The Secretome and N-Glycosylation Profiles of the Charophycean Green Alga, Penium margaritaceum, Resemble Those of Embryophytes
Abstract
:1. Introduction
2. Material and Methods
2.1. P. margaritaceum Growth Conditions
2.2. Collection of P. margaritaceum Cells and EPS
2.3. Algal Protein Extraction and LAC
2.4. EPS Protein Extraction
2.5. Trypsin Digestion and N-glycopeptide Enrichment
2.6. NanoLC-MS/MS (Liquid Chromatography-Tandem Mass Spectrometry) Analysis
2.7. Data Analysis and Interpretation
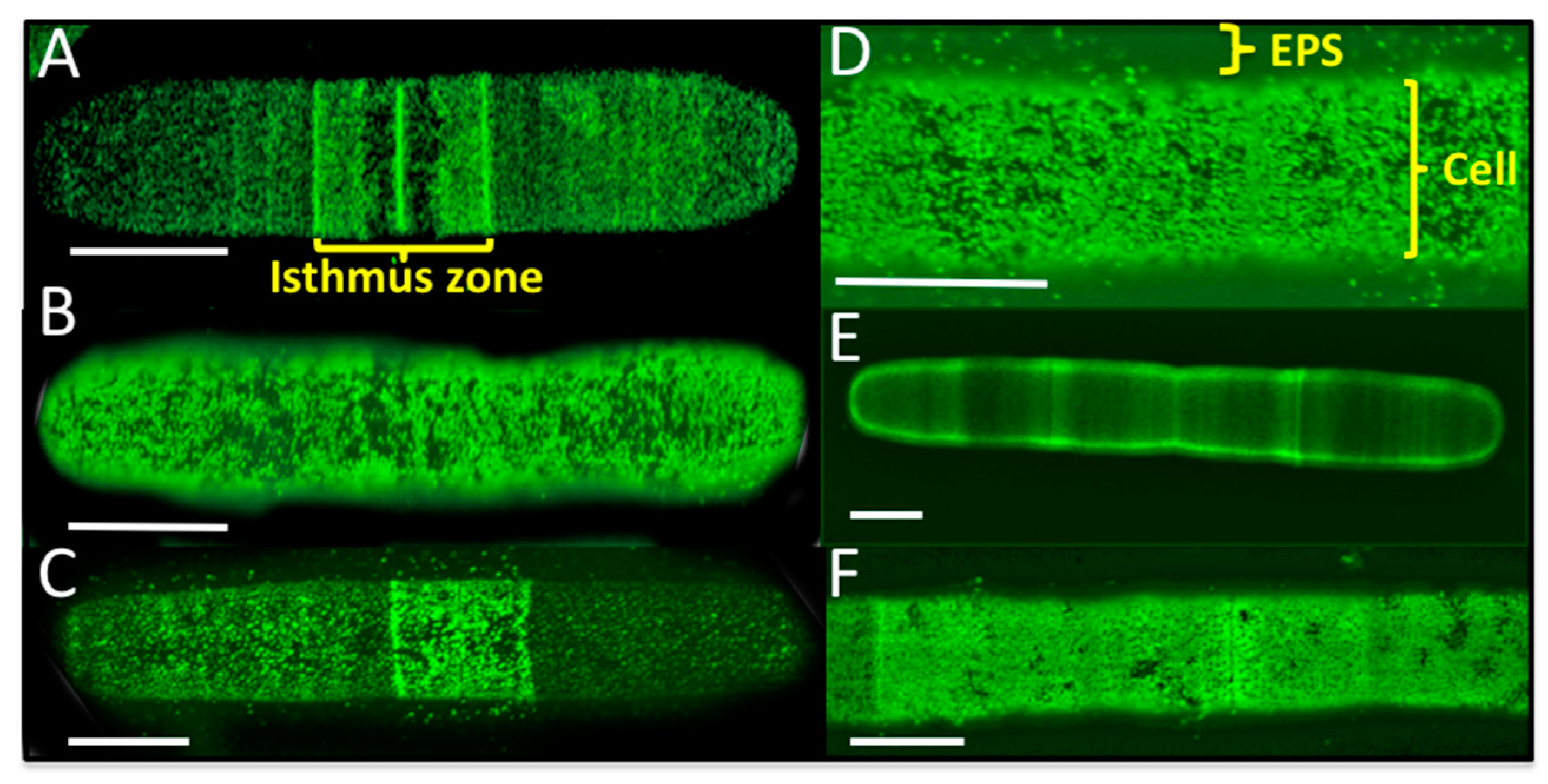
2.8. Live Cell and EPS Labeling of P. margaritaceum
2.9. Bioinformatic Analysis
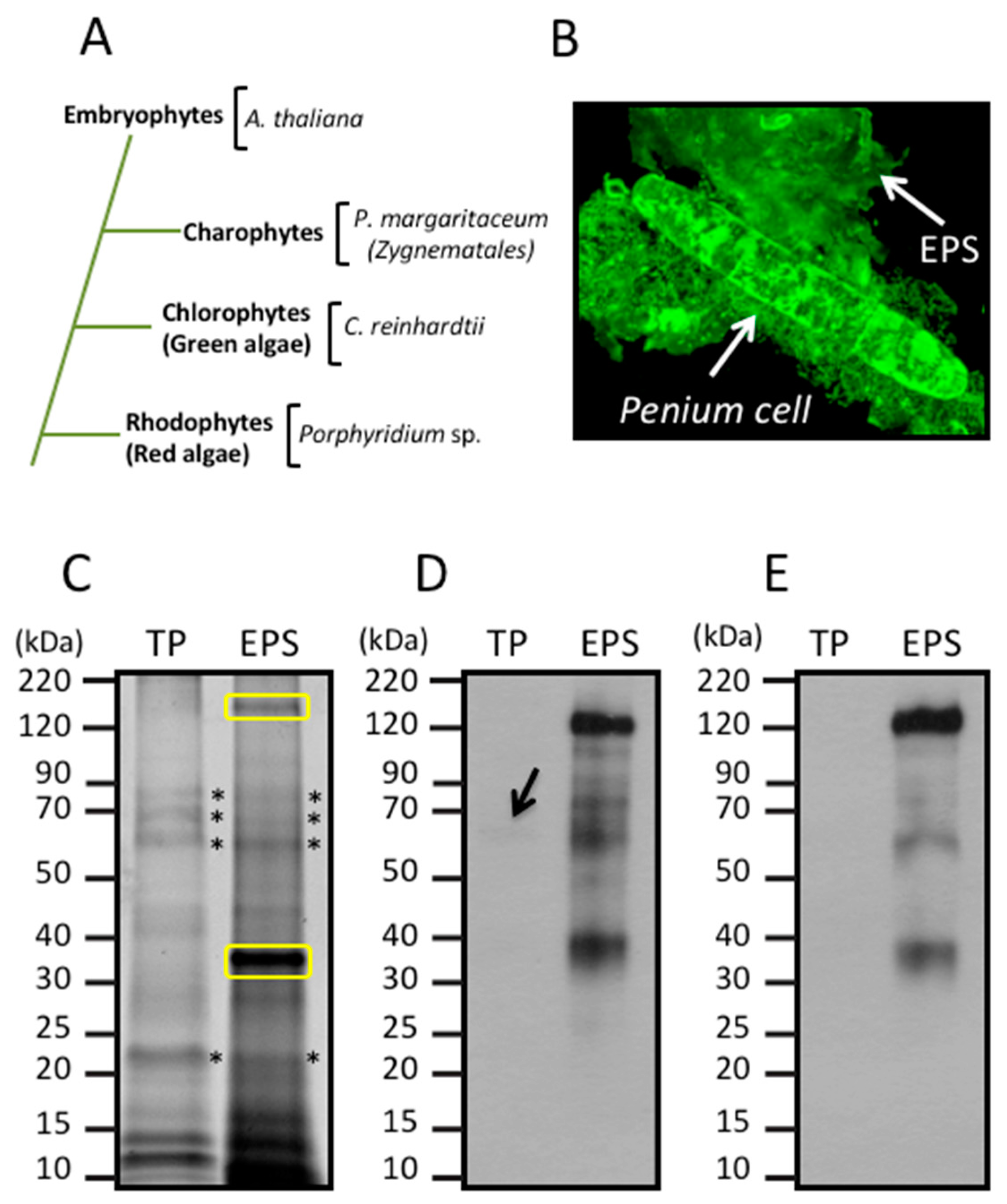
3. Results
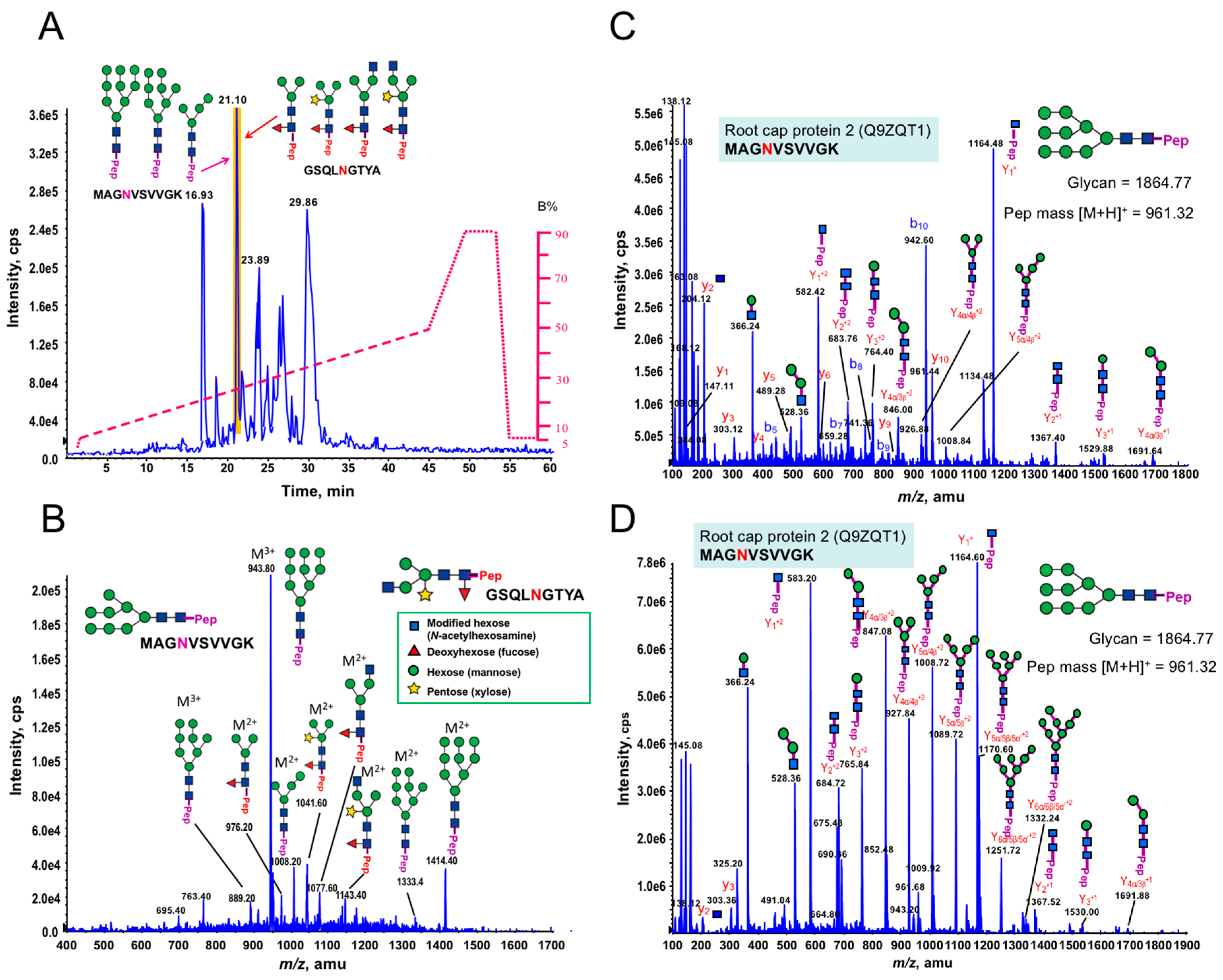
3.1. Glycan Structures of P. margaritaceum Extracellular N-glycoproteins
3.2. Secretome Profiling by LAC and nanoLC-MS/MS
3.3. Identification of Proteins Associated with the EPS
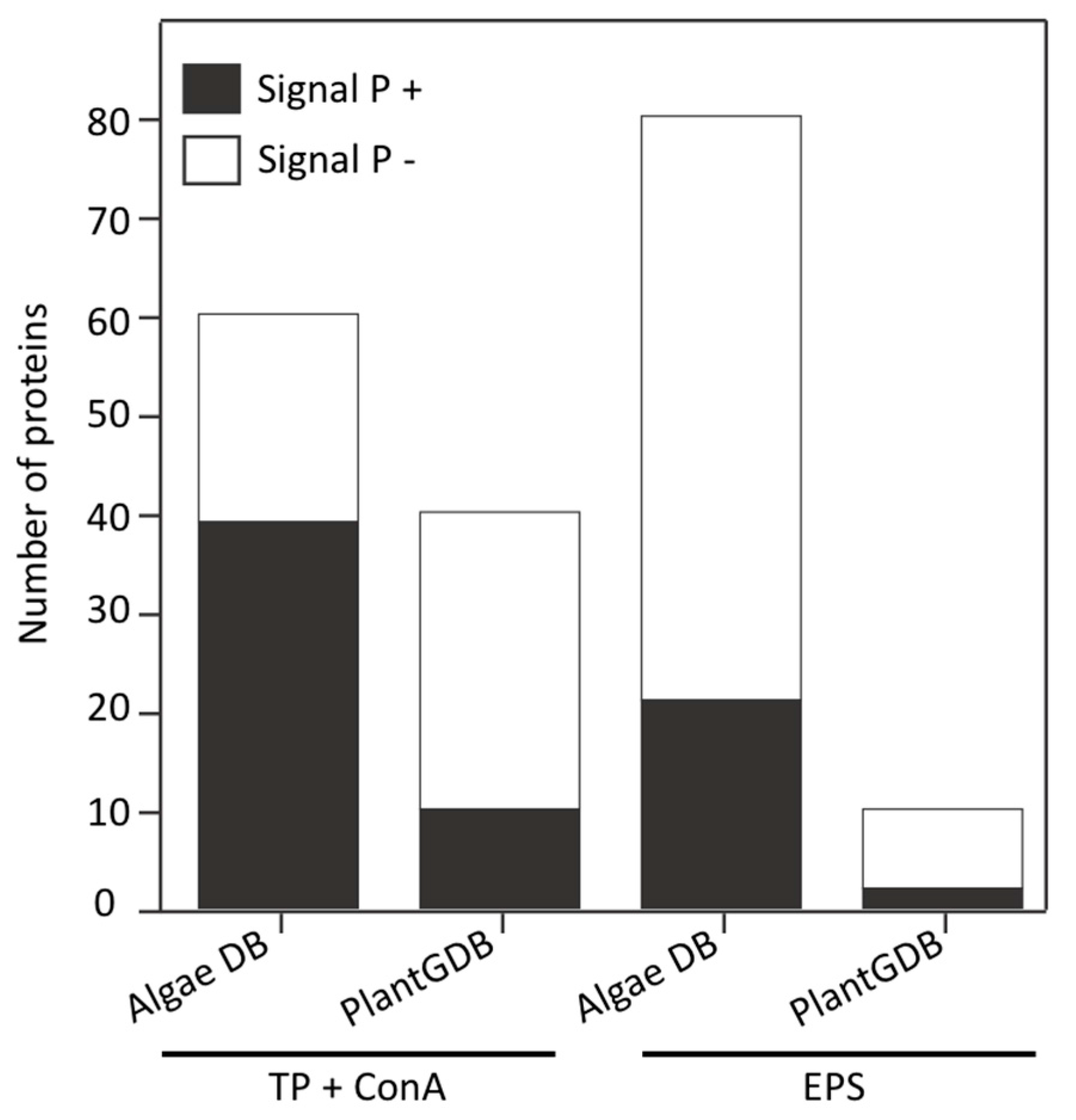
3.4. Land Plant Cell Wall Protein Homologs in the Cell Wall of P. margaritaceum
3.5. Structural Determination of N-glycosylation Patterns in the P. margaritaceum Secretome
3.6. The P. margaritaceum Secretome and N-Glycosylation Patterns Are Closely Related to That of Embryophytes
4. Discussion
4.1. Proteomic Analysis of P. margaritaceum Using an RNA-Sequence Database as Reference
4.2. The Secretome of P. margaritaceum is Closely Related to that of Embryophytes
4.3. The EPS is Tightly Associated with the Cell Wall in P. margaritaceum
4.4. N-Glycosylation Patterns Are Conserved between P. margaritaceum and Land Plants
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Klis, F.M.; Sosinska, G.J.; de Groot, P.W.; Brul, S. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res. 2009, 9, 1013–1028. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.K.; Jwa, N.S.; Lebrun, M.H.; Job, D.; Rakwal, R. Plant secretome: Unlocking secrets of the secreted proteins. Proteomics 2010, 10, 799–827. [Google Scholar] [CrossRef] [PubMed]
- Sorgo, A.G.; Heilmann, C.J.; Brul, S.; de Koster, C.G.; Klis, F.M. Beyond the wall: Candida albicans secret(e)s to survive. FEMS Microbiol. Lett. 2013, 338, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Wardhan, V.; Kumar, A.; Rathi, D.; Pandey, A.; Chakraborty, S.; Chakraborty, N. Secretome analysis of chickpea reveals dynamic extracellular remodeling and identifies a Bet v1-like protein, CaRRP1 that participates in stress response. Sci. Rep. 2015, 5, 18427. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-T.; Jeon, J.; Choi, J.; Cheong, K.; Song, H.; Choi, G.; Kang, S.; Lee, Y.-H. Kingdom-wide analysis of fungal small secreted proteins (SSPs) reveals their potential role in host association. Front. Plant Sci. 2016, 7, 186. [Google Scholar] [CrossRef] [PubMed]
- Zielinska, D.F.; Gnad, F.; Schropp, K.; Wisniewski, J.R.; Mann, M. Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Mol. Cell 2012, 46, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Meissner, F.; Scheltema, R.A.; Mollenkopf, H.J.; Mann, M. Direct proteomic quantification of the secretome of activated immune cells. Science 2013, 340, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Smeekens, J.M.; Xiao, H.; Wu, R. Global analysis of secreted proteins and glycoproteins in Saccharomyces cerevisiae. J. Proteome Res. 2017, 16, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Hervé, V.; Duruflé, H.; San Clemente, H.; Albenne, C.; Balliau, T.; Zivy, M.; Dunand, C.; Jamet, E. An enlarged cell wall proteome of Arabidopsis thaliana rosettes. Proteomics 2016, 16, 3183–3187. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Celma, J.; Ceballos-Laita, L.; Grusak, M.A.; Abadía, J.; López-Millán, A.