tRNA Modification Detection Using Graphene Nanopores: A Simulation Study
Abstract
:1. Introduction
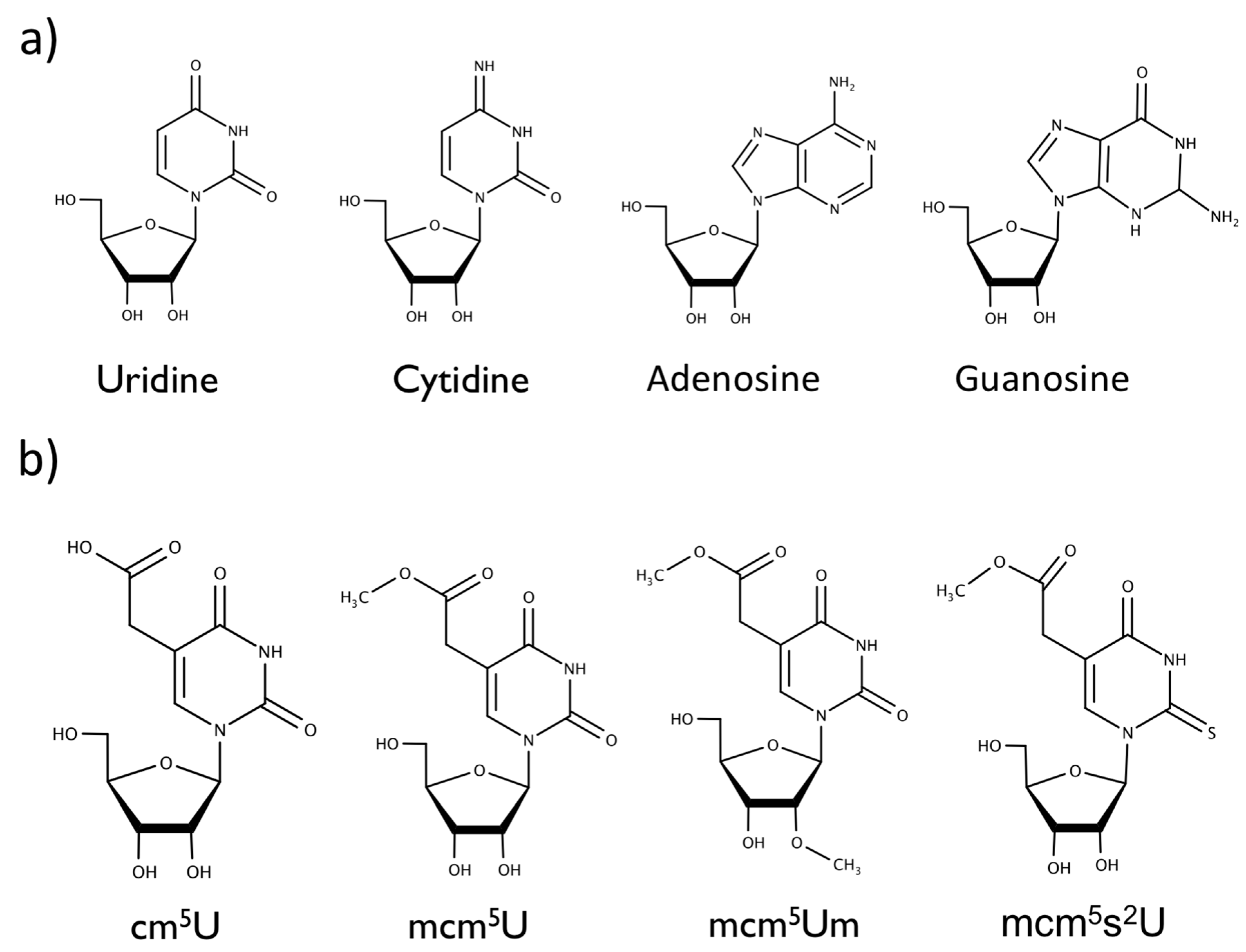
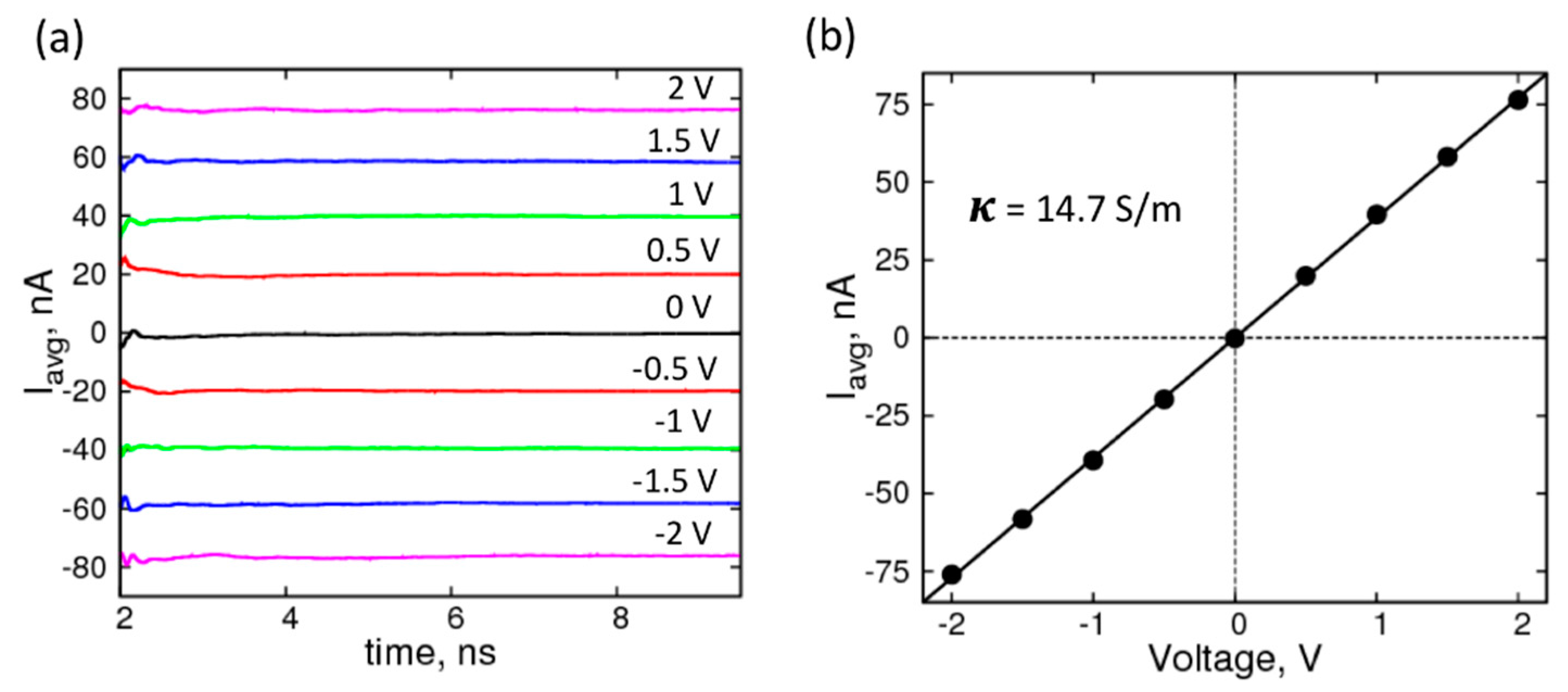
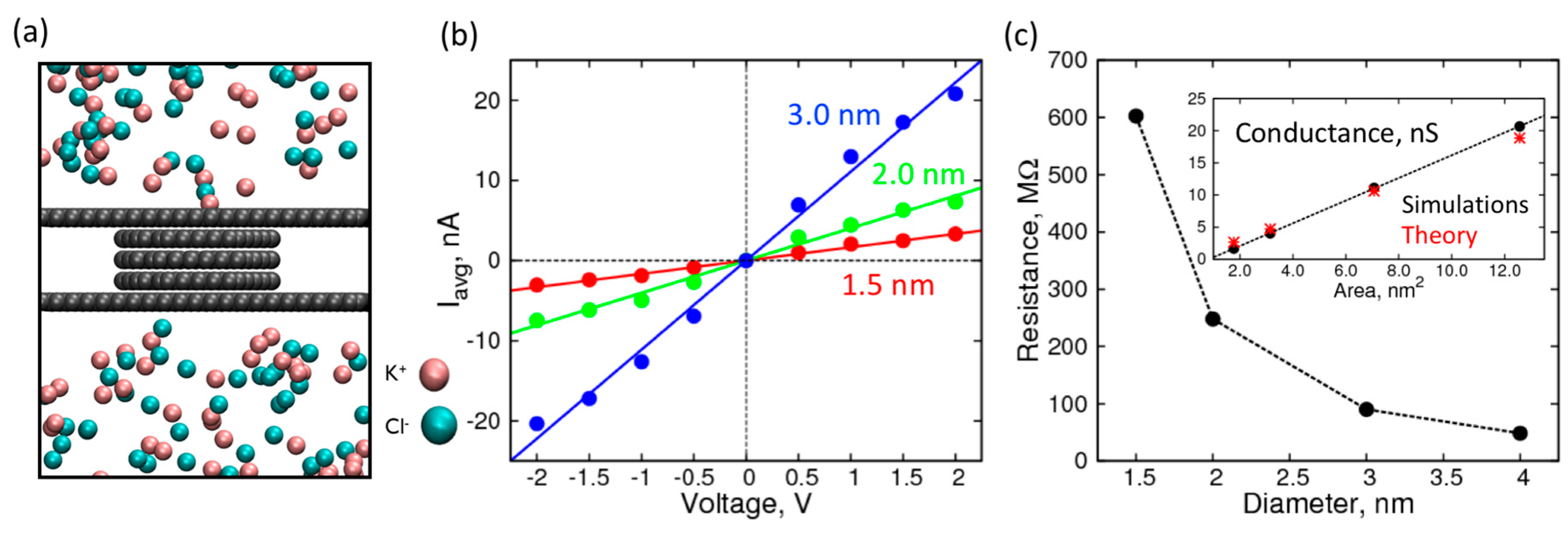
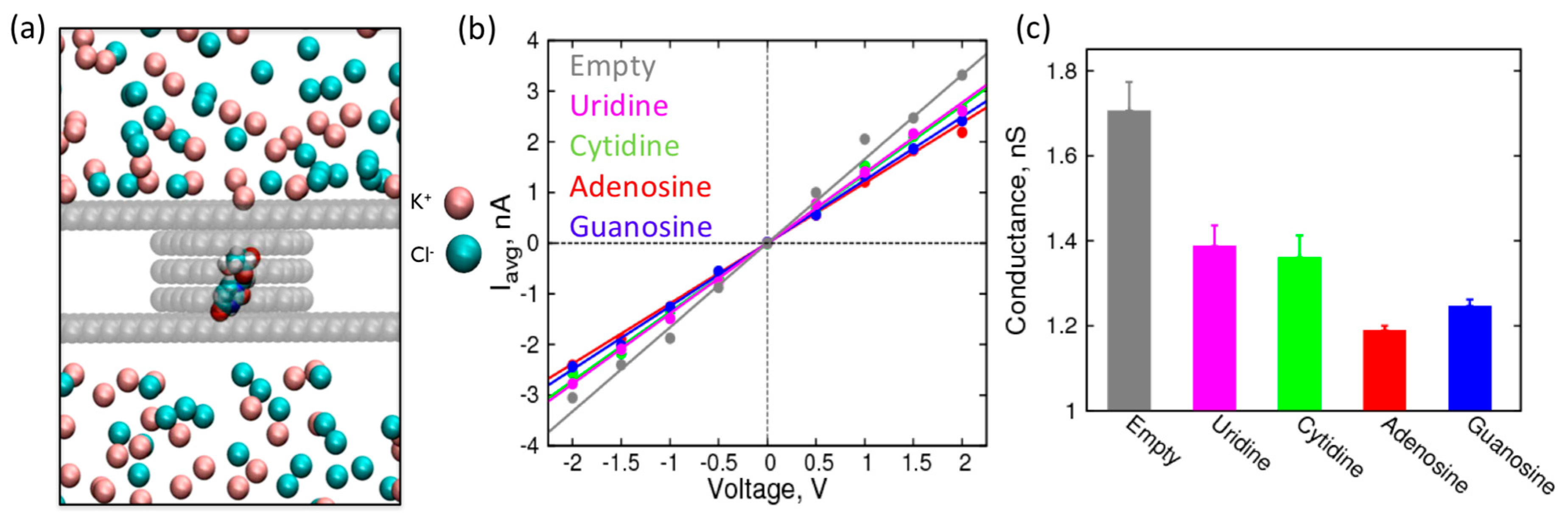
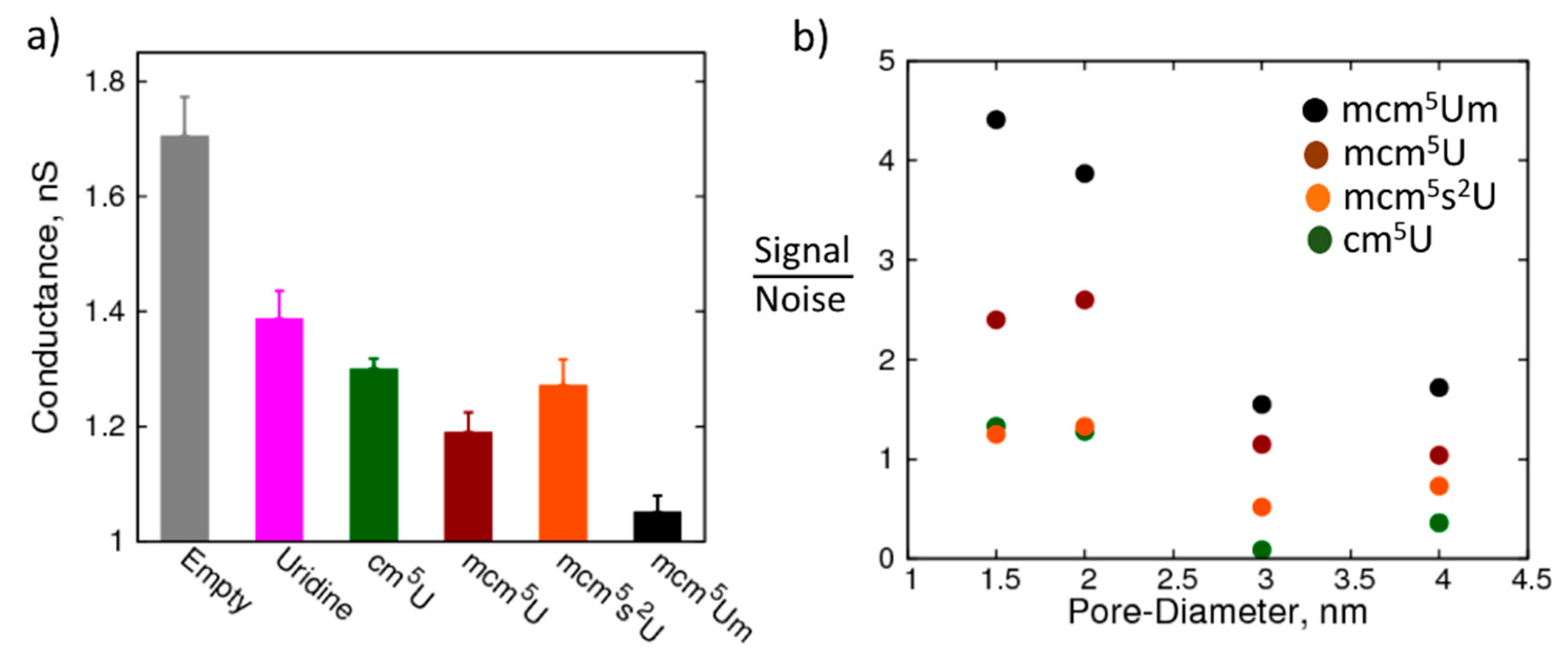
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gustilo, E.M.; Vendeix, F.A.; Agris, P.F. tRNA’s modifications bring order to gene expression. Curr. Opin. Microbiol. 2008, 11, 134–140. [Google Scholar] [CrossRef]
- Hall, R.H. The nature of strctural modification of nucleic acids. In The Modified Nucleosides in Nucelic Acids; Columbia University Press: New York, NY, USA, 1971; pp. 1–16. [Google Scholar]
- Russell, P.S.; Limbach, A.P. Evaluating the reproducibility of quantifying modified nucleosides from ribonucleic acids by LC–UV–MS. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 923–924, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Songe-Møller, L.; Born, E.V.D.; Leihne, V.; Vågbø, C.B.; Kristoffersen, T.; Krokan, H.E.; Kirpekar, F.; Falnes, P.L.Ø.; Klungland, A. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell. Biol. 2010, 30, 1814–1827. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.T.; Carlson, B.A.; Anderson, C.B.; Hatfield, D.L. Translational redefinition of UGA codons is regulated by selenium availability. J. Biol. Chem. 2013, 288, 19401–19413. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, S.; Bruynooghe, Y.; Denecker, G.; Huffel, S.V.; Tinton, S.; Beyaert, R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell 2000, 5, 597–605. [Google Scholar] [CrossRef]
- Coppinger, R.J.; Diamond, A.M. Selenium deficiency and human disease. In Selenium: Its Molecula Biology and Human Health; Hatfield, D.L., Ed.; Springer: New York, NY, USA, 2011; pp. 219–233. [Google Scholar]
