Affinity for Bivalent Metal Ions
4. Materials and Methods
4.1. Cloning of Orb2A Mutants
- Orb2A88Y2G NcoI GCCCCCTTCACCATGGGCAACAAATTTG
- Orb2A88Y2G XhoI ATAAGCTCGAGCGATCCTCCGCCTCCTCCACC
- Orb2A88WTsense GAAGGAGATATACCATGTACAACAAATTTGTTAATTTCATTTGCGGTG
- Orb2A88 WT antisense ACCGCAAATGAAATTAACAAATTTGTTGTACATGGTATATCTCCTTC
- Orb2A87 No polyhistidine-tag sense GAGGAGGCGGAGGATAGCTCGAGCACC
- Orb2A87 No polyhistidine-tag antisense GGTGCTCGAGCTATCCTCCGCCTCCTC
- Orb2A87 H29A sense AGCTCCACCAGCAACAGGCTCAACAACAGCATCAGC
- Orb2A87 H29A antisense GCTGATGCTGTTGTTGAGCCTGTTGCTGGTGGAGCT
- Orb2A87 H46A sense AACAGCAGCAACAGCTCGCTCAGCACCAACAGCAAC
- Orb2A87 H46A antisense GTTGCTGTTGGTGCTGAGCGAGCTGTTGCTGCTGTT
- Orb2A87 H60A sense GAATCTGAGTGCCCTGGCCCATCATCACCAGCAG
- Orb2A87 H60A antisense CTGCTGGTGATGATGGGCCAGGGCACTCAGATTC
- Orb2A87 H61Y sense CTGAGTGCCCTGCACTATCATCACCAGCAGC
- Orb2A87 H61Y antisense GCTGCTGGTGATGATAGTGCAGGGCACTCAG
4.2. Protein Expression
4.3. Protein Purification
4.4. Affinity Chromatography
4.5. ITC Measurements
4.6. Circular Dichroism
4.7. Thioflavin T Fluorescence Assay
4.8. Electron Microscopy
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ivshina, M.; Lasko, P.; Richter, J.D. Cytoplasmic Polyadenylation Element Binding Proteins in Development, Health, and Disease. Annu. Rev. Cell Dev. Biol. 2014, 30, 393–415. [Google Scholar] [CrossRef] [PubMed]
- Si, K.; Choi, Y.-B.; White-Grindley, E.; Majumdar, A.; Kandel, E.R. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell 2010, 140, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Si, K.; Giustetto, M.; Etkin, A.; Hsu, R.; Janisiewicz, A.M.; Miniaci, M.C.; Kim, J.-H.; Zhu, H.; Kandel, E.R. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in Aplysia. Cell 2003, 115, 893–904. [Google Scholar] [CrossRef]
- Raveendra, B.L.; Siemer, A.B.; Puthanveettil, S.V.; Hendrickson, W.A.; Kandel, E.R.; McDermott, A.E. Characterization of prion-like conformational changes of the neuronal isoform of Aplysia CPEB. Nat. Struct. Mol. Biol. 2013, 20, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Si, K.; Lindquist, S.; Kandel, E.R. A neuronal isoform of the Aplysia CPEB has prion-like properties. Cell 2003, 115, 879–891. [Google Scholar] [CrossRef]
- Mastushita-Sakai, T.; White-Grindley, E.; Samuelson, J.; Seidel, C.; Si, K. Drosophila Orb2 targets genes involved in neuronal growth, synapse formation, and protein turnover. Proc. Natl. Acad. Sci. USA 2010, 107, 11987–11992. [Google Scholar] [CrossRef] [PubMed]
- Keleman, K.; Krüttner, S.; Alenius, M.; Dickson, B.J. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat. Neurosci. 2007, 10, 1587–1593. [Google Scholar] [CrossRef] [PubMed]
