Image-Based Analysis to Dissect Vertical Distribution and Horizontal Asymmetry of Conspecific Root System Interactions in Response to Planting Densities, Nutrients and Root Exudates in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
2.1. Capturing Root Systems Using a Portable Imaging Platform
2.2. Vertical Distribution of Root Systems
2.3. Root Circular Distributions of the Central Plant
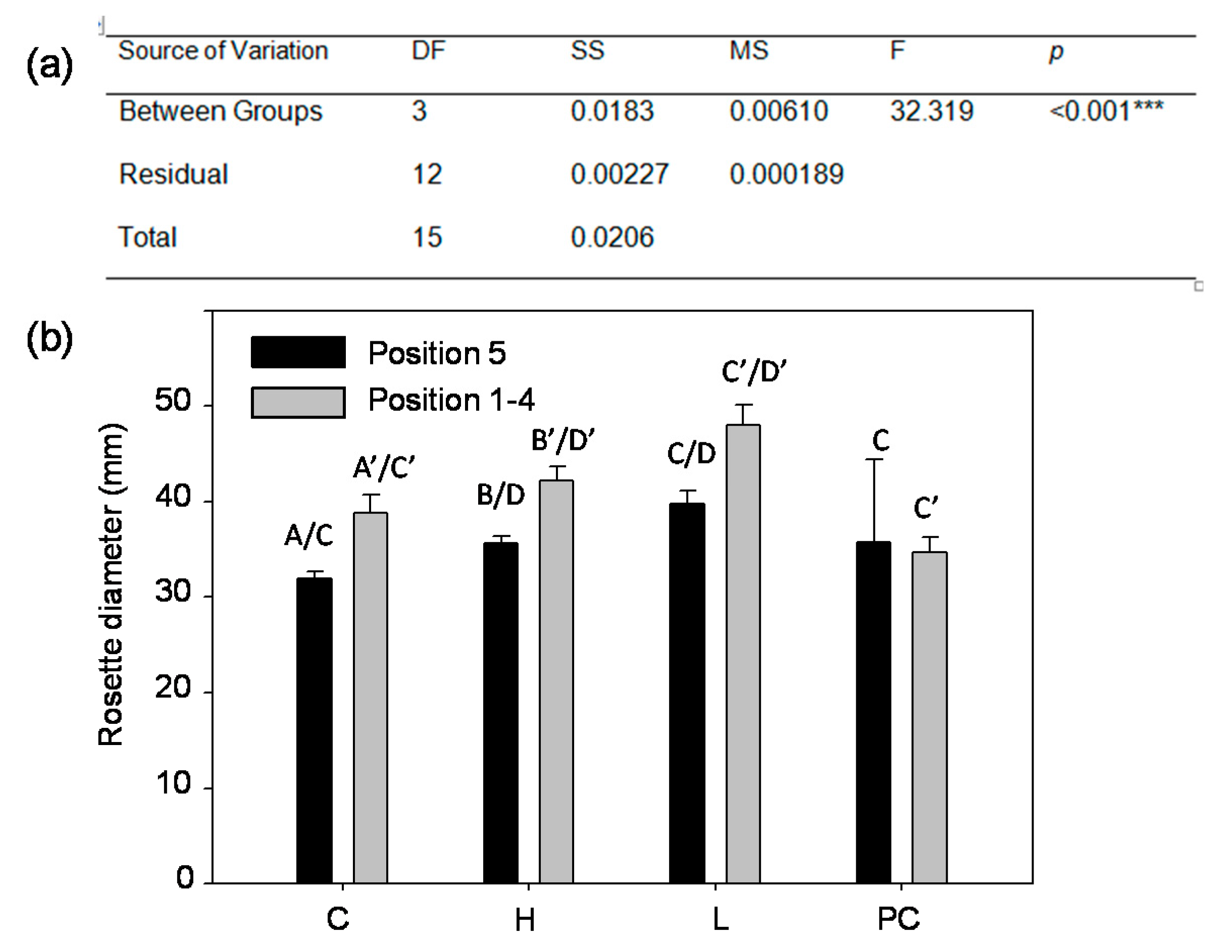
2.4. Aboveground Rosette Size at 28 Days after Planting
2.5. Diffusion of Inorganic Nitrogen Nutrients through Gel Media and Utilization of the Nutrients at 28 Days after Planting
3. Discussion
4. Materials and Methods
4.1. Seeds, Seed Germination, Conditioned Media Preparation, and Treatment Setup
4.2. Fluorescence Image Acquisition and Processing
4.3. Analysis of Inorganic Nitrogen Contents in the Gel Media and their Diffusion
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Grime, J. Plant Strategies and Vegetation Processes; John Wiley and Sons: Chichester, UK, 1979. [Google Scholar]
- Tilman, D. Resource Competition and Community Structure; Princeton University Press: Princeton, NJ, USA, 1982. [Google Scholar]
- Hacker, S.D.; Gaines, S.D. Some implications of direct positive interactions for community species diversity. Ecology 1997, 78, 1990–2003. [Google Scholar] [CrossRef]
- Tow, P.; Lazenby, A. Competition and Succession in Pastures—Some Concepts and Question; CABI Publishing: Wallingford, UK, 2001; Volume 1, pp. 1–14. [Google Scholar]
- Bruno, J.F.; Stachowicz, J.J.; Bertness, M.D. Inclusion of facilitation into ecological theory. Trends Ecol. Evol. 2003, 18, 119–125. [Google Scholar] [CrossRef]
- Clements, F.; Weaver, J.; Hanson, H. Plant Competition: An Analysis of Community Functions; Carnegie Institution: Washington, DC, USA, 1929. [Google Scholar]
- Clements, F.E. Competition in Plant Societies; Carnegie Institution of Washington: Washington, DC, USA, 1933. [Google Scholar]
- Grace, J.; Tilman, D. Perspectives on Plant Competition; Academic Press: San Diego, CA, USA, 1990. [Google Scholar]
- Keddy, P.A. Competition, 2nd ed.; Kluwer: Dordrecht, The Netherlands, 2001. [Google Scholar]
- Callaway, R.M. Positive Interactions and Interdependence in Plant Communities; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Soliveres, S.; Maestre, F.T. Plant-plant interactions, environmental gradients and plant diversity: A global synthesis of community-level studies. Perspect. Plant Ecol. Evol. Syst. 2014, 16, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.A.; Reich, P.B.; Palik, B.J. Untangling positive and negative biotic interactions: Views from above and below ground in a forest ecosystem. Ecology 2010, 91, 3641–3655. [Google Scholar] [CrossRef] [PubMed]
- Craine, J.M.; Dybzinski, R. Mechanisms of plant competition for nutrients, water and light. Funct. Ecol. 2013, 27, 833–840. [Google Scholar] [CrossRef]
- Skálová, H.; Jarošík, V.; Dvořáčková, Š.; Pyšek, P. Effect of intra- and interspecific competition on the performance of native and invasive species of impatiens under varying levels of shade and moisture. PLoS ONE 2013, 8, e62842. [Google Scholar] [CrossRef] [PubMed]
- Pierik, R.; de Wit, M. Shade avoidance: Phytochrome signalling and other aboveground neighbour detection cues. J. Exp. Bot. 2014, 65, 2815–2824. [Google Scholar] [CrossRef] [PubMed]
- Casper, B.B.; Jackson, R.B. Plant competition underground. Annu. Rev. Ecol. Syst. 1997, 28, 545–570. [Google Scholar] [CrossRef]
- Hawkes, C.V.; Casper, B.B. Lateral root function and root overlap among mycorrhizal and nonmycorrhizal herbs in a florida shrubland, measured using rubidium as a nutrient analog. Am. J. Bot. 2002, 89, 1289–1294. [Google Scholar] [CrossRef] [PubMed]
- Cahill, J.; Kembel, S.; Gustafson, D. Differential genetic influences on competitive effect and response in Arabidopsis thaliana. J. Ecol. 2005, 93, 958–967. [Google Scholar] [CrossRef]
- Alwerdt, J.L.; Gibson, D.J.; Ebbs, S.D.; Wood, A.J. Intraspecific interactions in Arabidopsis thaliana and the stomatal mutants tmm1–1 and sdd1–2. Biol. Plant. 2006, 50, 205–209. [Google Scholar] [CrossRef]
