A Novel Three-Stage Filter-Wrapper Framework for miRNA Subset Selection in Cancer Classification
Abstract
:1. Introduction
- In most classification algorithms, the complexity of the algorithm depends on the dimensions of data. So, in order to reduce the memory and computational time, we are interested in removing the irrelevant and redundant miRNAs from the dataset.
- When it has been determined that a miRNA does not have a positive effect on the cancer diagnosis, the time and biochemical cost required to extract this miRNA can be saved. It should be noted that this can significantly accelerate the process of the cancer diagnosis.
- When it is possible to represent the data without losing the information in a small number of miRNAs, they can be plotted visually and this can help extracting the useful knowledge about cancer.
- When data are represented by fewer miRNAs, it is easier to analyze the processes that generate the data and this helps to extract useful knowledge about cancer.
- A novel three-stage feature subset selection framework is proposed. In the first stage of the proposed framework, an algorithm consisting of multiple filters are used to evaluate the relevance of each miRNA with the class label, to rank miRNAs according to their relevance value, to select the top-ranked miRNAs obtained from each filter algorithms, and finally to generate a miRNAs pool. In the second stage, another algorithm is used to rank the miRNAs of miRNA pool and then to weight the probability of selecting each miRNA. In the third stage, a wrapper algorithm tries to find an optimal subset from the weighed miRNAs of miRNA pool which gives us the most information about cancer. The third stage provides a situation in which top-ranked miRNAs of miRNA pool have a higher chance of being selected than low-ranked miRNAs. To the best of our knowledge, no empirical research has been conducted on the using feature importance obtained by the ensemble of filters to weight the feature probabilities in the wrapper algorithms.
- A novel method is proposed to assign different threshold value to each miRNA according to the importance of that miRNA in the dataset. For this purpose, the knowledge extracted about the miRNA importance is used to adjust the value of threshold parameter in CSO for each miRNA. In the best of our knowledge, the value of threshold parameter in all previous works is considered the same for all features. It should be noted that when we set the same threshold values for all the miRNAs, the algorithm does not initially distinguish between relevant and irrelevant miRNAs and therefore requires a lot of time to find the optimal miRNAs subset.
2. Fundamental Backgrounds and Literature Review
2.1. Competitive Swarm Optimization (CSO)
2.2. Literature Review
3. The Proposed Framework
| Algorithm 1. General outline of proposed algorithm. |
| // first stage: miRNA pool generation |
| Calculate the rank of each miRNA by every filter algorithms; |
| Construct miRNA pool by high-ranked miRNAs; |
| // second stage: miRNA weights calculation |
| Calculate the rank of each miRNA of miRNA pool by every filter algorithms; |
| Calculate the miRNA weights for each member of miRNA pool; |
| // third stage: search to find the best miRNA subset of miRNA pool |
| t = 0; |
| Randomly generate the initial swarm ; |
| Evaluate the initial swarm with the evaluation function; |
| While stopping criterion is not satisfied Do |
| Randomly partition the swarm into two groups; Perform pairwise competitions between particles from each group; Directly transmit the winner particles to the swarm of next iteration; Update the position of each loser particle by learning from its corresponding winner particle; Evaluate the swarm with the evaluation function; |
| t = t + 1; |
| End while |
| Output: The best solution found. |
3.1. Generation the miRNA Pool
3.2. Weighting the miRNAs of miRNA Pool
3.3. Solution Representation
3.4. Fuzzy Balancer to Balance between Exploration and Exploitation
- If iter is Low and diversity is Low, then = VHigh.
- If iter is Low and diversity is Med, then = High.
- If iter is Low and diversity is High, then = Med.
- If iter is Med and diversity is Low, then = High.
- If iter is Med and diversity is Med, then = Med.
- If iter is Med and diversity is High, then = Low.
- If iter is High and diversity is Low, then = Med.
- If iter is High and diversity is Med, then = Low.
- If iter is High and diversity is High, then = VLow.