-F. Plant fluid proteomics: Delving into the xylem sap, phloem sap and apoplastic fluid proteomes. Biochim. Biophys. Acta 2016, 1864, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Calderan-Rodrigues, M.J.; Jamet, E.; Douché, T.; Bonassi, M.B.R.; Cataldi, T.R.; Fonseca, J.G.; San Clemente, H.; Pont-Lezica, R.; Labate, C.A. Cell wall proteome of sugarcane stems: Comparison of a destructive and a non-destructive extraction method showed differences in glycoside hydrolases and peroxidases. BMC Plant Biol. 2016, 16, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen-Kim, H.; San Clemente, H.; Balliau, T.; Zivy, M.; Dunand, C.; Albenne, C.; Jamet, E. Arabidopsis thaliana root cell wall proteomics: Increasing the proteome coverage using a combinatorial peptide ligand library and description of unexpected Hyp in peroxidase amino acid sequences. Proteomics 2016, 16, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Wang, Y.; Lee, K.H.; Park, Z.Y.; Park, J.; Wu, J.; Kwon, S.J.; Lee, Y.H.; Agrawal, G.K.; Rakwal, R.; et al. In-depth insight into in vivo apoplastic secretome of rice-Magnaporthe oryzae interaction. J. Proteom. 2013, 78, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Lee, S.E.; Agrawal, G.K.; Rakwal, R.; Park, S.; Wang, Y.; Kim, S.T. Understanding the plant-pathogen interactions in the context of proteomics-generated apoplastic proteins inventory. Front. Plant Sci. 2015, 6, 352. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Waffenschmidt, S.; Small, L.; Goodenough, U. Between-species analysis of short-repeat modules in cell wall and sex-related hydroxyproline-rich glycoproteins of Chlamydomonas. Plant Physiol. 2007, 144, 1813–1826. [Google Scholar] [CrossRef] [PubMed]
- Baba, M.; Suzuki, I.; Shiraiwa, Y. Proteomic analysis of high-CO2-inducible extracellular proteins in the unicellular green alga, Chlamydomonas reinhardtii. Plant Cell Physiol. 2011, 52, 1302–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathieu-Rivet, E.; Scholz, M.; Arias, C.; Dardelle, F.; Schulze, S.; Le Mauff, F.; Teo, G.; Hochmal, A.K.; Blanco-Rivero, A.; Loutelier-Bourhis, C.; et al. Exploring the N-glycosylation pathway in Chlamydomonas reinhardtii unravels novel complex structures. Mol. Cell. Proteom. 2013, 12, 3160–3183. [Google Scholar] [CrossRef] [PubMed]
- Voigt, J.; Stolarczyk, A.; Zych, M.; Malec, P.; Burczyk, J. The cell-wall glycoproteins of the green alga Scenedesmus obliquus. The predominant cell-wall polypeptide of Scenedesmus obliquus is related to the cell-wall glycoprotein gp3 of Chlamydomonas reinhardtii. Plant Sci. 2014, 215–216, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.; Grief, C.; Hills, G.J.; Shaw, P.J. Cell wall glycoproteins: Structure and function. J. Cell Sci. 1985, 2, 105–127. [Google Scholar] [CrossRef]
- Stanley, M.S.; Perry, R.M.; Callow, J.A. Analysis of expressed sequence tags from the green alga Ulva linza (chlorophyta). J. Phycol. 2005, 41, 1219–1226. [Google Scholar] [CrossRef]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, F.; Aebi, M. Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 2011, 21, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Evolutionary forces shaping the golgi glycosylation machinery: Why cell surface glycans are universal to living cells. Cold Spring Harb. Perspect. Biol. 2011, 3, a005462. [Google Scholar] [CrossRef] [PubMed]
- Schiller, B.; Hykollari, A.; Yan, S.; Paschinger, K.; Wilson, I.B.H. Complicated N-linked glycans in simple organisms. Biol. Chem. 2012, 393, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Baiet, B.; Burel, C.; Saint-Jean, B.; Louvet, R.; Menu-Bouaouiche, L.; Kiefer-Meyer, M.C.; Mathieu-Rivet, E.; Lefebvre, T.; Castel, H.; Carlier, A.; et al. N-glycans of Phaeodactylum tricornutum diatom and functional characterization of its N-acetylglucosaminyltransferase I enzyme. J. Biol. Chem. 2011, 286, 6152–6164. [Google Scholar] [CrossRef] [PubMed]