- Combs, G.F. Selenium as a cancer preventive agent. In Selenium: Its Molecular Biology and Role in Human Health; Hatfield, D.L., Ed.; Springer: New York, NY, USA, 2011; pp. 205–217. [Google Scholar]
- Ischiropoulos, H.; Beckman, J.S. Oxidative stress and nitration in neurodegeneration: Cause, effect, or association? J. Clin. Investig. 2003, 111, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Storz, P. Reactive oxygen species in tumor progression. Front. Biosci. 2005, 10, 1881–1896. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, A.V.; Hatfield, D.L.; Gladyshev, V.N. Eukaryotic selenoproteins and selenoproteomes. Biochim. Biophys. Acta 2009, 1790, 1424–1428. [Google Scholar] [CrossRef] [PubMed]
- Jameson, R.R.; Diamond, A.M. A regulatory role for Sec tRNA[Ser]Sec in selenoprotein synthesis. RNA 2004, 10, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- The RNA Institute. The RNA Modification Database. Available online: http://mods.rna.albany.edu/mods/modifications/search (accessed on 10 November 2016).
- Chan, C.T.Y.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS ONE 2010, 6, e1001247. [Google Scholar] [CrossRef] [Green Version]
- Begley, U.; Sosa, M.S.; Avivar-Valderas, A.; Patil, A.; Endres, L.; Estrada, Y.; Chan, C.T.Y.; Su, D.; Dedon, P.C.; Aguirre-Ghiso, J.A.; et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-a. EMBO Mol. Med. 2013, 5, 366–383. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Dyavaiah, M.; Joseph, F.; Rooney, J.P.; Chan, C.T.Y.; Dedon, P.C.; Begley, T.J. Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response. Cell Cycle 2012, 11, 3656–3665. [Google Scholar] [CrossRef]
- Hall, R.H. Methods for isolation of modified nucleosides. In The Modified Nucleosides in Nucleic Acids; Columbia University Press: New York, NY, USA, 1971; pp. 209–255. [Google Scholar]