3.5. Particle Evaluation
3.6. Algorithmic Details and Flowchart
| Algorithm 2. Outline of proposed algorithm for feature selection in classification. |
| // first stage: miRNA pool generation |
| Calculate the rank of each miRNA by every filter algorithms; |
| Construct miRNA pool by high-ranked miRNAs; |
| // second stage: miRNA weights calculation |
| Calculate the rank of each miRNA of miRNA pool by every filter algorithms; |
| Calculate the miRNA weights for each member of miRNA pool and threshold vector |
| ; |
| // third stage: search to find the best miRNA subset of miRNA pool |
| t = 0; |
| For i = 1 to N Do |
| Randomly generate the initial solution ; |
| Randomly generate the initial velocity ; |
| End for |
| While stopping criteria is not satisfied Do |
| ; |
| While Do |
| Randomly choose two different particles and from ; |
| /* algorithmic steps of is shown in Algorithm (3) */ |
| If |
| ; |
| ; |
| Else |
| ; |
| ; |
| End if |
| For each dimension d of update by Eq. (3); |
| For each dimension d of update by Eq. (4); |
| ; |
| ; |
| End while |
| t = t + 1; |
| End while |
| Output: Best solution found. |
| Algorithm 3. Outline of algorithm. |
| Inputs: vector and vector . |
| ; |
| For d = 1 to n Do |
| If |
| ; |
| End if |
| End for |
| Cost = the cost of k-NN by 10-fold cross-validation; |
| Output: Cost of . |
4. Experimental Study
4.1. Dataset Properties and Experimental Settings
4.2. Results and Comparisons
5. Conclusions and Future Work
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Gheyas, I.A.; Smith, L.S. Feature subset selection in large dimensionality domains. Pattern Recognit. 2010, 43, 5–13. [Google Scholar] [CrossRef]
- Garey, M.R.; Johnson, D.S. A Guide to the Theory of NP-Completeness, 1st ed.; WH Freemann: New York, NY, USA, 1979; ISBN 978-0716710455. [Google Scholar]
- Xue, B.; Zhang, M.; Browne, W.N.; Yao, X. A survey on evolutionary computation approaches to feature selection. IEEE. Trans. Evolut. Comput. 2016, 20, 606–626. [Google Scholar] [CrossRef]
- Pudil, P.; Novovičová, J.; Kittler, J. Floating search methods in feature selection. Pattern Recogn. Lett. 1994, 15, 1119–1125. [Google Scholar] [CrossRef]
- Alpaydin, E. Introduction to Machine Learning, 3rd ed.; MIT Press: Cambridge, MA, USA, 2014; ISBN 978-8120350786. [Google Scholar]
- Talbi, E.G. Metaheuristics: From Design to Implementation; John Wiley & Sons: Hoboken, NJ, USA, 2009; ISBN 978-0470278581. [Google Scholar]
- Dowlatshahi, M.B.; Nezamabadi-Pour, H.; Mashinchi, M. A discrete gravitational search algorithm for solving combinatorial optimization problems. Inf. Sci. 2014, 258, 94–107. [Google Scholar] [CrossRef]