- Ford, K.L.; Zeng, W.; Heazlewood, J.L.; Bacic, A. Characterization of protein N-glycosylation by tandem mass spectrometry using complementary fragmentation techniques. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Levy-Ontman, O.; Arad, S.M.; Harvey, D.J.; Parsons, T.B.; Fairbanks, A.; Tekoah, Y. Unique N-glycan moieties of the 66-kDa cell wall glycoprotein from the red microalga Porphyridium sp. J. Biol. Chem. 2011, 286, 21340–21352. [Google Scholar] [CrossRef] [PubMed]
- Colombino, L.F.; Bosmann, H.B.; McLean, R.J. Cell surface localization of the sialyltransferase ectoenzyme system during the Chlamydomonas mating reaction. Exp. Cell Res. 1978, 112, 25–30. [Google Scholar] [CrossRef]
- Mamedov, T.; Yusibov, V. Green algae Chlamydomonas reinhardtii possess endogenous sialylated N-glycans. FEBS Open Biol. 2011, 1, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-May, E.; Hucko, S.; Howe, K.J.; Zhang, S.; Sherwood, R.W.; Thannhauser, T.W.; Rose, J.K. A Comparative study of lectin affinity based plant N-glycoproteome profiling using tomato fruit as a model. Mol. Cell. Proteom. 2014, 13, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Lewis, L.A.; McCourt, R.M. Green algae and the origin of land plants. Am. J. Bot. 2004, 91, 1535–1556. [Google Scholar] [CrossRef] [PubMed]
- Becker, B.; Marin, B. Streptophyte algae and the origin of embryophytes. Ann. Bot. 2009, 103, 999–1004. [Google Scholar] [CrossRef] [PubMed]
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; De Clerck, O. Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef]
- Wodniok, S.; Brinkmann, H.; Glöckner, G.; Heidel, A.J.; Philippe, H.; Melkonian, M.; Becker, B. Origin of land plants: Do conjugating green algae hold the key? BMC Evol. Biol. 2011, 11, 104. [Google Scholar] [CrossRef] [PubMed]
- Zhong, B.; Sun, L.; Penny, D. The origin of land plants: A phylogenomic perspective. Evol. Bioinform. 2015, 11, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Timme, R.E.; Delwiche, C.F. Uncovering the evolutionary origin of plant molecular processes: Comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol. 2010, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, I.; Pettolino, F.A.; Bacic, A.; Ralph, J.; Lu, F.; O’Neill, M.A.; Fei, Z.; Rose, J.K.; Domozych, D.S.; Willats, W.G. The charophycean green algae provide insights into the early origins of plant cell walls. Plant J. 2011, 68, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Maruyama, F.; Fujisawa, T.; Togashi, T.; Yamamoto, N.; Seo, M.; Sato, S.; Yamada, T.; Mori, H.; Tajima, N.; et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 2014, 5, 3978. [Google Scholar] [CrossRef] [PubMed]
- Domozych, D.S.; Kort, S.; Benton, S.; Yu, T. The extracellular polymeric substance of the green alga Penium margaritaceum and its role in biofilm formation. Biofilms 2005, 2, 129–144. [Google Scholar] [CrossRef]
- Domozych, D.S. Exopolymer production by the green alga Penium margaritaceum: Implication for biofilm residency. Int. J. Plant Sci. 2007, 168, 763–774. [Google Scholar] [CrossRef]
- Domozych, D.S.; Serfis, A.; Kiemle, S.N.; Gretz, M.R. The structure and biochemistry of charophycean cell walls: I. Pectins of Penium margaritaceum. Protoplasma 2007, 230, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Domozych, D.S.; Brechka, B.; Britton, A.; Toso, M. Cell wall growth and modulation dynamics in a model unicellular green alga-Penium margaritaceum: Live cell labeling with monoclonal antibodies. J. Bot. 2011, 1–8. [Google Scholar] [CrossRef]
- Eder, M.; Tenhaken, R.; Driouich, A.; Lutz-Meindl, U. Occurrence and characterization of arabinogalactan-like proteins and hemicelluloses in Micrasterias (Streptophyta). J. Phycol. 2008, 44, 1221–1234. [Google Scholar] [CrossRef] [PubMed]