- Cahill, J.; Lamb, E. Interactions between root and shoot competition and plant traits. Hortic Sci. 2007, 42, 1110–1112. [Google Scholar]
- Novoplansky, A. Picking battles wisely: Plant behaviour under competition. Plant Cell Environ. 2009, 32, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Geisler, M.; Gibson, D.J.; Lindsey, K.J.; Millar, K.; Wood, A.J. Upregulation of photosynthesis genes, and down-regulation of stress defense genes, is the response of Arabidopsis thaliana shoots to intraspecific competition. Bot. Stud. 2012, 53, 85–96. [Google Scholar]
- Masclaux, F.; Bruessow, F.; Schweizer, F.; Gouhier-Darimont, C.; Keller, L.; Reymond, P. Transcriptome analysis of intraspecific competition in Arabidopsis thaliana reveals organ-specific signatures related to nutrient acquisition and general stress response pathways. BMC Plant Biol. 2012, 12, 227. [Google Scholar] [CrossRef] [PubMed]
- Bowsher, A.W.; Shetty, P.; Anacker, B.L.; Siefert, A.; Strauss, S.Y.; Friesen, M.L. Transcriptomic responses to conspecific and congeneric competition in co-occurring Trifolium. J. Ecol. 2017, 105, 602–615. [Google Scholar] [CrossRef]
- Poorter, H.; Niklas, K.J.; Reich, P.B.; Oleksyn, J.; Poot, P.; Mommer, L. Biomass allocation to leaves, stems and roots: Meta-analyses of interspecific variation and environmental control. New Phytol. 2012, 193, 30–50. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.G.; Ali, M.; Yang, S.; Parchami, N.; Bento, T.; Mazzella, A.; Oni, M.; Riley, M.C.; Schneider, K.; Massa, N. Kin recognition is a nutrient-dependent inducible phenomenon. Plant Signal. Behav. 2016, 11, e1224045. [Google Scholar] [CrossRef] [PubMed]
- Semchenko, M.; Saar, S.; Lepik, A. Plant root exudates mediate neighbour recognition and trigger complex behavioural changes. New Phytol. 2014, 204, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Dudley, S.A.; Murphy, G.P.; File, A.L. Kin recognition and competition in plants. Funct. Ecol. 2013, 27, 898–906. [Google Scholar] [CrossRef]
- Falik, O.; Reides, P.; Gersani, M.; Novoplansky, A. Root navigation by self inhibition. Plant Cell Environ. 2005, 28, 562–569. [Google Scholar] [CrossRef]
- Falik, O.; Reides, P.; Gersani, M.; Novoplansky, A. Self/non-self discrimination in roots. J. Ecol. 2003, 91, 525–531. [Google Scholar] [CrossRef]
- Hess, L.; De Kroon, H. Effects of rooting volume and nutrient availability as an alternative explanation for root self/non-self discrimination. J. Ecol. 2007, 95, 241–251. [Google Scholar] [CrossRef]
- Semchenko, M.; Hutchings, M.J.; John, E.A. Challenging the tragedy of the commons in root competition: Confounding effects of neighbour presence and substrate volume. J. Ecol. 2007, 95, 252–260. [Google Scholar] [CrossRef]
- Enquist, B.J.; Niklas, K.J. Global allocation rules for patterns of biomass partitioning in seed plants. Science 2002, 295, 1517–1520. [Google Scholar] [CrossRef] [PubMed]
- De Dorlodot, S.; Forster, B.; Pagès, L.; Price, A.; Tuberosa, R.; Draye, X. Root system architecture: Opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 2007, 12, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Deak, K.I.; Malamy, J. Osmotic regulation of root system architecture. Plant J. 2005, 43, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Malamy, J. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 2005, 28, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Osmont, K.S.; Sibout, R.; Hardtke, C.S. Hidden branches: Developments in root system architecture. Annu. Rev. Plant Biol. 2007, 58, 93–113. [Google Scholar] [CrossRef] [PubMed]
- Gruber, B.D.; Giehl, R.F.H.; Friedel, S.; von Wirén, N. Plasticity of the arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013, 163, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Giehl, R.F.H.; Gruber, B.D.; von Wirén, N. It’s time to make changes: Modulation of root system architecture by nutrient signals. J. Exp. Bot. 2014, 65, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Giehl, R.F.H.; von Wirén, N. Root nutrient foraging. Plant Physiol. 2014, 166, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Kellermeier, F.; Armengaud, P.; Seditas, T.J.; Danku, J.; Salt, D.E.; Amtmann, A. Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 2014, 26, 1480–1496. [Google Scholar] [CrossRef] [PubMed]