- Han, K.H.; Kim, J.H. Quantum-inspired evolutionary algorithm for a class of combinatorial optimization. IEEE. Trans. Evolut. Comput. 2002, 6, 580–593. [Google Scholar] [CrossRef]
- Banks, A.; Vincent, J.; Anyakoha, C. A review of particle swarm optimization. Part II: hybridisation, combinatorial, multicriteria and constrained optimization, and indicative applications. Nat. Comput. 2008, 7, 109–124. [Google Scholar] [CrossRef]
- Cheng, R.; Jin, Y. A competitive swarm optimizer for large scale optimization. IEEE. Trans. Cybern. 2015, 45, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Dowlatshahi, M.B.; Rezaeian, M. Training spiking neurons with gravitational search algorithm for data classification. In Proceedings of the Swarm Intelligence and Evolutionary Computation (CSIEC), Bam, Iran, 9–11 March 2016; pp. 53–58. [Google Scholar]
- Dowlatshahi, M.B.; Derhami, V. Winner Determination in Combinatorial Auctions using Hybrid Ant Colony Optimization and Multi-Neighborhood Local Search. J. AI Data Min. 2017, 5, 169–181. [Google Scholar]
- Xue, B.; Zhang, M.; Browne, W.N. Multi-objective particle swarm optimisation (PSO) for feature selection. In Proceedings of the 14th Annual Conference on Genetic and Evolutionary Computation Conference (GECCO), Philadelphia, PA, USA, 7–11 July 2012; pp. 81–88. [Google Scholar]
- Zhang, Y.; Gong, D.W.; Cheng, J. Multi-objective particle swarm optimization approach for cost-based feature selection in classification. IEEE/ACM. Trans. Comput. Biol. Bioinform. 2017, 14, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Siedlecki, W.; Sklansky, J. A note on genetic algorithms for large-scale feature selection. Pattern Recogn. Lett. 1989, 10, 335–347. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, S.; Zeng, X. Research of multi-population agent genetic algorithm for feature selection. Expert. Syst. Appl. 2009, 36, 11570–11581. [Google Scholar] [CrossRef]
- Kabir, M.M.; Shahjahan, M.; Murase, K. A new local search based hybrid genetic algorithm for feature selection. Neurocomputing 2011, 74, 2914–2928. [Google Scholar] [CrossRef]
- Eroglu, D.Y.; Kilic, K. A novel Hybrid Genetic Local Search Algorithm for feature selection and weighting with an application in strategic decision making in innovation management. Inf. Sci. 2017, 405, 18–32. [Google Scholar] [CrossRef]
- Phan, A.V.; Le Nguyen, M.; Bui, L.T. Feature weighting and SVM parameters optimization based on genetic algorithms for classification problems. Appl. Intell. 2017, 46, 455–469. [Google Scholar] [CrossRef]