- Su, D.; Chan, C.T.Y.; Gu, C.; Lim, K.S.; Chionh, Y.H.; McBee, M.E.; Russell, B.S.; Babu, I.R.; Begley, T.J.; Dedon, P.C. Quantitative analysis of tRNA modifications by HPLC-coupled mass spectrometry. Nat. Protoc. 2014, 9, 828–841. [Google Scholar] [CrossRef] [PubMed]
- Mishima, E.; Jinno, D.; Akiyama, Y.; Itoh, K.; Nankumo, S.; Shima, H.; Kikuchi, K.; Takeuchi, Y.; Elkordy, A.; Suzuki, T.; et al. Immuno-northern blotting: Detection of RNA modifications by using antibodies against modified nucleosides. PLoS ONE 2015, 10, e0143756. [Google Scholar] [CrossRef] [PubMed]
- Kariko, K.; Muramatsu, H.; Ludwig, J.N.; Weissman, D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011, 39, e142. [Google Scholar] [CrossRef]
- Siwy, Z.S.; Davenport, M. Graphene opens up to DNA. Nat. Nanotechnol. 2010, 5, 697–698. [Google Scholar] [CrossRef]
- Gu, L.-Q.; Shim, J.W. Single molecule sensing by nanopores and nanopore devices. Analyst 2010, 135, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Luan, B.; Peng, H.; Polonsky, S.; Rossnagel, S.; Stolovitzky, G.; Martyna, G. Base-by-base ratcheting of single stranded DNA through a solid-state nanopore. Phys. Rev. Lett. 2010, 104, 238103. [Google Scholar] [CrossRef] [PubMed]
- Merchant, C.A.; Healy, K.; Wanunu, M.; Ray, V.; Peterman, N.; Bartel, J.; Fishbein, M.D.; Venta, K.; Luo, Z.; Johnson, A.T.C.; et al. DNA translocation thorugh graphene nanopores. Nano Lett. 2010, 10, 2915–2921. [Google Scholar] [CrossRef] [PubMed]
- Fyta, M.; Melchionna, S.; Succi, S. Translocation of biomolecules through solid-state nanopores: Theory meets experiments. J. Polym. Sci. Part B Polym. Phys. 2011, 49, 985–1011. [Google Scholar] [CrossRef]