- Paerl, H.W.; Pinckney, J.L. A mini-review of microbial consortia: Their roles in aquatic production and biogeochemical cycling. Microb. Ecol. 1996, 31, 225–247. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ari, E.T. Not just slime: Beneath the slippery exterior of a microbial biofilm lies a remarkably organized community of organisms. BioScience 1999, 49, 689–695. [Google Scholar] [CrossRef]
- Christensen, B.E. Physical and chemical properties of extracellular polysaccharides associated with biofilms and related systems. In Microbial Extracellular Polymeric Substances; Wingender, J., Neu, T.R., Flemming, H.C., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; Chapter 8; pp. 143–154. [Google Scholar]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, I.W. Biofilm exopolysaccharides: A strong and sticky framework. Microbiology 2001, 147, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, I.W. The biofilm matrix: An immobilized but dynamic microbial environment. Trends Microbiol. 2001, 9, 222–227. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. Relevance of microbial extracellular polymeric substances (EPSs)—Part I: Structural and ecological aspects. Water Sci. Technol. 2001, 43, 1–8. [Google Scholar] [PubMed]
- Niklas, K.J. The cell walls that bind the tree of life. BioScience 2004, 54, 831–841. [Google Scholar] [CrossRef]
- Elster, J.; Lukavsky, J.; Harding, K.; Benson, E.E.; Day, J.G. Deployment of the encapsulation-dehydration protocol to cryopreserve polar microalgae held at the Czech Republic Academy of Sciences Institute of Botany. CryoLetters 2008, 29, 27–28. [Google Scholar] [PubMed]
- Sørensen, I.; Fei, Z.; Andreas, A.; Willats, W.G.T.; Domozych, D.S.; Rose, J.K.C. Stable transformation and reverse genetic analysis of Penium margaritaceum: A platform for studies of charophyte green algae, the immediate ancestors of land plants. Plant J. 2014, 77, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Freeze, H.H.; Stanley, P.; Marth, J.D.; Bertozzi, C.R.; Hart, G.W.; Etzler, M.E. Symbol nomenclature for glycan representation. Proteomics 2009, 9, 5398–5399. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Giboulot, A.; Zivy, M.; Valot, B.; Jamet, E.; Albenne, C. Combining various strategies to increase the coverage of the plant cell wall glycoproteome. Phytochemistry 2010, 72, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Lombard, J. The multiple evolutionary origins of the eukaryotic N-glycosylation pathway. Biol. Direct 2016, 11, 36. [Google Scholar] [CrossRef] [PubMed]
- Domozych, D.; Lietz, A.; Patten, M.; Singer, E.; Tinaz, B.; Raimundo, S.C. Imaging the dynamics of cell wall polymer deposition in the unicellular model plant, Penium margaritaceum. In Light Microscopy: Methods and Protocols; Methods in Molecular Biology; Markaki, Y., Hartmann, H., Eds.; Springer: New York, NY, USA, 2017; Volume 1563, pp. 91–105. [Google Scholar]
- Katoh, H.; Ikeuchi, M. Targeted disruption of psbX and biochemical characterization of photosystem II complex in the thermophilic cyanobacterium Synechococcus elongatus. Plant Cell Physiol. 2001, 42, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Catalá, C.; Howe, K.J.; Hucko, S.; Rose, J.K.; Thannhauser, T.W. Towards characterization of the glycoproteome of tomato (Solanum lycopersicum) fruit using concanavalin A lectin affinity chromatography and LC-MALDI-MS/MS analysis. Proteomics 2011, 11, 1530–1544. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sherwood, R.W.; Yang, Y.; Fish, T.; Chen, W.; McCardle, J.A.; Jones, R.M.; Yusibov, V.; May, E.R.; Rose, J.K.; et al. Comparative characterization of the glycosylation profiles of an influenza hemagglutinin produced in plant and insect hosts. Proteomics 2012, 12, 1269–1288. [Google Scholar] [CrossRef] [PubMed]