- Derrac, J.; Garcia, S.; Herrera, F. A first study on the use of coevolutionary algorithms for instance and feature selection. Hybrid Artif. Intell. Syst. 2009, 5572, 557–564. [Google Scholar]
- Oh, I.; Lee, J.S.; Moon, B.R. Hybrid genetic algorithm for feature selection. IEEE. Trans. Pattern Anal. Mach. Intell. 2004, 26, 1424–1437. [Google Scholar] [PubMed]
- Huang, C.L.; Dun, J.F. A distributed PSO–SVM hybrid system with feature selection and parameter optimization. Appl. Soft. Comput. 2008, 8, 1381–1391. [Google Scholar] [CrossRef]
- Chuang, L.Y.; Yang, C.H.; Li, J.C. Chaotic maps based on binary particle swarm optimization for feature selection. Appl. Soft. Comput. 2011, 11, 239–248. [Google Scholar] [CrossRef]
- Gu, S.; Cheng, R.; Jin, Y. Feature selection for high-dimensional classification using a competitive swarm optimizer. Soft Comput. 2016, 1–12. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, R.; Yu, Z.; Hu, R.; Tang, J.; Gui, C.; Yuan, Y. A BPSO-SVM algorithm based on memory renewal and enhanced mutation mechanisms for feature selection. Appl. Soft. Comput. 2017, 58, 176–192. [Google Scholar] [CrossRef]
- Bello, R.; Gomez, Y.; Garcia, M.M.; Nowe, A. Two-step particle swarm optimization to solve the feature selection problem. In Proceedings of the Seventh International Conference on Intelligent Systems Design and Applications (ISDA 2007), Rio de Janeiro, Brazil, 20–24 October 2007; pp. 691–696. [Google Scholar]
- Tanaka, K.; Kurita, T.; Kawabe, T. Selection of import vectors via binary particle swarm optimization and cross-validation for kernel logistic regression. In Proceedings of the 2007 International Joint Conference on Neural Networks, Orlando, FL, USA, 12–17 August 2007; pp. 12–17. [Google Scholar]
- Wang, X.; Yang, J.; Teng, X.; Xia, W.; Jensen, R. Feature selection based on rough sets and particle swarm optimization. Pattern Recognit. Lett. 2007, 28, 459–471. [Google Scholar] [CrossRef] [Green Version]
- Sivagaminathan, R.K.; Ramakrishnan, S. A hybrid approach for feature subset selection using neural networks and ant colony optimization. Expert. Syst. Appl. 2007, 33, 49–60. [Google Scholar] [CrossRef]
- Kashef, S.; Nezamabadi-pour, H. An advanced ACO algorithm for feature subset selection. Neurocomputing 2014, 147, 271–279. [Google Scholar] [CrossRef]
- Ghosh, A.; Datta, A.; Ghosh, S. Self-adaptive differential evolution for feature selection in hyperspectral image data. Appl. Soft. Comput. 2013, 3, 1969–1977. [Google Scholar] [CrossRef]