- Venkatesan, B.M.; Bashir, R. Nanopore sensors for nucleic acid analysis. Nat. Nanotechnol. 2011, 6, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Rosenstein, J.K.; Wanunu, M.; Merchant, C.A.; Drndic, M.; Sherpard, K.L. Intergrated nanopore sensing platform with sub-microsecond temporal resolution. Nat. Methods 2012, 9, 487–492. [Google Scholar] [CrossRef]
- Haque, F.; Li, J.; Wu, H.-C.; Liang, X.-J.; Guo, P. Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA. Nano Today 2013, 8, 56–74. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Derek, S.; McMullan, C.; Branton, D.; Aziz, M.J.; Golovchenko, J. Ion-beam sculpting at nanometre length scales. Nature 2001, 412, 166–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, B.; Li, J.; Xu, D. Porous biomimetic membranes: Fabrication, properties and future applications. Phys. Chem. Chem. Phys. 2011, 13, 10584–10592. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.; Rivera, J.A.; Bashir, R. Electron beam induced local crystallization of HfO2 nanopores for biosensing applications. Nanoscale 2013, 5, 10887–10893. [Google Scholar] [CrossRef] [PubMed]
- Deamer, D.; Akeson, M.; Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016, 34, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Wendell, D.; Jing, P.; Geng, J.; Subramaniam, V.; Lee, T.J.; Montemagno, C.; Guo, P. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat. Nanotechnol. 2009, 4, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Abu-Shumays, R.; Akeson, M.; Bernick, D.L. Capture, unfolding, and detection of individual tRNA molecules using a nanopore device. Front. Bioeng. Biotechnol. 2015, 3, 91. [Google Scholar] [CrossRef] [PubMed]