- Jamet, E.; Canut, H.; Boudart, G.; Pont-Lezica, R.F. Cell wall proteins: A new insight through proteomics. Trends Plant Sci. 2006, 11, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Nakai, K.; Horton, P. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999, 24, 34–36. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Tardif, M.; Atteia, A.; Specht, M.; Cogne, G.; Rolland, N.; Brugiere, S.; Hippler, M.; Ferro, M.; Bruley, C.; Peltier, G.; et al. PredAlgo: A new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 2012, 29, 3625–3639. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Hirokawa, T.; Boon-Chieng, S.; Mitaku, S. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 1998, 14, 378–379. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Stanley, P.; Hart, G.; Aebi, M.; Darvill, A.; Kinoshita, T.; Packer, N.H.; Prestegard, J.J.; et al. Essentials of Glycobiology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Minic, Z.; Jamet, E.; Negroni, L.; Arsene der Garabedian, P.; Zivy, M.; Jouanin, L. A sub-proteome of Arabidopsis thaliana mature stems trapped on Concanavalin A is enriched in cell wall glycoside hydrolases. J. Exp. Bot. 2007, 58, 2503–2512. [Google Scholar] [CrossRef] [PubMed]
- Martiniere, A.; Gayral, P.; Hawes, C.; Runions, J. Building bridges: Formin1 of Arabidopsis forms a connection between the cell wall and the actin cytoskeleton. Plant J. 2011, 66, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Held, M.A.; Tan, L.; Kamyab, A.; Hare, M.; Shpak, E.; Kieliszewski, M.J. Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross-linked in vitro. J. Biol. Chem. 2004, 279, 55474–55482. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.; Egelund, J.; Schultz, C.J.; Bacic, A. Arabinogalactan-proteins: Key regulators at the cell surface? Plant Physiol. 2010, 153, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Eberhard, S.; Pattathil, S.; Warder, C.; Glushka, J.; Yuan, C.H.; Hao, Z.Y.; Zhu, X.; Avci, U.; Miller, J.S.; et al. An arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 2013, 25, 270–287. [Google Scholar] [CrossRef] [PubMed]
- Knox, J.P.; Linstead, P.J.; Peart, J.; Cooper, C.; Roberts, K. Developmentally regulated epitopes of cell-surface arabinogalactan proteins and their relation to root-tissue pattern-formation. Plant J. 1991, 1, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, M.; Beven, A.; Donovan, N.; Neill, S.J.; Peart, J.; Roberts, K.; Knox, J.P. Localization of cell-wall proteins in relation to the developmental anatomy of the carrot root Apex. Plant J. 1994, 5, 237–246. [Google Scholar] [CrossRef]
- Yates, E.A.; Knox, J.P. Investigations into the occurrence of plant-cell surface epitopes in exudate gums. Carbohydr. Polym. 1994, 24, 281–286. [Google Scholar] [CrossRef]
- Yates, E.A.; Valdor, J.F.; Haslam, S.M.; Morris, H.R.; Dell, A.; Mackie, W.; Knox, J.P. Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology 1996, 6, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Showalter, A.M. Arabinogalactan-proteins: Structure, expression and function. Cell. Mol. Life Sci. 2001, 58, 1399–1417. [Google Scholar] [CrossRef] [PubMed]
- Laine, A.C.; Gomord, V.; Faye, L. Xylose-specific antibodies as markers of subcompartmentation of terminal glycosylation in the golgi apparatus of sycamore cells. FEBS Lett. 1991, 295, 179–184. [Google Scholar] [CrossRef]
- Velasquez, S.M.; Ricardi, M.M.; Dorosz, J.G.; Fernandez, P.V.; Nadra, A.D.; Pol-Fachin, L.; Egelund, J.; Gille, S.; Harholt, J.; Ciancia, M.; et al. O-Glycosylated cell wall proteins are essential in root hair growth. Science 2011, 332, 1401–1403. [Google Scholar] [CrossRef] [PubMed]