- Stepien, B.K.; Oppitz, C.; Gerlach, D.; Dag, U.; Novatchkova, M.; Krüttner, S.; Stark, A.; Keleman, K. RNA-binding profiles of Drosophila CPEB proteins Orb and Orb2. Proc. Natl. Acad. Sci. USA 2016, 113, E7030–E7038. [Google Scholar] [CrossRef] [PubMed]
- Krüttner, S.; Stepien, B.; Noordermeer, J.N.; Mommaas, M.A.; Mechtler, K.; Dickson, B.J.; Keleman, K. Drosophila CPEB Orb2A Mediates Memory Independent of Its RNA-Binding Domain. Neuron 2012, 76, 383–395. [Google Scholar] [CrossRef] [PubMed]
- White-Grindley, E.; Li, L.; Mohammad Khan, R.; Ren, F.; Saraf, A.; Florens, L.; Si, K. Contribution of Orb2A Stability in Regulated Amyloid-Like Oligomerization of Drosophila Orb. PLoS Biol. 2014, 12, e1001786. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.R.; Li, L.; Pérez-Sánchez, C.; Saraf, A.; Florens, L.; Slaughter, B.D.; Unruh, J.R.; Si, K. Amyloidogenic Oligomerization Transforms Drosophila Orb2 from a Translation Repressor to an Activator. Cell 2015, 163, 1468–1483. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, A.; Cesario, W.C.; White-Grindley, E.; Jiang, H.; Ren, F.; Khan, M.R.; Li, L.; Choi, E.M.-L.; Kannan, K.; Guo, F.; et al. Critical Role of Amyloid-like Oligomers of Drosophila Orb2 in the Persistence of Memory. Cell 2012, 148, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Hervás, R.; Li, L.; Majumdar, A.; Fernández-Ramírez, M.C.; Unruh, J.R.; Slaughter, B.D.; Galera-Prat, A.; Santana, E.; Suzuki, M.; Nagai, Y.; et al. Molecular Basis of Orb2 Amyloidogenesis and Blockade of Memory Consolidation. PLoS Biol. 2016, 14, e1002361. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, S.A.; Bajakian, T.H.; Soria, M.A.; Falk, A.S.; Service, R.J.; Langen, R.; Siemer, A.B. Identification and Structural Characterization of the N-terminal Amyloid Core of Orb2 isoform A. Sci. Rep. 2016, 6, 38265. [Google Scholar] [CrossRef] [PubMed]
- Soria, M.A.; Cervantes, S.A.; Bajakian, T.H.; Siemer, A.B. The functional amyloid Orb2A binds to lipid membranes. Biophys. J. 2017, 113, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.-B.; Yang, N.; Sun, H. Metal-binding properties of an Hpn-like histidine-rich protein. Chem. Eur. J. 2011, 17, 5852–5860. [Google Scholar] [CrossRef] [PubMed]