- Souza, N.D. Protein nanopores to detect DNA methylation. Nat. Methods 2014, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Bell, N.A.W.; Hernandez-Anisa, S.M.; Engst, C.R.; Liedl, T.; Keyser, U.F. Ionic current detection of DNA origami nanostructures with nanocapillaries. In Nanopores for Bioanalytical Applications: Proceedings of the International Conference; Edel, J., Albrecht, T., Eds.; RSC Publishing: Piccadilly, London, UK, 2012; pp. 1–5. [Google Scholar]
- Oxford Nanopore Technologies. Solid-State Nanopores. Available online: https://nanoporetech.com/science-technology/introduction-to-nanopore-sensing/solid-state-nanopores (accessed on 12 July 2016).
- Venkatesan, B.M.; Rashid, B. Solid-state nanopore sensors for nucleic acid analysis. In Nanopores: Sensing and Fundamental Biological Applications; Iqbal, S.M., Basir, R., Eds.; Springer: New York, NY, USA, 2011; pp. 1–34. [Google Scholar]
- Schneider, G.G.F.; Kowalczyk, S.W.; Calado, V.E.; Pandraud, G.G.; Zandbergen, H.W.; Vandersypen, L.M.K.; Dekker, C. DNA translocation through graphene nanopores. Nano Lett. 2010, 10, 3163–3167. [Google Scholar] [CrossRef] [PubMed]
- Garaj, S.; Hubbard, W.; Reina, A.; Kong, J.; Branton, D.; Golovchenko, J.A. Graphene as a subnanometre trans-electrode membrane. Nature 2010, 467, 190–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comer, J.; Dimitrov, V.; Zhao, Q.; Timp, G.; Aksimentiev, A. Microscopic mechanics of hairpin DNA translocation through synthetic nanopores. Biophys. J. 2009, 96, 593–608. [Google Scholar] [CrossRef] [PubMed]
- Comer, J.; Aksimentiev, A. DNA sequence-dependent ionic currents in ultra-small solid-state nanopores. Nanoscale 2016, 8, 9600–9613. [Google Scholar] [CrossRef] [PubMed]
- Wells, D.B.; Belkin, M.; Comer, J.; Aksimentiev, A. Assessing graphene nanopores for sequencing DNA. Nano Lett. 2012, 12, 4117–4123. [Google Scholar] [CrossRef] [PubMed]