- Champagne, A.; Boutry, M. Proteomics of nonmodel plant species. Proteomics 2013, 13, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Casado, G.; Covey, P.A.; Bedinger, P.A.; Mueller, L.A.; Thannhauser, T.W.; Zhang, S.; Fei, Z.; Giovannoni, J.J.; Rose, J.K. Enabling proteomic studies with RNA-Seq: The proteome of tomato pollen as a test case. Proteomics 2012, 12, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Witters, E.; Laukens, K.; Deckers, P.; Van Dongen, W.; Esmans, E.; Van Onckelen, H. Fast liquid chromatography coupled to electrospray tandem mass spectrometry peptide sequencing for cross-species protein identification. Rapid Commun. Mass Spectrom. 2003, 17, 2188–2194. [Google Scholar] [CrossRef] [PubMed]
- Bräutigam, A.; Shrestha, R.P.; Whitten, D.; Wilkerson, C.G.; Carr, K.M.; Froehlich, J.E.; Weber, A.P. Low-coverage massively parallel pyrosequencing of cDNAs enables proteomics in non-model species: Comparison of a species-specific database generated by pyrosequencing with databases from related species for proteome analysis of pea chloroplast envelopes. J. Biotechnol. 2008, 136, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.C.; Beynon, R.J.; Hubbard, S.J. Cross species proteomics. Methods Mol. Biol. 2010, 604, 123–135. [Google Scholar] [PubMed]
- Friso, G.; van Wijk, K.J. Posttranslational protein modifications in plant metabolism. Plant Physiol. 2015, 169, 1469–1487. [Google Scholar] [PubMed]
- Neubert, P.; Halim, A.; Zauser, M.; Essig, A.; Joshi, H.J.; Zatorska, E.; Larsen, I.S.B.; Loibl, M.; Castells-Ballester, J.; Aebi, M.; et al. Mapping the O-Mannose glycoproteome in Saccharomyces cerevisiae. Mol. Cell. Proteom. 2016, 15, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Chen, F.; Linskens, H.F.; Cresti, M. Distribution of unesterified and esterified pectins in cell walls of pollen tubes of flowering plants. Sex. Plant Reprod. 1994, 7, 145–152. [Google Scholar] [CrossRef]
- Li, Y.Q.; Zhang, H.Q.; Pierson, E.S.; Huang, F.Y.; Linskens, H.F.; Hepler, P.K.; Cresti, M. Enforced growth-rate fluctuation causes pectin ring formation in the cell wall of Lilium longiflorum pollen tubes. Planta 1996, 200, 41–49. [Google Scholar] [CrossRef]
- Nuñez, A.; Fishman, M.L.; Fortis, L.L.; Cooke, P.H.; Hotchkiss, A.T., Jr. Identification of extensin protein associated with sugar beet pectin. J. Agric. Food Chem. 2009, 57, 10951–10958. [Google Scholar] [CrossRef] [PubMed]
- MacMillan, C.P.; Mansfield, S.D.; Stachurski, Z.H.; Evans, R.; Southerton, S.G. Fasciclin-like arabinogalactan proteins: Specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J. 2010, 62, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.L.; Kibble, N.A.; Bacic, A.; Schultz, C.J. A fasciclin-like arabinogalactan-protein (FLA) mutant of Arabidopsis thaliana, fla1, shows defects in shoot regeneration. PLoS ONE 2011, 6, e25154. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Showalter, A.M.; Egelund, J.; Hernandez-Sanchez, A.; Doblin, M.S.; Bacic, A. Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front Plant Sci. 2012, 3, 140. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.Q.; Gong, S.; Xu, W.L.; Li, W.; Li, P.; Zhang, C.J.; Li, D.D.; Zheng, Y.; Li, F.G.; Li, X.B. A fasciclin-like arabinogalactan protein, GhFLA1, is involved in fiber initiation and elongation of cotton. Plant Physiol. 2013, 161, 1278–1290. [Google Scholar] [CrossRef] [PubMed]
- Kiemle, S.N.; Domozych, D.S.; Gretz, M.R. The extracellular polymeric substances of desmids (Conjugatophyceae, Streptophyta): Chemistry, structural analyses and implications in wetland biofilms. Phycologia 2007, 46, 617–627. [Google Scholar] [CrossRef]