- Kiba, T.; Feria-Bourrellier, A.-B.; Lafouge, F.; Lezhneva, L.; Boutet-Mercey, S.; Orsel, M.; Bréhaut, V.; Miller, A.; Daniel-Vedele, F.; Sakakibara, H.; et al. The Arabidopsis nitrate transporter nrt2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 2012, 24, 245–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiba, T.; Krapp, A. Plant nitrogen acquisition under low availability: Regulation of uptake and root architecture. Plant Cell Physiol. 2016, 57, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.G. Nitrogen signalling pathways shaping root system architecture: An update. Curr. Opin. Plant Biol. 2014, 21, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, Z.; Amtmann, A. Food for thought: How nutrients regulate root system architecture. Curr. Opin. Plant Biol. 2017, 39, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Cahill, J.F.; McNickle, G.G.; Haag, J.J.; Lamb, E.G.; Nyanumba, S.M.; St. Clair, C.C. Plants integrate information about nutrients and neighbors. Science 2010, 328, 1657. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Aggarwal, P.; Robbins, N.E.; Sturrock, C.J.; Thompson, M.C.; Tan, H.Q.; Tham, C.; Duan, L.; Rodriguez, P.L.; Vernoux, T.; et al. Plant roots use a patterning mechanism to position lateral root branches toward available water. Proc. Natl. Acad. Sci. USA 2014, 111, 9319–9324. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- De-la-Peña, C.; Loyola-Vargas, V.M. Biotic interactions in the rhizosphere: A diverse cooperative enterprise for plant productivity. Plant Physiol. 2014, 166, 701–719. [Google Scholar] [CrossRef] [PubMed]
- Barlow, P.W. The root cap: Cell dynamics, cell differentiation and cap function. J. Plant Growth Regul. 2002, 21, 261–286. [Google Scholar] [CrossRef]
- Driouich, A.; Durand, C.; Cannesan, M.-A.; Percoco, G.; Vicré-Gibouin, M. Border cells versus border-like cells: Are they alike? J. Exp. Bot. 2010, 61, 3827–3831. [Google Scholar] [CrossRef] [PubMed]
- Durand, C.; Vicré-Gibouin, M.; Follet-Gueye, M.L.; Duponchel, L.; Moreau, M.; Lerouge, P.; Driouich, A. The organization pattern of root border-like cells of arabidopsis is dependent on cell wall homogalacturonan. Plant Physiol. 2009, 150, 1411–1421. [Google Scholar] [CrossRef] [PubMed]
- Kumpf, R.P.; Nowack, M.K. The root cap: A short story of life and death. J. Exp. Bot. 2015, 66, 5651–5662. [Google Scholar] [CrossRef] [PubMed]
- Badri, D.V.; Loyola-Vargas, V.M.; Broeckling, C.D.; De-la-Peña, C.; Jasinski, M.; Santelia, D.; Martinoia, E.; Sumner, L.W.; Banta, L.M.; Stermitz, F.; et al. Altered profile of secondary metabolites in the root exudates of Arabidopsis atp-binding cassette transporter mutants. Plant Physiol. 2008, 146, 762–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strehmel, N.; Böttcher, C.; Schmidt, S.; Scheel, D. Profiling of secondary metabolites in root exudates of arabidopsis thaliana. Phytochemistry 2014, 108, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Armengaud, P. Ez-rhizo software: The gateway to root architecture analysis. Plant Signal. Behav. 2009, 4, 139–141. [Google Scholar] [CrossRef] [PubMed]
- French, A.; Ubeda-Tomás, S.; Holman, T.J.; Bennett, M.J.; Pridmore, T. High-throughput quantification of root growth using a novel image-analysis tool. Plant Physiol. 2009, 150, 1784–1795. [Google Scholar] [CrossRef] [PubMed]
- Iyer-Pascuzzi, A.S.; Symonova, O.; Mileyko, Y.; Hao, Y.; Belcher, H.; Harer, J.; Weitz, J.S.; Benfey, P.N. Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol. 2010, 152, 1148–1157. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.T.; MacCurdy, R.B.; Jung, J.K.; Shaff, J.E.; McCouch, S.R.; Aneshansley, D.J.; Kochian, L.V. Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol. 2011, 156, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Lopez, L.; Shantharaj, D.; Liu, L.; Baisb, H.; Yu, J. Robust image-based 3-d modeling of root architecture. In Proceedings of the Computer Graphics International 2011, Ottawa, ON, Canada, 12–15 June 2011. [Google Scholar]