- Wanunu, M.; Cohen-Karni, D.; Johnson, R.R.; Fields, L.; Benner, J.; Peterman, N.; Zheng, Y.; Klein, M.L.; Drndic, M. Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J. Am. Chem. Soc. 2011, 133, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Carson, S.; Wilson, J.; Aksimentiev, A.; Weigele, P.R.; Wanunu, M. Hydroxymethyluracil modifications enhance the flexibility and hydrophilicity of double-stranded DNA. Nucleic Acid Res. 2016, 44, 2085–2092. [Google Scholar] [CrossRef] [PubMed]
- Aksimentiev, A.; Schulten, K. Imaging a-hemolysin with molecular dynamics: Ionic conductance, osmotic permeability, and the electrostatic potential map. Biophys. J. 2005, 88, 3745–3761. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Muzard, J.; Payet, L.; Mathe, J.; Bockelmann, U.; Aksimentiev, A.; Viasnoff, V. Rectification of the current in α-hemolysin pore depends on the cation type: The alkali series probed by molecular dynamics simulations and experiments. J. Phys. Chem. 2011, 115, 4255–4264. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Wanunu, M.; Bell, D.C.; Meller, A. Rapid fabrication of uniformly sized nanopores and nanopore arrays for parallel DNA analysis. Adv. Mater. 2006, 18, 3149–3153. [Google Scholar] [CrossRef]
- Ranganathan, S.V.; Halvorsen, K.; Myers, C.A.; Robertson, N.M.; Yigit, M.V.; Chen, A.A. Complex thermodynamic behavior of single-stranded nucleic acid adsorption to graphene surfaces. Langmuir 2016, 32, 6028–6034. [Google Scholar] [CrossRef] [PubMed]
- Michealson, A.M. Chemistry of nucleosides. In The Chemistry of Nucleosides and Nucleotides; Academic Press Inc.: London, UK, 1963; pp. 4–97. [Google Scholar]