- Shunmugapriya, P.; Kanmani, S. A hybrid algorithm using ant and bee colony optimization for feature selection and classification (AC-ABC Hybrid). Swarm Evol. Comput. 2017, 36, 27–36. [Google Scholar] [CrossRef]
- Zorarpacı, E.; Özel, S.A. A hybrid approach of differential evolution and artificial bee colony for feature selection. Expert Syst. Appl. 2016, 62, 91–103. [Google Scholar] [CrossRef]
- Vieira, S.; Sousa, J.; Runkler, T. Multi-criteria ant feature selection using fuzzy classifiers. In Swarm Intelligence for Multi-objective Problems in Data Mining, vol. 242 of Studies in Computational Intelligence; Springer: Berlin, Germany, 2009; pp. 19–36. [Google Scholar]
- Xue, B.; Zhang, M.; Browne, W.N. Particle swarm optimization for feature selection in classification: A multi-objective approach. IEEE. Trans. Cybern. 2013, 43, 1656–1671. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Fu, W.; Zhang, M. Multi-objective feature selection in classification: A differential evolution approach. In Proceedings of the Asia-Pacific Conference on Simulated Evolution and Learning, Dunedin, New Zealand, 15–18 December 2014; pp. 516–528. [Google Scholar]
- Sahu, B.; Mishra, D. A novel feature selection algorithm using particle swarm optimization for cancer microarray data. Procedia Eng. 2012, 38, 27–31. [Google Scholar] [CrossRef]
- Oreski, S.; Oreski, G. Genetic algorithm-based heuristic for feature selection in credit risk assessment. Expert. Syst. Appl. 2014, 41, 2052–2064. [Google Scholar] [CrossRef]
- Tan, F.; Fu, X.Z.; Zhang, Y.Q.; Bourgeois, A. A genetic algorithm based method for feature subset selection. Soft Comput. 2008, 12, 111–120. [Google Scholar] [CrossRef]
- Battiti, R. Using mutual information for selecting features in supervised neural net learning. IEEE. Trans. Neural Netw. 1994, 5, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Moody, J. Data visualization and feature selection: New algorithms for non-gaussian data. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 1999; pp. 687–693. [Google Scholar]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. IEEE. Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Jakulin, A. Machine Learning Based on Attribute Interactions. Ph.D. Thesis, University of Ljubljana, Ljubljana, Slovenia, 2005. [Google Scholar]
- Lin, D.; Tang, X. Conditional infomax learning: An integrated framework for feature extraction and fusion. In Proceedings of the 9th European Conference on Computer Vision, Graz, Austria, 7–13 May 2006; pp. 68–82. [Google Scholar]