- Galkovskyi, T.; Mileyko, Y.; Bucksch, A.; Moore, B.; Symonova, O.; Price, C.; Topp, C.; Iyer-Pascuzzi, A.; Zurek, P.; Fang, S.; et al. Gia roots: Software for the high throughput analysis of plant root system architecture. BMC Plant Biol. 2012, 12, 116. [Google Scholar] [CrossRef] [PubMed]
- Wells, D.M.; French, A.P.; Naeem, A.; Ishaq, O.; Traini, R.; Hijazi, H.; Bennett, M.J.; Pridmore, T.P. Recovering the dynamics of root growth and development using novel image acquisition and analysis methods. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1517–1524. [Google Scholar] [CrossRef] [PubMed]
- Iyer-Pascuzzi, A.; Zurek, P.; Benfey, P. High-throughput, noninvasive imaging of root systems. In Plant Organogenesis; De Smet, I., Ed.; Humana Press: New York, NY, USA, 2013; Volume 959, pp. 177–187. [Google Scholar]
- Ristova, D.; Rosas, U.; Krouk, G.; Ruffel, S.; Birnbaum, K.D.; Coruzzi, G.M. Rootscape: A landmark-based system for rapid screening of root architecture in Arabidopsis. Plant Physiol. 2013, 161, 1086–1096. [Google Scholar] [CrossRef] [PubMed]
- Topp, C.N.; Iyer-Pascuzzi, A.S.; Anderson, J.T.; Lee, C.-R.; Zurek, P.R.; Symonova, O.; Zheng, Y.; Bucksch, A.; Mileyko, Y.; Galkovskyi, T.; et al. 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc. Natl. Acad. Sci. USA 2013, 110, E1695–E1704. [Google Scholar] [CrossRef] [PubMed]
- Jeudy, C.; Adrian, M.; Baussard, C.; Bernard, C.; Bernaud, E.; Bourion, V.; Busset, H.; Cabrera-Bosquet, L.; Cointault, F.; Han, S.; et al. Rhizotubes as a new tool for high throughput imaging of plant root development and architecture: Test, comparison with pot grown plants and validation. Plant Methods 2016, 12, 31. [Google Scholar] [CrossRef] [PubMed]
- Metzner, R.; Eggert, A.; van Dusschoten, D.; Pflugfelder, D.; Gerth, S.; Schurr, U.; Uhlmann, N.; Jahnke, S. Direct comparison of mri and X-ray ct technologies for 3d imaging of root systems in soil: Potential and challenges for root trait quantification. Plant Methods 2015, 11, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Dusschoten, D.; Metzner, R.; Kochs, J.; Postma, J.A.; Pflugfelder, D.; Bühler, J.; Schurr, U.; Jahnke, S. Quantitative 3d analysis of plant roots growing in soil using magnetic resonance imaging. Plant Physiol. 2016, 170, 1176–1188. [Google Scholar] [CrossRef] [PubMed]
- Schulz, H.; Postma, J.; van Dusschoten, D.; Scharr, H.; Behnke, S. Plant root system analysis from mri images. In Computer Vision, Imaging and Computer Graphics. Theory and Application; Csurka, G., Kraus, M., Laramee, R., Richard, P., Braz, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 359, pp. 411–425. [Google Scholar]
- Rellán-Álvarez, R.; Lobet, G.; Lindner, H.; Pradier, P.-L.; Sebastian, J.; Yee, M.-C.; Geng, Y.; Trontin, C.; LaRue, T.; Schrager-Lavelle, A.; et al. Glo-roots: An imaging platform enabling multidimensional characterization of soil-grown root systems. eLife 2015, 4, e07597. [Google Scholar] [CrossRef] [PubMed]
- Rellán-Álvarez, R.; Lobet, G.; Lindner, H.; Pradier, P.-L.M.; Yee, M.-C.; Sebastian, J.; Geng, Y.; Trontin, C.; LaRue, T.; Schrager Lavelle, A.; et al. Multidimensional mapping of root responses to soil environmental cues using a luminescence-based imaging system. bioRxiv 2015, 016931. [Google Scholar] [CrossRef]
- Postma, J.A.; Kuppe, C.; Owen, M.R.; Mellor, N.; Griffiths, M.; Bennett, M.J.; Lynch, J.P.; Watt, M. Opensimroot: Widening the scope and application of root architectural models. New Phytol. 2017, 215, 1274–1286. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P.; Nielsen, K.L.; Davis, R.D.; Jablokow, A.G. Simroot: Modelling and visualization of root systems. Plant Soil 1997, 188, 139–151. [Google Scholar] [CrossRef]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, 8, 679–698. [Google Scholar] [CrossRef] [PubMed]
- MathWorks, T.M., Inc. Matlab—The Language of Technical Computing. Available online: http://www.mathworks.com/products/matlab/ (accessed on 12 October 2016).