- Hellen, C.U.T.; Sarnow, P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001, 15, 1593–1612. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Vanquelef, E.; Simon, S.; Marquant, G.; Garcia, E.; Klimerak, G.; Delepine, J.C.; Cieplak, P.; Dupradeau, F.-Y. R.E.D. Server: A web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucl. Acids Res. 2011, 39, W511–W517. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.A.; García, A.E. High-resolution reversible folding of hyperstable RNA tetraloops using molecular dynamics simulations. Proc. Natl. Acad. Sci USA 2013, 110, 16820–16825. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.A.; Pappu, R.V. Quantitative characterization of ion pairing and cluster formation in strong 1:1 electrolytes. J. Phys. Chem. B 2007, 111, 6469–6478. [Google Scholar] [CrossRef]
- Werder, T.; Walther, J.H.; Jaffe, R.L.; Halicioglu, T.; Koumoutsakos, P. On the water-carbon interaction for use in molecular dynamics simulations of graphite and carbon nanotubes. J. Phys. Chem. 2003, 107, 1345–1352. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Towns, J.; Cockerill, T.; Dahan, M.; Foster, I.; Gaither, K.; Grimshaw, A.; Hazlewood, V.; Lathrop, S.; Lifka, D.; Peterson, G.D.; et al. XSEDE: Accelerating scientific discovery. Comput. Sci. Eng. 2014, 16, 62–74. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onanuga, K.; Begley, T.J.; Chen, A.A.; Ranganathan, S.V. tRNA Modification Detection Using Graphene Nanopores: A Simulation Study. Biomolecules 2017, 7, 65. https://doi.org/10.3390/biom7030065
Onanuga K, Begley TJ, Chen AA, Ranganathan SV. tRNA Modification Detection Using Graphene Nanopores: A Simulation Study. Biomolecules. 2017; 7(3):65. https://doi.org/10.3390/biom7030065
Chicago/Turabian StyleOnanuga, Khadijah, Thomas J. Begley, Alan A. Chen, and Srivathsan V. Ranganathan. 2017. "tRNA Modification Detection Using Graphene Nanopores: A Simulation Study" Biomolecules 7, no. 3: 65. https://doi.org/10.3390/biom7030065