- Van Gisbergen, P.A.; Li, M.; Wu, S.Z.; Bezanilla, M. Class II formin targeting to the cell cortex by binding PI(3,5)P(2) is essential for polarized growth. J. Cell Biol. 2012, 198, 235–250. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, C.; Wang, J.; Ren, Z.; Staiger, C.J.; Ren, H. A processive Arabidopsis formin modulates actin filament dynamics in association with profilin. Mol. Plant 2016, 9, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Balshüsemann, D.; Jaenicke, L. The oligosaccharides of the glycoprotein pheromone of Volvox-Carteri F Nagariensis Iyengar (Chlorophyceae). Eur. J. Biochem. 1990, 192, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Chalovich, J.M.; Eisenberg, E. High-sensitivity analytical approaches for the structural characterization of glycoproteins. Chem. Rev. 2005, 257, 2432–2437. [Google Scholar]


| Unigene ID a | Name | UAN b | Species | PSORT c | N-Glyco d | Experiment |
|---|---|---|---|---|---|---|
| Cell wall associated proteins | ||||||
| Penium13014-0R | Probable endo-1,3(4)-beta-glucanase Aspergillus niger | A2QBQ3 | Aspergillus niger | Extr | 5 | ConA |
| PU01918-0R | Leucine-rich repeat extensin-like protein 1 | Q9LJ64 | Arabidopsis thaliana | Extr | 3 | ConA |
| Penium10692-1R | Arabinogalactan protein | Q9M7I5 | Zea mays | Extr | 0 | ConA |
| Penium51331-0F | Fasciclin-like arabinogalactan protein | G7ILU2 | Medicago truncatula | Extr | 3 | ConA |
| Penium05518-1F | Structural constituent of cell-wall, putative | B9S9J6 | Ricinus communis | Extr | 1 | EPS |
| PU11354-0R | Putative uncharacterized protein (Bacterial cellulose synthase subunit) | E6TGN8 | Mycobacterium sp. | Ext | 5 | ConA |
| PU02505-0R | Formin-like protein 2 | Q7XUV2 | Oryza sativa subsp. japonica | Plas | 2 | EPS |
| Proteolysis | ||||||
| corb_UMD_Coleochaete_c15397_c_s | Peptidase S1 and S6 | A2Q336 | Medicago truncatula | Extr | 3 | ConA |
| Penium21547-0F | Cysteine protease, putative | B9R8S7 | Ricinus communis | Extr | 1 | ConA |
| Penium50713-1F | Intramembrane protease RasP/YluC | A0A0H4WXM9 | Myxococcus sp. | Plas | 1 | EPS |
| corb_UMD_Coleochaete_c16755_c_s | Propeptide PepSY amd peptidase M4 | E8WAR7 | Streptomyces flavogriseus | Extr | 1 | ConA |
| PU17998-0F | Secreted serine protease | B1VSD5 | Streptomyces griseus subsp. griseus | Extr | 3 | ConA |
| PU16802-1F | Cysteine proteinase 2 | Q10717 | Zea mays | Vacu | 2 | ConA |
| PU11399-1F | Membrane protease subunit, stomatin/prohibitin | K6FM44 | Desulfovibrio magneticus | Extr | 1 | ConA |
| Penium04469-1F | Serine carboxypeptidase S28 family protein | D7MUL9 | Arabidopsis lyrata subsp. lyrata | Vacu | 6 | ConA |
| Oxidoreductases | ||||||
| PU21449-0R | Superoxide dismutase | G9M4K4 | Pogonatum inflexum | Extr | 0 | EPS |
| Penium04764-0F | Ferredoxin thioredoxin reductase catalytic beta chain family | D7L839 | Arabidopsis lyrata subsp. lyrata | Extr | 0 | EPS |
| PU02432-1F | Epimerase/dehydrogenase | D7FJ06 | Ectocarpus siliculosus | Extr | 0 | EPS |
| Penium24287-2R | Protein disulfide isomerase | Q9FEG4 | Triticum durum | Vacu | 1 | ConA and EPS |
| PU08835-1F | Protein disulfide isomerase S-2 | E3W9C1 | Glycine max | ER | 0 | EPS |
| PU15966-1R | Protein disulfide isomerase-like 1-3 | Q8VX13 | Arabidopsis thaliana | ER | 8 | ConA |
| PU13392-0F | Microneme protein, putative | A0A086JBX3 | Toxoplasma gondii | Golg | 8 | ConA |
| PU14524-1R | Glutaredoxin-C4 | Q8LFQ6 | Arabidopsis thaliana | Vacu | 0 | EPS |
| Miscellaneous | ||||||
| PU00748-1F | GDSL esterase/lipase | Q9M2R9 | Arabidopsis thaliana | Extr | 3 | ConA and EPS |
| Penium53827-1R | Glucosidase II beta subunit | B9SBM9 | Ricinus communis | Extr | 0 | ConA |
| Penium51452-0R | Root cap protein 1 | B6TV36 | Zea mays | Extr | 1 | EPS |
| Penium50864-2R | Alpha amylase, catalytic domain protein | A7A7M5 | Bifidobacterium adolescentis | Chlo | 8 | EPS |