- MIT. Matlab: R2016b. Available online: http://www.mathworks.com/products/matlab/ (accessed on 9 October 2016).
- Gersani, M.; Brown, J.; O’Brien, E.; Maina, G.; Abramsky, Z. Tragedy of the commons as a result of root competition. J. Ecol. 2001, 89, 660–669. [Google Scholar] [CrossRef]
- Maina, G.; Brown, J.; Gersani, M. Intra-plant versus inter-plant root competition in beans: Avoidance, resource matching or tragedy of the commons. Plant Ecol. 2002, 160, 235–247. [Google Scholar] [CrossRef]
- Gruntman, M.; Novoplansky, A. Physiologically mediated self/non-self discrimination in roots. Proc. Natl. Acad. Sci. USA 2004, 101, 3863–3867. [Google Scholar] [CrossRef] [PubMed]
- Boyes, D.C.; Zayed, A.M.; Ascenzi, R.; McCaskill, A.J.; Hoffman, N.E.; Davis, K.R.; Gorlach, J. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 2001, 13, 1499–1510. [Google Scholar] [CrossRef] [PubMed]
- Kjemtrup, S.; Boyes, D.C.; Christensen, C.; McCaskill, A.J.; Hylton, M.; Davis, K. Growth stage-based phenotypic profiling of plants. Methods Mol. Biol. 2003, 236, 427–442. [Google Scholar] [PubMed]
- Waisel, Y.; Eshel, A.; Beeckman, T.; Kafkafi, U. Plant Roots: The Hidden Half, 3rd ed.; Marcerl Dekker, Inc.: New York, NY, USA; Basel, Switzerland, 2002; p. 1644. [Google Scholar]
- Eshel, A.; Beeckman, T. Plant Roots: The Hidden Half, 4th ed.; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Ohto, M.-A.; Onai, K.; Furukawa, Y.; Aoki, E.; Araki, T.; Nakamura, K. Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol. 2001, 127, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Purves, D.W.; Law, R. Fine-scale spatial structure in a grassland community: Quantifying the plant’s-eye view. J. Ecol. 2002, 90, 121–129. [Google Scholar] [CrossRef]
- Mutic, J.J.; Wolf, J.B. Indirect genetic effects from ecological interactions in Arabidopsis thaliana. Mol. Ecol. 2007, 16, 2371–2381. [Google Scholar] [CrossRef] [PubMed]