- Meyer, P.; Bontempi, G. On the use of variable complementarity for feature selection in cancer classification. In Proceedings of the Evolutionary Computation and Machine Learning in Bioinformatics, Budapest, Hungary, 10–12 April 2006; pp. 91–102. [Google Scholar]
- Han, J.; Pei, J.; Kamber, M. Data Mining: Concepts and Techniques, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2011; ISBN 978-9380931913. [Google Scholar]
- Dowlatshahi, M.B.; Nezamabadi-Pour, H. GGSA: a grouping gravitational search algorithm for data clustering. Eng. Appl. Artif. Intell. 2014, 36, 114–121. [Google Scholar] [CrossRef]
- Črepinšek, M.; Liu, S.H.; Mernik, M. Exploration and exploitation in evolutionary algorithms: A survey. ACM Comput. Surv. 2013, 45, 1–35. [Google Scholar] [CrossRef]
- Rafsanjani, M.K.; Dowlatshahi, M.B. A Gravitational search algorithm for finding near-optimal base station location in two-tiered WSNs. In Proceedings of the 3rd International Conference on Machine Learning and Computing, Singapore, 26–28 February 2011; pp. 213–216. [Google Scholar]
- Dowlatshahi, M.B.; Derhami, V.; Nezamabadi-pour, H. Ensemble of Filter-Based Rankers to Guide an Epsilon-Greedy Swarm Optimizer for High-Dimensional Feature Subset Selection. Information 2017, 8, 152. [Google Scholar] [CrossRef]
- Li, H.; Wang, J.; Du, H.; Karimi, H.R. Adaptive sliding mode control for Takagi-Sugeno fuzzy systems and its applications. IEEE. Trans. Fuzzy Syst. 2017, in press. [Google Scholar] [CrossRef]
- Van Blerkom, M.L. Measurement and Statistics for Teachers; Taylor & Francis: Didcot, UK, 2017. [Google Scholar]
- Abbasifard, M.R.; Ghahremani, B.; Naderi, H. A survey on nearest neighbor search methods. Int. J. Comput. Appl. 2014, 95, 39–52. [Google Scholar]
- Gene Expression Omnibus (GEO). Available online: https://www.ncbi.nlm.nih.gov/geo/ (accessed on 19 October 2017).
- Han, X.; Chang, X.; Quan, L.; Xiong, X.; Li, J.; Zhang, Z.; Liu, Y. Feature subset selection by gravitational search algorithm optimization. Inf. Sci. 2014, 281, 128–146. [Google Scholar] [CrossRef]
- Barani, F.; Mirhosseini, M.; Nezamabadi-pour, H. Application of binary quantum-inspired gravitational search algorithm in feature subset selection. Appl. Intell. 2017, 47, 304–318. [Google Scholar] [CrossRef]
- Derrac, J.; García, S.; Molina, D.; Herrera, F. A practical tutorial on the use of nonparametric statistical tests as a methodology for comparing evolutionary and swarm intelligence algorithms. Swarm Evol. Comput. 2011, 1, 3–18. [Google Scholar] [CrossRef]