| corb_Contig1457 | Protein phosphatase 2C | G7I7K2 | Medicago truncatula | Plas | 1 | EPS |
| PU00133-0R | Endonuclease 5 | F4JJL3 | Arabidopsis thaliana | Extr | 4 | ConA and EPS |
| PU14522-2F | PXN-FBPL | Q2LK77 | Xenopus laevis | Vacu | 5 | ConA |
| Penium15798-2R | Alpha/beta-type gliadin | Q41632 | Triticum urartu | Vacu | 0 | ConA |
| Penium42317-1F | Pollen coat oleosin-glycine rich protein | Q6V5D9 | Olimarabidopsis pumila | Extr | 0 | ConA |
| PU21202-1R | BURP domain-containing protein 7 | Q60E34 | Oryza sativa subsp. japonica | Extr | 0 | ConA |
| PU12863-2F | Putative ABC-type transport system | J2K1X5 | Rhizobium sp. CF080 | Extr | 1 | ConA |
| PU00769-1R | Endoplasmin homolog | Q9STX5 | Arabidopsis thaliana | ER | 3 | ConA |
| Penium03560-1F | Serine protease inhibitor | Q32TF4 | Argopecten irradians | Extr | 1 | ConA |
| PU08363-0R | Carbonic anhydrase | H1XWM8 | Caldithrix abyssi DSM 13497 | Extr | 3 | EPS |
| Penium02868-2F | Peritrophin-1 | E2ADF6 | Camponotus floridanus | Extr | 6 | ConA |
| PU21572-2F | Putative cuticle protein | C0H6H4 | Bombyx mori | Extr | 1 | ConA |
| PU26029-0R | Olfactory receptor 5AK2 | Q8NH90 | Homo sapiens | Plas | 2 | ConA |
| PU32535-2R | Type III secretion protein SpaR/YscT/HrcT | A1TJC4 | Acidovorax citrulli | Plas | 1 | ConA |
| Unknown function | ||||||
| Penium54960-0F | Uncharacterized protein | G7ME15 | Macaca mulatta | Plas | 12 | ConA |
| PU20188-1R | Putative uncharacterized protein | D7SHJ2 | Vitis vinifera | Extr | 0 | ConA |
| PU23729-1F | Predicted protein | A9TWI4 | Physcomitrella patens subsp. patens | Extr | 8 | ConA |
| PU11719-0R | Putative uncharacterized protein | D8U3A6 | Volvox carteri | Extr | 0 | ConA |
| Penium17901-2F | Putative uncharacterized protein | B6SPS4 | Zea mays | Plas | 0 | ConA |
| Penium42683-0F | Predicted protein | A9RQJ8 | Physcomitrella patens subsp. patens | Extr | 1 | ConA and EPS |
| Penium15957-1F | Uncharacterized protein | L7UB03 | Myxococcus stipitatus DSM 14675 | Extr | 0 | EPS |
| PU16053-0F | Putative uncharacterized protein | A3CD48 | Oryza sativa subsp. japonica (Rice) | Plas | 0 | ConA |
| Penium33821-0F | Putative uncharacterized | G7E6T5 | Mixia osmundae | Plas | 3 | ConA |
| PU00167-0R | Putative uncharacterized protein | F1YT29 | Acetobacter pomorum | Plas | 3 | EPS |
| PU08479-1R | Uncharacterized protein | L2G3N6 | Colletotrichum gloeosporioides | ER | 0 | ConA |
| Unigene | Protein | UAN a | E-Value | Species | Peptide | Pep Mass [M + H]+ | N-Glycan Structures b |
|---|---|---|---|---|---|---|---|
| PU00576-0F | Receptor of activated protein kinase C 1 | A8J8Y1 | 4 × 10−26 | Chlamydomonas reinhardtii | GSQLNGTYA | 910.32 |  |
| PU00894-1F | Root cap protein 2 | Q9ZQT1 | 3 × 10−11 | Zea mays | MAGNVSVVGK | 961.32 |  |
| 687.52 |  | ||||||
| 771.48 |  | ||||||
| 815.40 |  | ||||||
| 815.40 |  | ||||||
| 815.40 |  | ||||||
| 815.48 |  |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-May, E.; Sørensen, I.; Fei, Z.; Zhang, S.; Domozych, D.S.; Rose, J.K.C. The Secretome and N-Glycosylation Profiles of the Charophycean Green Alga, Penium margaritaceum, Resemble Those of Embryophytes. Proteomes 2018, 6, 14. https://doi.org/10.3390/proteomes6020014
Ruiz-May E, Sørensen I, Fei Z, Zhang S, Domozych DS, Rose JKC. The Secretome and N-Glycosylation Profiles of the Charophycean Green Alga, Penium margaritaceum, Resemble Those of Embryophytes. Proteomes. 2018; 6(2):14. https://doi.org/10.3390/proteomes6020014
Chicago/Turabian StyleRuiz-May, Eliel, Iben Sørensen, Zhangjun Fei, Sheng Zhang, David S. Domozych, and Jocelyn K. C. Rose. 2018. "The Secretome and N-Glycosylation Profiles of the Charophycean Green Alga, Penium margaritaceum, Resemble Those of Embryophytes" Proteomes 6, no. 2: 14. https://doi.org/10.3390/proteomes6020014