- Craine, J.M.; Fargione, J.; Sugita, S. Supply pre-emption, not concentration reduction, is the mechanism of competition for nutrients. New Phytol. 2005, 166, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Risueno, M.A.; Van Norman, J.M.; Moreno, A.; Zhang, J.; Ahnert, S.E.; Benfey, P.N. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 2010, 329, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.L.; Ishikawa, H.; Estelle, M.A. Responses of Arabidopsis roots to auxin studied with high temporal resolution: Comparison of wild type and auxin-response mutants. Planta 1994, 194, 215–222. [Google Scholar] [CrossRef]
- Vermeer, J.E.M.; Geldner, N. Lateral root initiation in Arabidopsis thaliana: A force awakens. F1000Prime Rep. 2015, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Vermeer, J.E.M.; von Wangenheim, D.; Barberon, M.; Lee, Y.; Stelzer, E.H.K.; Maizel, A.; Geldner, N. A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 2014, 343, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, M.; ten Tusscher, K.H. Periodic lateral root priming: What makes it tick? Plant Cell 2017, 29, 432–444. [Google Scholar] [CrossRef] [PubMed]
- MacGregor, D.R.; Deak, K.I.; Ingram, P.A.; Malamy, J.E. Root system architecture in Arabidopsis grown in culture is regulated by sucrose uptake in the aerial tissues. Plant Cell 2008, 20, 2643–2660. [Google Scholar] [CrossRef] [PubMed]
- Pigliucci, M.; Kolodynska, A. Phenotypic plasticity and integration in response to flooded conditions in natural accessions of Arabidopsis thaliana (l.) heynh (brassicaceae). Ann. Bot. 2002, 90, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Rosas, U.; Cibrian-Jaramillo, A.; Ristova, D.; Banta, J.A.; Gifford, M.L.; Fan, A.H.; Zhou, R.W.; Kim, G.J.; Krouk, G.; Birnbaum, K.D.; et al. Integration of responses within and across Arabidopsis natural accessions uncovers loci controlling root systems architecture. Proc. Natl. Acad. Sci. USA 2013, 110, 15133–15138. [Google Scholar] [CrossRef] [PubMed]
- Drew, M.C. Comparison of the effects of a localised supply of phosphate, nitrate, ammonium and potassium on the growth of the seminal root system, and the shoot, in barley. New Phytol. 1975, 75, 479–490. [Google Scholar] [CrossRef]