- Chiera, N.M.; Rowinska-Zyrek, M.; Wieczorek, R.; Guerrini, R.; Witkowska, D.; Remelli, M.; Kozlowski, H. Unexpected impact of the number of glutamine residues on metal complex stability. Metallomics 2013, 5, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Simon, J.D. Insights into the thermodynamics of copper association with amyloid-β, α-synuclein and prion proteins. Metallomics 2011, 3, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Tõugu, V.; Tiiman, A.; Palumaa, P. Interactions of Zn(II) and Cu(II) ions with Alzheimer’s amyloid-β peptide. Metal ion binding, contribution to fibrillization and toxicity. Metallomics 2011, 3, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Volz, J.; Uwe Bosch, F.; Wunderlin, M.; Schuhmacher, M.; Melchers, K.; Bensch, K.; Steinhilber, W.; Schäfer, K.P.; Tóth, G.; Penke, B.; et al. Molecular characterization of metal-binding polypeptide domains by electrospray ionization mass spectrometry and metal chelate affinity chromatography. J. Chromatogr. A 1998, 800, 29–37. [Google Scholar] [CrossRef]
- Arnold, F.H. Metal-affinity separations: A new dimension in protein processing. Biotechnology 1991, 9, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Ueda, E.K.M.; Gout, P.W.; Morganti, L. Current and prospective applications of metal ion-protein binding. J. Chromatogr. A 2003, 988, 1–23. [Google Scholar] [CrossRef]
- LeVine, H. Thioflavine T interaction with amyloid β-sheet structures. Amyloid 1995, 2, 1–6. [Google Scholar] [CrossRef]
- Groenning, M.; Olsen, L.; van de Weert, M.; Flink, J.M.; Frokjaer, S.; Jørgensen, F.S. Study on the binding of Thioflavin T to β-sheet-rich and non-β-sheet cavities. J. Struct. Biol. 2007, 158, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Pandey, N.K.; Singha Roy, A.; Tripathy, D.R.; Dinda, A.K.; Dasgupta, S. Prolonged Glycation of Hen Egg White Lysozyme Generates Non Amyloidal Structures. PLoS ONE 2013, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.L.; Hulett, M.D.; Parish, C.R. Histidine-rich glycoprotein: A novel adaptor protein in plasma that modulates the immune, vascular and coagulation systems. Immunol. Cell Biol. 2005, 83, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Battistoni, A.; Pacello, F.; Mazzetti, A.P.; Capo, C.; Kroll, J.S.; Langford, P.R.; Sansone, A.; Donnarumma, G.; Valenti, P.; Rotilio, G. A histidine-rich metal binding domain at the N terminus of Cu, Zn-superoxide dismutases from pathogenic bacteria. A novel strategy for metal chaperoning. J. Biol. Chem. 2001, 276, 30315–30325. [Google Scholar] [CrossRef] [PubMed]
- Ranieri-Raggi, M.; Moir, A.J.G.; Raggi, A. The role of histidine-proline-rich glycoprotein as zinc chaperone for skeletal muscle AMP deaminase. Biomolecules 2014, 4, 474–497. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.H.; Wanker, E.E.; Andrade-Navarro, M.A. Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res. 2012, 40, 4273–4287. [Google Scholar] [CrossRef] [PubMed]