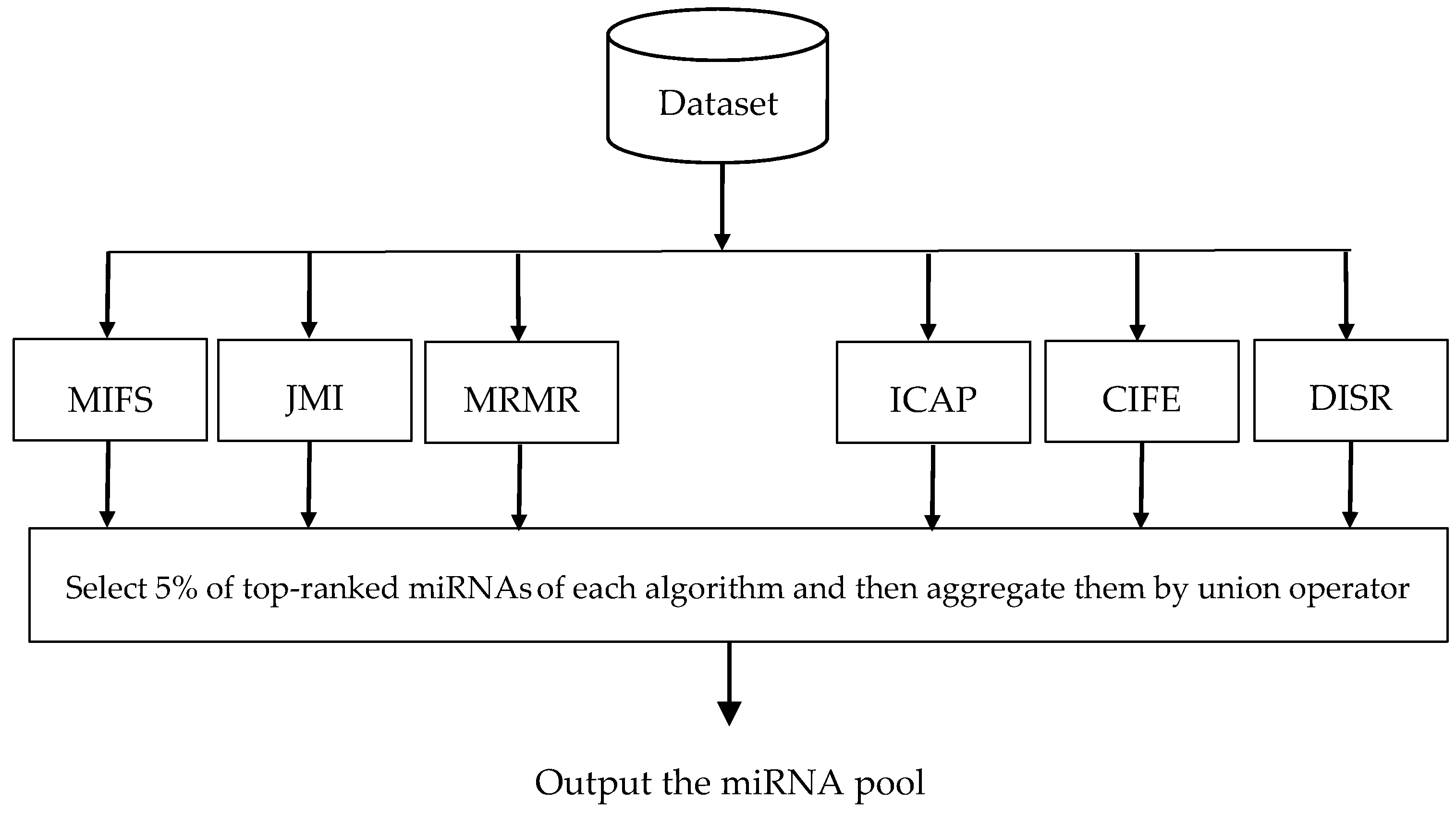
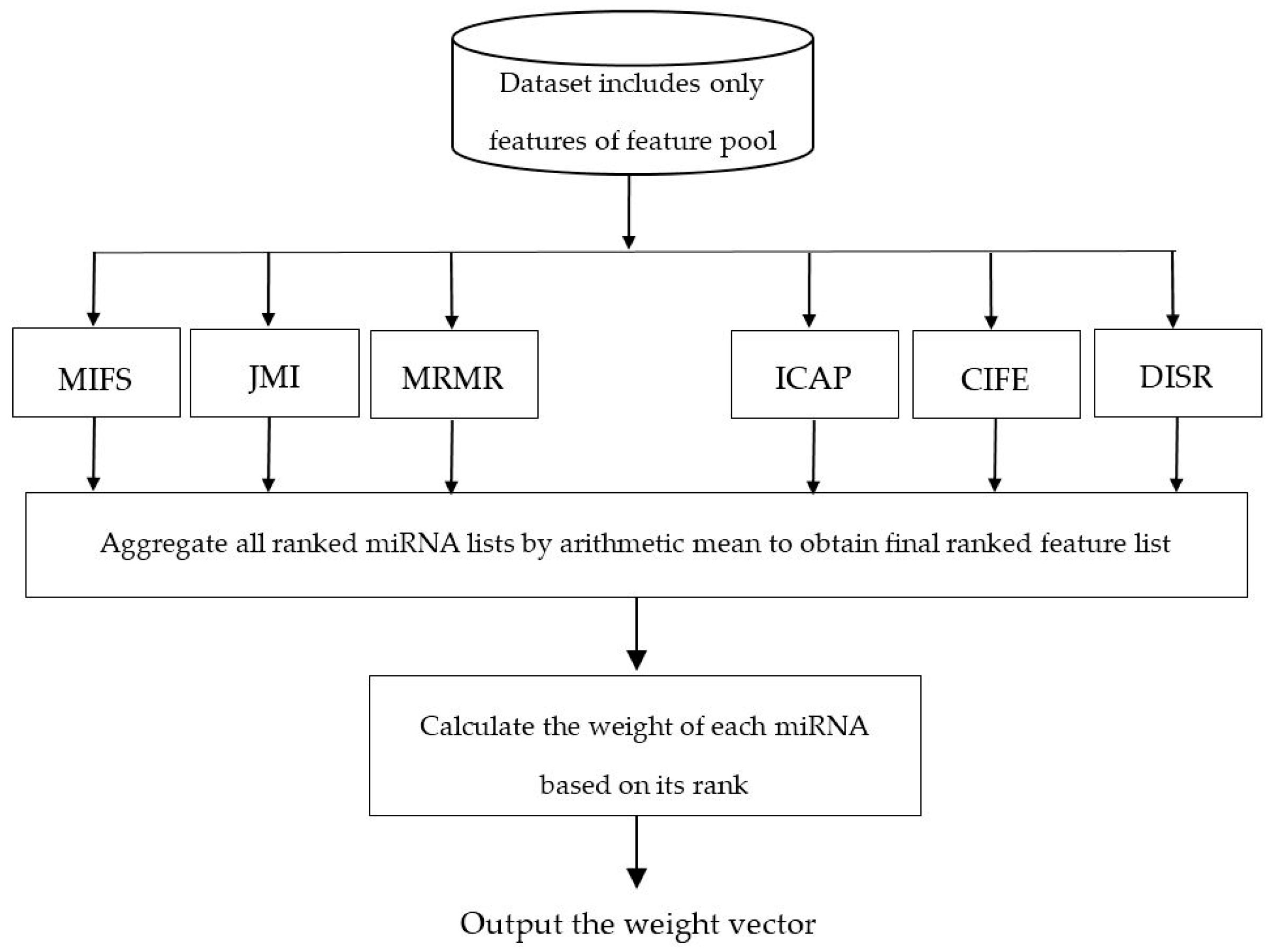
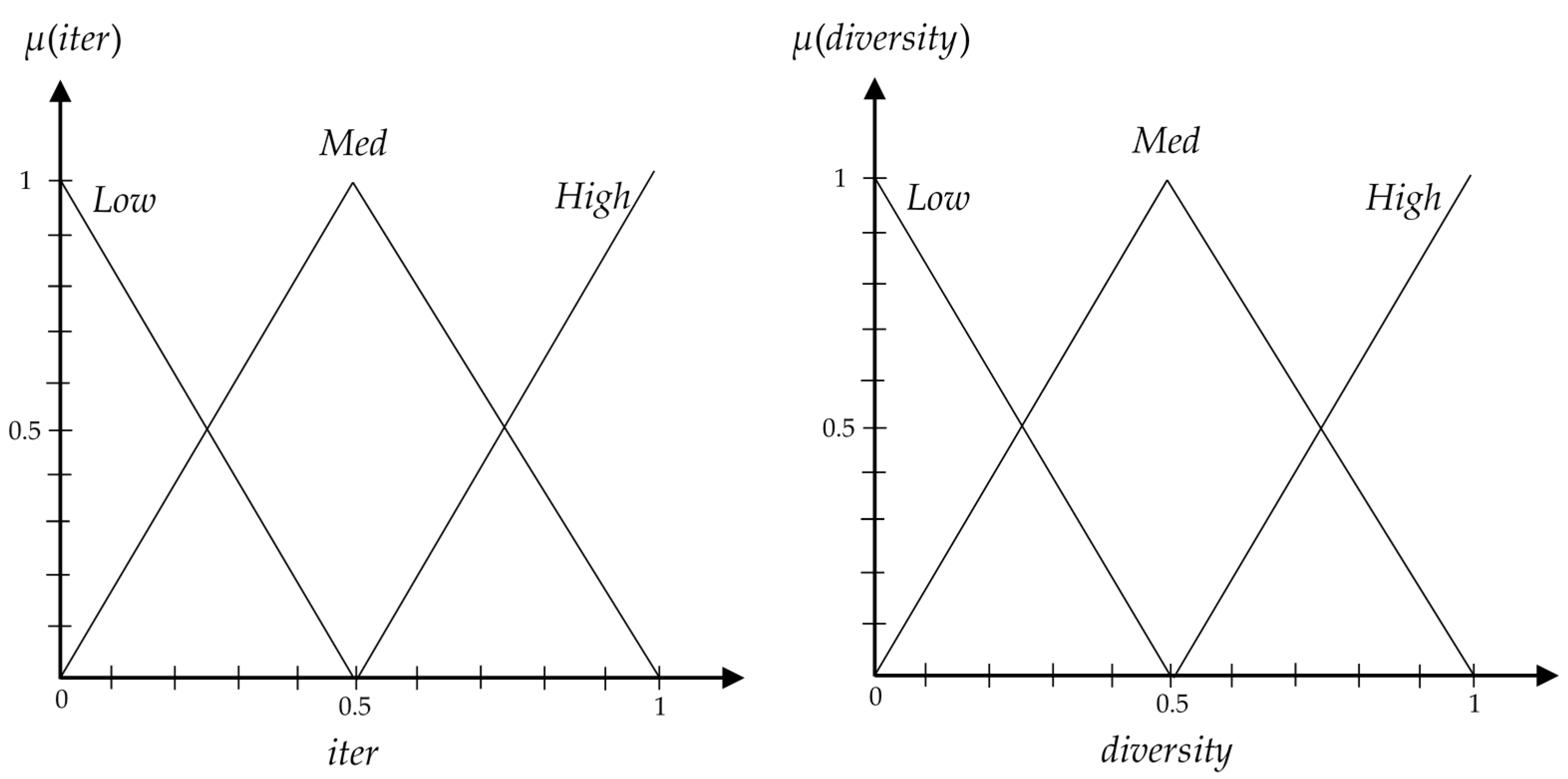
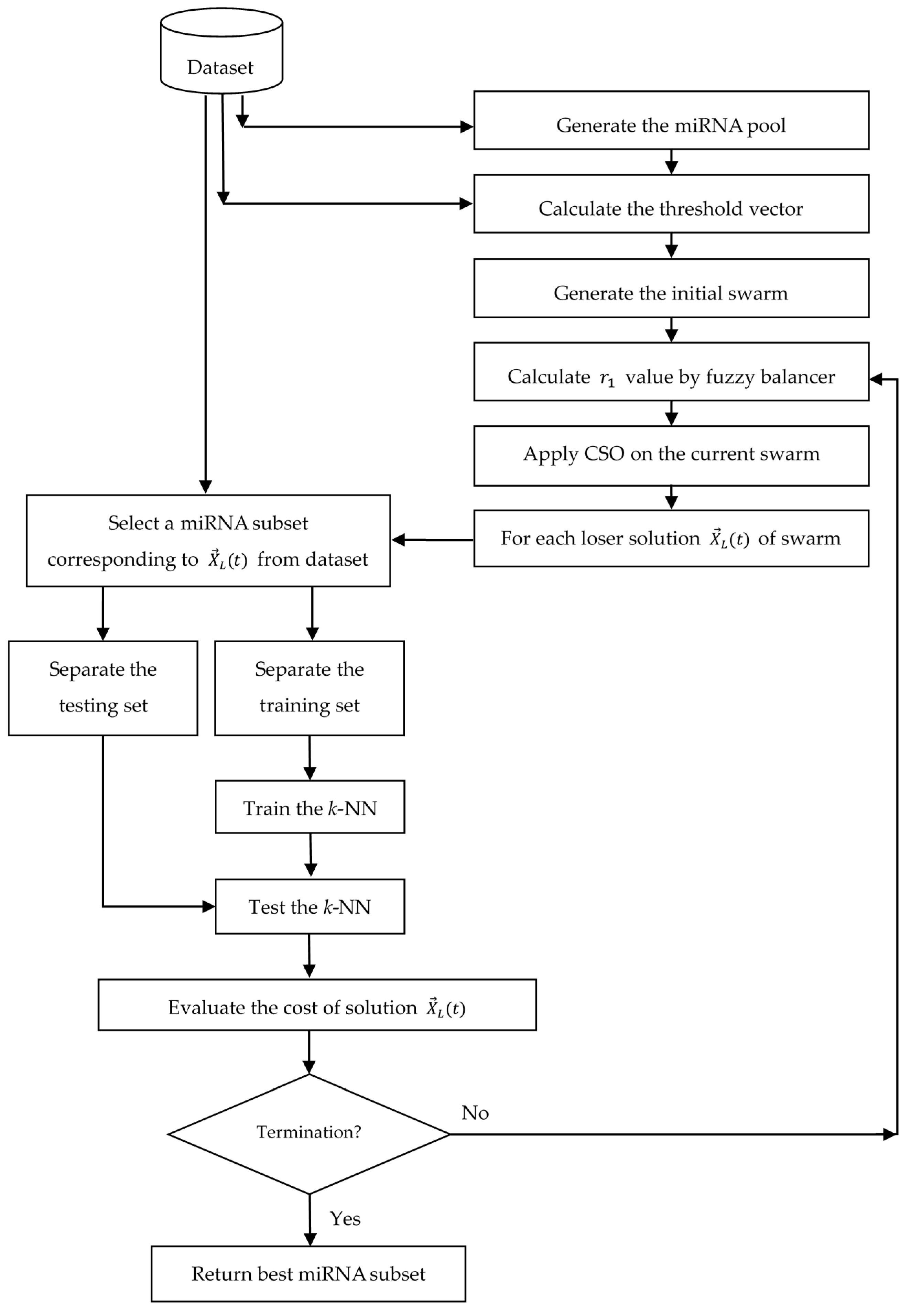
| Dataset | No. of miRNAs | No. of Normal Patients | No. of Cancer Patients | No. of Classes |
|---|---|---|---|---|
| Lung | 866 | 19 | 17 | 2 |
| Nasopharyngeal | 887 | 19 | 31 | 2 |
| Melanoma | 864 | 22 | 35 | 2 |
| Dataset | Lung | Nasopharyngeal | Melanoma |
|---|---|---|---|
| Proposed algorithm | 0.1931 | 0.3257 | 0.1605 |
| (0.0159) | (0.0317) | (0.0179) | |
| GA | 0.3312 + | 0.4623 + | 0.2981 + |
| (0.0393) | (0.0396) | (0.0296) | |
| CSO | 0.2607 + | 0.3393 ≈ | 0.2350 + |
| (0.0236) | (0.0352) | (0.0284) | |
| PSO | 0.3515 + | 0.4625 + | 0.3173 + |
| (0.0442) | (0.0342) | (0.0342) | |
| DE | 0.3227 + | 0.3902 + | 0.3092 + |
| (0.0371) | (0.0325) | (0.0307) | |
| ABC-DE | 0.3073 + | 0.3721 + | 0.3217 + |
| (0.0481) | (0.0362) | (0.0374) | |
| ACOFS | 0.3406 + | 0.3916 + | 0.3686 + |
| (0.0361) | (0.0403) | (0.0391) | |
| ACO-ABC | 0.3204 + | 0.3993 + | 0.2730 + |
| (0.0382) | (0.0381) | (0.0311) | |
| GSA | 0.3372 + | 0.3931 + | 0.3511 + |
| (0.0423) | (0.0350) | (0.0368) | |
| BQIGSA | 0.3007 + | 0.3384 ≈ | 0.2761 + |
| (0.0299) | (0.0301) | (0.0273) | |
| Xue1-PSO | 0.3603 + | 0.4321 + | 0.3154 + |
| (0.0490) | (0.0314) | (0.0491) | |
| Xue2-PSO | 0.3249 + | 0.3791 + | 0.2920 + |
| (0.0405) | (0.0424) | (0.0301) | |
| Xue3-PSO | 0.3618 + | 0.4412 + | 0.3064 + |
| (0.0506) | (0.0376) | (0.0429) | |
| Xue4-PSO | 0.3242 + | 0.3918 + | 0.2796 + |
| (0.0345) | (0.0350) | (0.0307) | |
| 2S-GA | 0.2785 + | 0.3341 ≈ | 0.2532 + |
| (0.0272) | (0.0366) | (0.0291) | |
| 2S-HGA | 0.2804 + | 0.3393 ≈ | 0.2699 + |
| (0.0316) | (0.0348) | (0.0254) | |
| 2S-PSO | 0.2523 + | 0.3478 + | 0.2382 + |
| (0.0205) | (0.0351) | (0.0232) | |
| Better | 16 | 13 | 16 |
| Worse | 0 | 0 | 0 |
| Similar | 0 | 3 | 0 |
| Dataset | Lung | Nasopharyngeal | Melanoma |
|---|---|---|---|
| Proposed algorithm | 0.8852 | 0.6949 | 0.9217 |
| (0.0317) | (0.0317) | (0.0253) | |
| GA | 0.7021 + | 0.5731 + | 0.7326 + |
| (0.0437) | (0.0482) | (0.0491) | |
| CSO | 0.7419 + | 0.5874 + | 0.7147 + |
| (0.0379) | (0.0391) | (0.0572) | |
| PSO | 0.7112 + | 0.5730 + | 0.7492 + |
| (0.0512) | (0.0462) | (0.0420) | |
| DE | 0.6826 + | 0.5592 + | 0.7718 + |
| (0.0417) | (0.0418) | (0.0325) | |
| ABC-DE | 0.6991 + | 0.5973 + | 0.7930 + |
| (0.0454) | (0.0427) | (0.0458) | |
| ACOFS | 0.6592 + | 0.6881 ≈ | 0.7504 + |
| (0.0459) | (0.0425) | (0.0429) | |
| ACO-ABC | 0.6811 + | 0.5347 + | 0.8271 + |
| (0.0357) | (0.0452) | (0.0342) | |