- Kapulnik, Y.; Delaux, P.-M.; Resnick, N.; Mayzlish-Gati, E.; Wininger, S.; Bhattacharya, C.; Séjalon-Delmas, N.; Combier, J.-P.; Bécard, G.; Belausov, E.; et al. Strigolactones affect lateral root formation and root-hair elongation in arabidopsis. Planta 2011, 233, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Koltai, H. Strigolactones are regulators of root development. New Phytol. 2011, 190, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Ruyter-Spira, C.; Kohlen, W.; Charnikhova, T.; van Zeijl, A.; van Bezouwen, L.; de Ruijter, N.; Cardoso, C.; Lopez-Raez, J.A.; Matusova, R.; Bours, R.; et al. Physiological effects of the synthetic strigolactone analog gr24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiol. 2011, 155, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.C.; Koch, K.E.; Suzuki, M.; Wu, S.; Latshaw, S.; Petruff, T.; Goulet, C.; Klee, H.J.; McCarty, D.R. Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol. 2012, 160, 1303–1317. [Google Scholar] [CrossRef] [PubMed]
- Micallef, S.A.; Shiaris, M.P.; Colón-Carmona, A. Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J. Exp. Bot. 2009, 60, 1729–1742. [Google Scholar] [CrossRef] [PubMed]
- Tinker, P.B.; Nye, P.H. Solute Movement in the Rhizosphere; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Ohkubo, Y.; Tanaka, M.; Tabata, R.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 2017, 3, 17029. [Google Scholar] [CrossRef] [PubMed]
- Domagalska, M.A.; Leyser, O. Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 2011, 12, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Puig, J.; Pauluzzi, G.; Guiderdoni, E.; Gantet, P. Regulation of shoot and root development through mutual signaling. Mol. Plant 2012, 5, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Giavasis, I.; Harvey, L.; McNeil, B. Gellan gum. Crit. Rev. Biotechnol. 2000, 20, 177–211. [Google Scholar] [CrossRef] [PubMed]
- Yokawa, K.; Kagenishi, T.; Kawano, T.; Mancuso, S.; Baluška, F. Illumination of arabidopsis roots induces immediate burst of ros production. Plant Signal. Behav. 2011, 6, 1460–1464. [Google Scholar] [CrossRef] [PubMed]
- Doi, K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Keeney, D.; Nelson, D. Nitrogen—Inorganic forms. In Methods of Soil Analysis, 2nd ed.; Page, A., Miller, R., Keeney, D., Eds.; American Society of Agronomy: Madison, WI, USA, 1982; Volume 9, Part 2; pp. 643–698. [Google Scholar]
- O’Dell, J. Determination of ammonia nitrogen by semi-automated colorimetry. In Method 350.1.; US Environmental Protection Agency: Cincinnati, OH, USA, 1993. [Google Scholar]