- Fretham, S.; Carlson, E.; Georgieff, M. The role of iron in learning and memory. Adv. Nutr. 2011, 2, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Kim, J. Effect of dietary iron loading on recognition memory in growing rats. PLoS ONE 2015, 10, e0120609. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Tamano, H. Significance of the degree of synaptic Zn2+ signaling in cognition. BioMetals 2016, 29, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.P. Metal ions and synaptic transmission: Think zinc. Proc. Natl. Acad. Sci. USA 1997, 94, 13386–13387. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hough, C.J.; Frederickson, C.J.; Sarvey, J.M. Induction of mossy fiber→CA3 long-term potentiation requires translocation of synaptically released Zn2+. J. Neurosci. 2001, 21, 8015–8025. [Google Scholar] [PubMed]
- Takeda, A. Insight into Glutamate Excitotoxicity from Synaptic Zinc Homeostasis. Int. J. Alzheimers. Dis. 2011, 2011, 491597. [Google Scholar] [CrossRef] [PubMed]
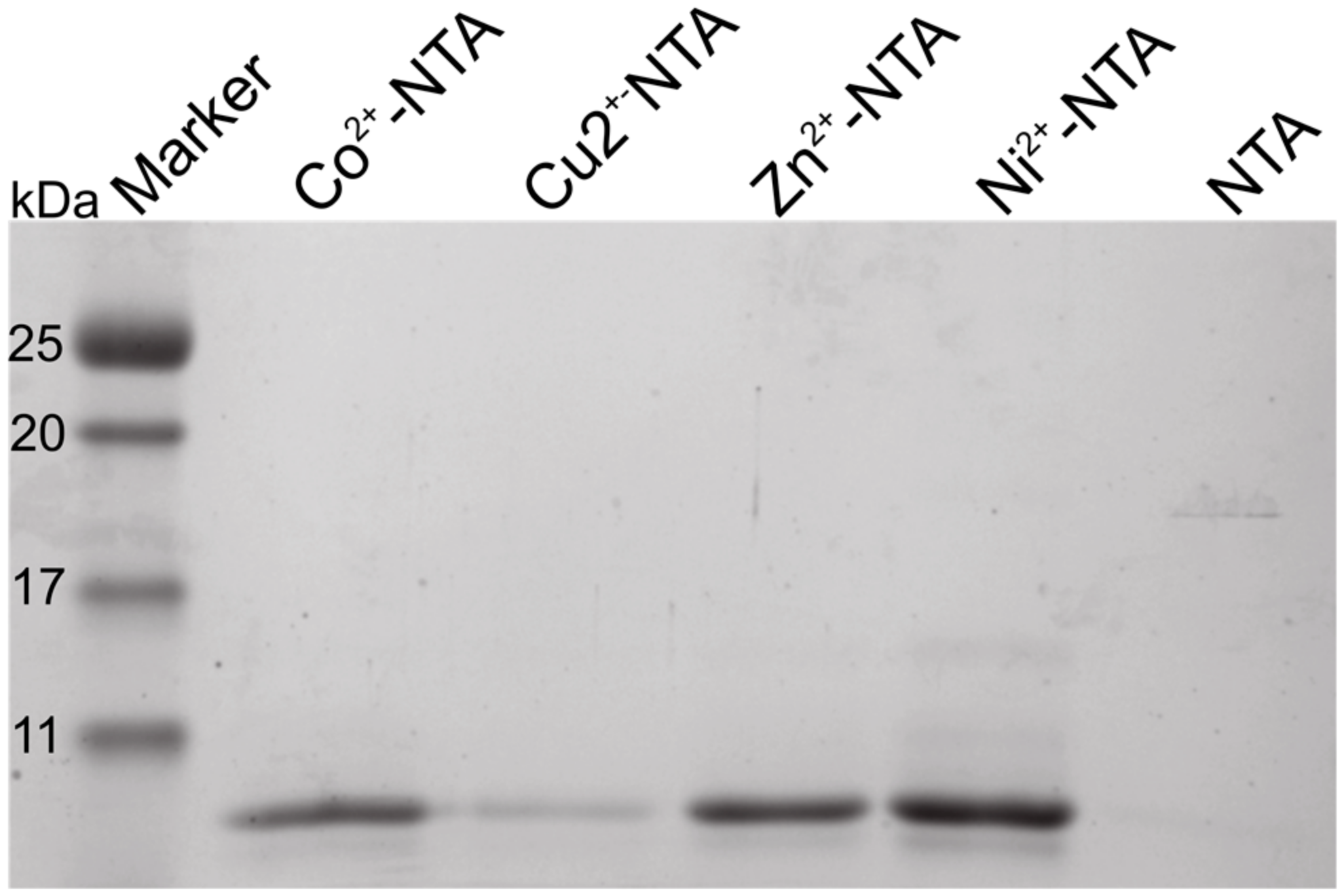

| Ligand | n (sites) | KD (M) | ΔH (kJ/mol) | ΔG (kJ/mol) | −TΔS (kJ/mol) |
|---|---|---|---|---|---|
| Ni2+ | 0.915 +/− 2.2·10−2 | 4.66e−6 +/− 758·10−9 | −69.4 +/− 3.57 | −31.0 | 38.4 |
| Zn2+ | 1.29 +/− 5.1·10−2 | 12.6e−6 +/− 2.13·10−6 | −37.8 +/− 3.53 | −28.5 | 9.38 |
| Cu2+ | Not determined * | - | - | - | - |
| Ca2+ | Not binding | - | - | - | - |
| Mg2+ | Not binding | - | - | - | - |
| Mutant | n (sites) | KD (M) | ΔH (kJ/mol) | ΔG (kJ/mol) | −TΔS (kJ/mol) |
|---|---|---|---|---|---|
| H29A | 0.412 +/− 1.1·10−2 | 8.21·10−6 +/− 805·10−9 | −61.3 +/− 3.48 | −29.5 | 31.7 |
| H46A | 0.715 +/− 6.8·10−2 | 4.14·10−6 +/− 809·10−9 | −41.9 +/− 6.12 | −31.3 | 10.7 |
| H60A | 0.652 +/− 6.8·10−2 | 2.49·10−6 +/− 1.33·10−6 | −41.9 +/− 8.06 | −32.5 | 9.41 |
| H61Y | Not binding * | - | - | - | - |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bajakian, T.H.; Cervantes, S.A.; Soria, M.A.; Beaugrand, M.; Kim, J.Y.; Service, R.J.; Siemer, A.B. Metal Binding Properties of the N-Terminus of the Functional Amyloid Orb2. Biomolecules 2017, 7, 57. https://doi.org/10.3390/biom7030057
Bajakian TH, Cervantes SA, Soria MA, Beaugrand M, Kim JY, Service RJ, Siemer AB. Metal Binding Properties of the N-Terminus of the Functional Amyloid Orb2. Biomolecules. 2017; 7(3):57. https://doi.org/10.3390/biom7030057
Chicago/Turabian StyleBajakian, Thalia H., Silvia A. Cervantes, Maria A. Soria, Maïwenn Beaugrand, Ji Yun Kim, Rachel J. Service, and Ansgar B. Siemer. 2017. "Metal Binding Properties of the N-Terminus of the Functional Amyloid Orb2" Biomolecules 7, no. 3: 57. https://doi.org/10.3390/biom7030057