- R-Core-Team. R: A Language and Environment for Statistical Computing, 3.4.0; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Pewsey, A.; Neuhäuser, M.; Ruxton, G. Circular Statistics in r; Oxford University Press: Oxford, UK, 2013. [Google Scholar]
- Littell, R.C.; Milliken, G.A.; Stroup, W.W.; Wolfinger, R.D.; Schabenberger, O. Sas® for Mixed Models, 2nd ed.; SAS Institute, Inc.: Cary, NC, USA, 2006. [Google Scholar]
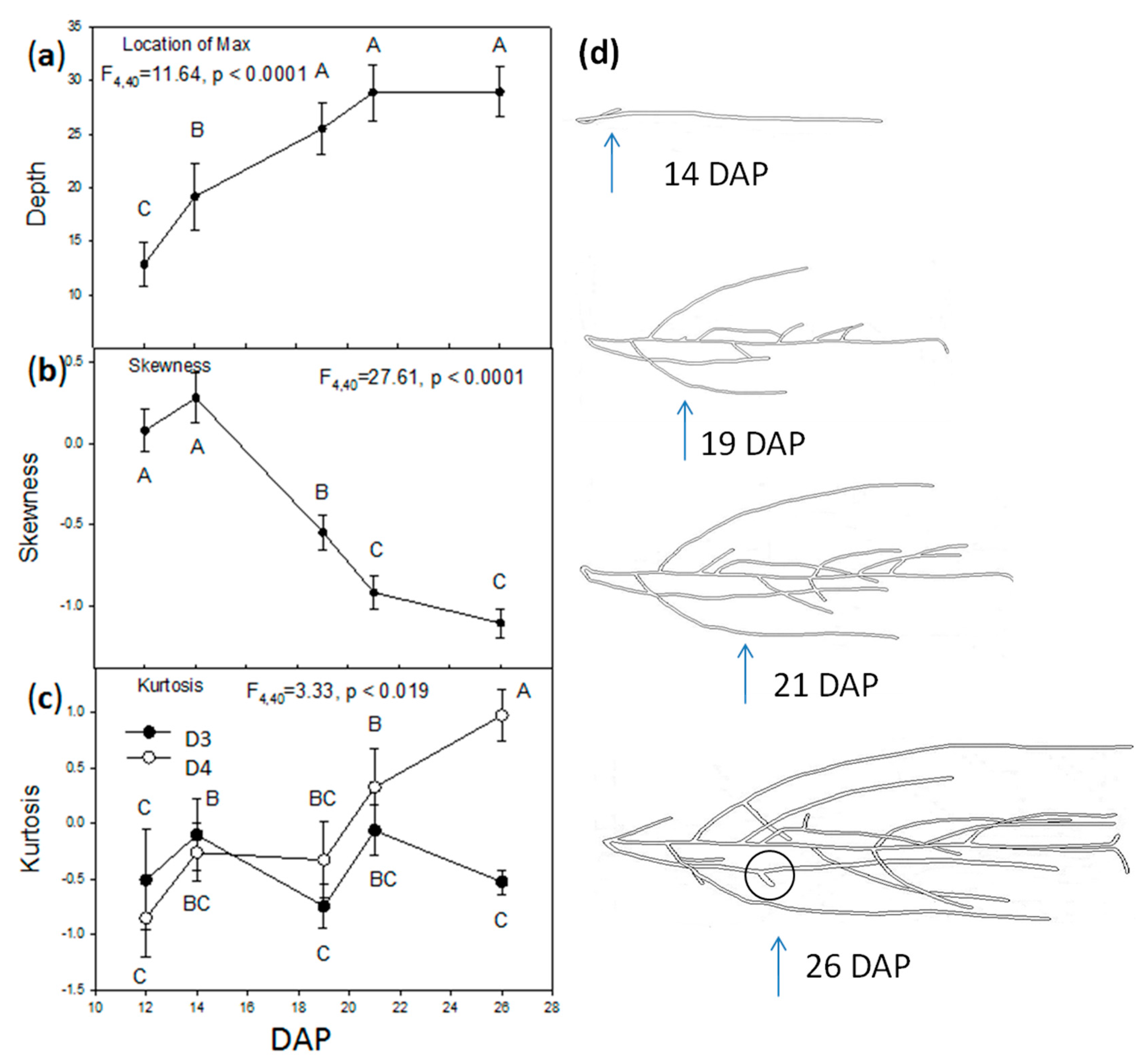
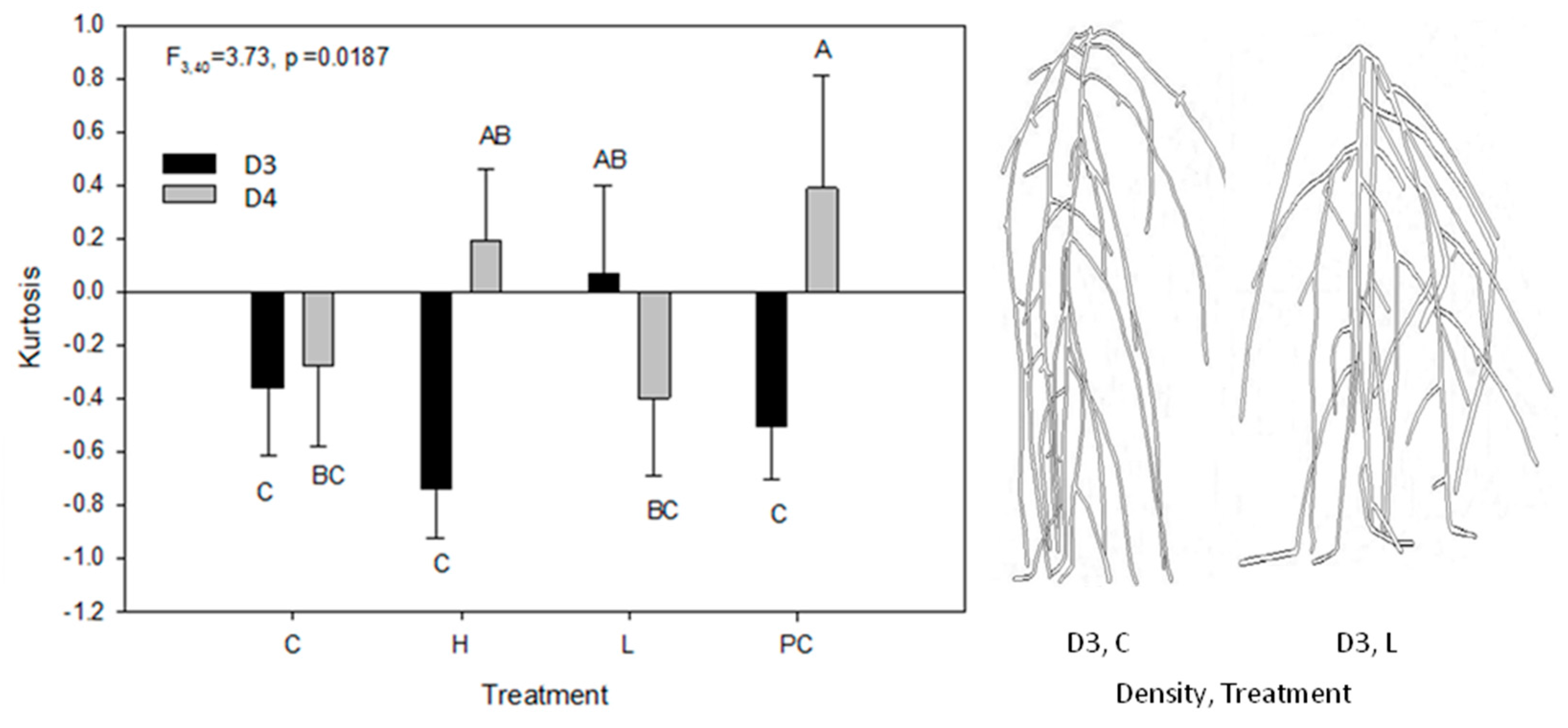
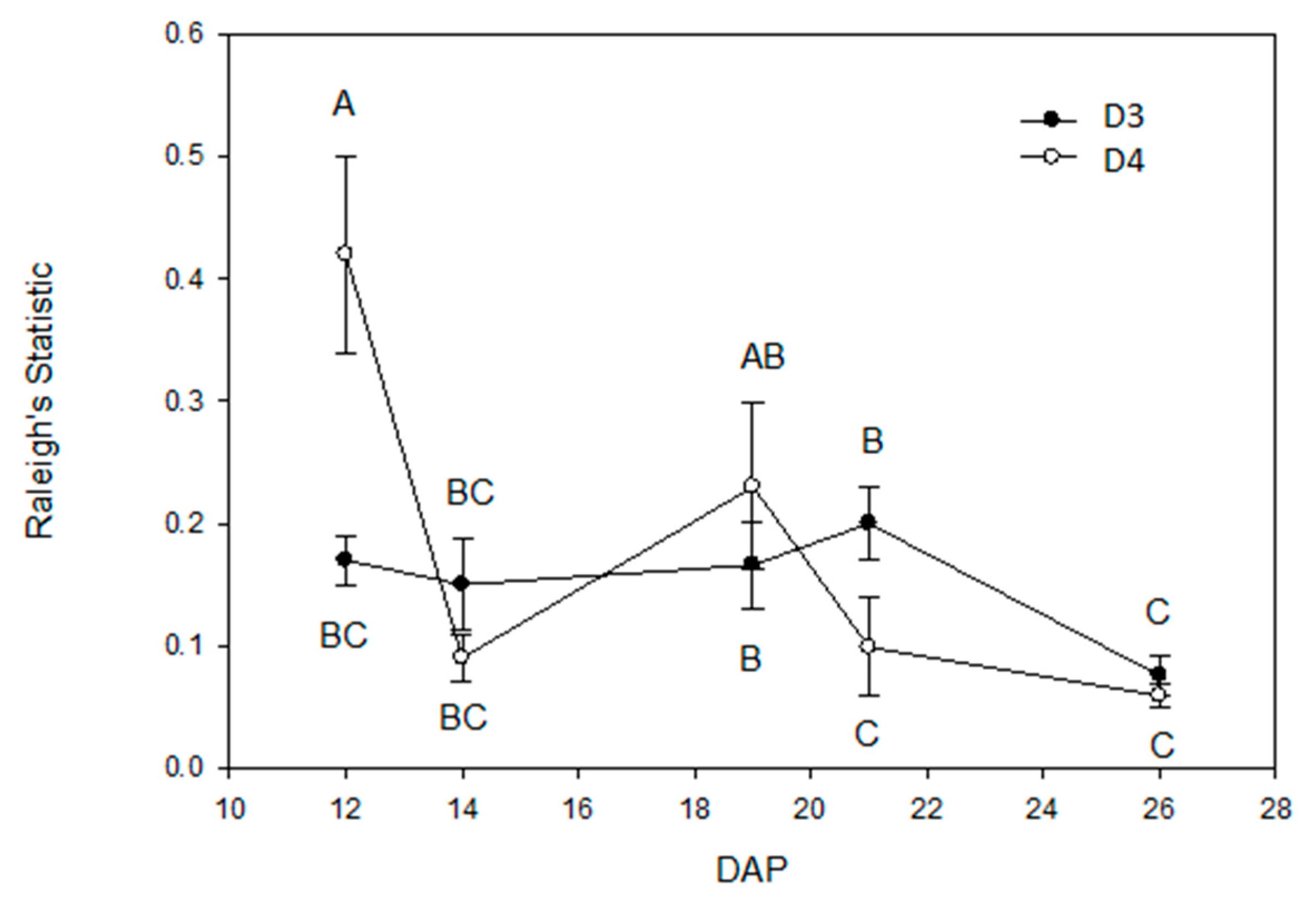
| Effect | DF | Location of Max (F) | Skewness (F) | Kurtosis (F) |
|---|---|---|---|---|
| Density | 1 | 0.34 | 10.09 ** | 4.21 * |
| DAP | 4 | 11.64 *** | 27.61 *** | 4.09 ** |
| Density * DAP | 4 | 0.71 | 0.26 | 3.33 * |
| Treatment | 3 | 1.99 | 1.30 | 0.45 |
| Density * Treatment | 3 | 0.21 | 0.66 | 3.73 * |
| DAP * Treatment | 1 | 0.78 | 0.73 | 1.18 |
| DAP * Density * Treatment | 12 | 0.80 | 1.17 | 1.80 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geisler-Lee, J.; Liu, X.; Rang, W.; Raveendiran, J.; Szubryt, M.B.; Gibson, D.J.; Geisler, M.; Cheng, Q. Image-Based Analysis to Dissect Vertical Distribution and Horizontal Asymmetry of Conspecific Root System Interactions in Response to Planting Densities, Nutrients and Root Exudates in Arabidopsis thaliana. Plants 2017, 6, 46. https://doi.org/10.3390/plants6040046
Geisler-Lee J, Liu X, Rang W, Raveendiran J, Szubryt MB, Gibson DJ, Geisler M, Cheng Q. Image-Based Analysis to Dissect Vertical Distribution and Horizontal Asymmetry of Conspecific Root System Interactions in Response to Planting Densities, Nutrients and Root Exudates in Arabidopsis thaliana. Plants. 2017; 6(4):46. https://doi.org/10.3390/plants6040046
Chicago/Turabian StyleGeisler-Lee, Jane, Xian Liu, Wei Rang, Jayanthan Raveendiran, Marisa Blake Szubryt, David John Gibson, Matt Geisler, and Qiang Cheng. 2017. "Image-Based Analysis to Dissect Vertical Distribution and Horizontal Asymmetry of Conspecific Root System Interactions in Response to Planting Densities, Nutrients and Root Exudates in Arabidopsis thaliana" Plants 6, no. 4: 46. https://doi.org/10.3390/plants6040046