| GSA | 0.6927 + | 0.5696 + | 0.7906 + |
| (0.0448) | (0.0406) | (0.0394) | |
| BQIGSA | 0.7284 + | 0.5872 + | 0.8985 ≈ |
| (0.0521) | (0.0397) | (0.0294) | |
| Xue1-PSO | 0.6268 + | 0.5303 + | 0.7892 + |
| (0.0457) | (0.0472) | (0.0426) | |
| Xue2-PSO | 0.6995 + | 0.5605 + | 0.8427 + |
| (0.0452) | (0.0403) | (0.0368) | |
| Xue3-PSO | 0.6817 + | 0.5482 + | 0.8244 + |
| (0.0481) | (0.0495) | (0.0401) | |
| Xue4-PSO | 0.7393 + | 0.5881 + | 0.8625 + |
| (0.0479) | (0.0573) | (0.0429) | |
| 2S-GA | 0.7692 + | 0.6822 ≈ | 0.8751 + |
| (0.0472) | (0.0415) | (0.0284) | |
| 2S-HGA | 0.7824 + | 0.6798 ≈ | 0.8901 + |
| (0.0385) | (0.0381) | (0.0372) | |
| 2S-PSO | 0.7924 + | 0.6402 + | 0.9063 ≈ |
| (0.0322) | (0.0493) | (0.0371) | |
| Better | 16 | 13 | 16 |
| Worse | 0 | 0 | 0 |
| Similar | 0 | 3 | 2 |
| Dataset | Lung | Nasopharyngeal | Melanoma |
|---|---|---|---|
| Proposed algorithm | 9.74 | 15.58 | 15.21 |
| (5.36) | (3.18) | (7.82) | |
| GA | 35.62 + | 292.26 + | 41.65 + |
| (18.52) | (16.02) | (19.23) | |
| CSO | 14.27 + | 15.22 ≈ | 20.52 + |
| (13.43) | (5.41) | (10.29) | |
| PSO | 30.44 + | 267.18 + | 37.93 + |
| (19.17) | (13.36) | (21.16) | |
| DE | 31.15 + | 225.48 + | 35.19 + |
| (15.66) | (12.15) | (18.80) | |
| ABC-DE | 47.91 + | 261.37 + | 42.01 + |
| (20.41) | (13.83) | (21.13) | |
| ACOFS | 50.14 + | 261.31 + | 40.24 + |
| (23.62) | (12.21) | (17.91) | |
| ACO-ABC | 32.81 + | 251.07 + | 36.74 + |
| (19.60) | (14.11) | (19.05) | |
| GSA | 52.83 + | 290.18 + | 41.10 + |
| (25.12) | (16.33) | (24.16) | |
| BQIGSA | 30.05 + | 242.36 + | 37.35 + |
| (17.91) | (15.13) | (16.87) | |
| Xue1-PSO | 29.37 + | 266.31 + | 36.13 + |
| (17.44) | (15.32) | (18.65) | |
| Xue2-PSO | 17.64 + | 31.06 + | 23.18 + |
| (21.49) | (8.42) | (15.75) | |
| Xue3-PSO | 28.14 + | 322.19 + | 34.92 + |
| (19.62) | (26.52) | (16.35) | |
| Xue4-PSO | 24.02 + | 273.21 + | 30.44 + |
| (16.71) | (32.04) | (17.32) | |
| 2S-GA | 15.67 + | 28.21 + | 22.47 + |
| (12.74) | (7.15) | (10.79) | |
| 2S-HGA | 17.05 + | 15.66 ≈ | 24.10 + |
| (14.25) | (5.73) | (12.32) | |
| 2S-PSO | 16.31 + | 23.14 + | 21.52 + |
| (13.27) | (7.35) | (11.26) | |
| Better | 16 | 14 | 16 |
| Worse | 0 | 0 | 0 |
| Similar | 0 | 2 | 0 |
| Lung | Nasopharyngeal | Melanoma |
|---|---|---|
| hsa-let-7d | hsa-miR-638 | hsa-miR-17 |
| hsa-miR-423-5p | hsa-miR-762 | hsa-miR-664 |
| hsa-let-7f | hsa-miR-1915 | hsa-miR-145 |
| hsa-miR-140-3p | hsa-miR-92a | hsa-miR-422a |
| hsa-miR-25 | hsa-miR-135a | hsa-miR-216a |
| hsa-miR-98 | hsa-miR-181a | hsa-miR-186 |
| hsa-miR-195 | hsa-miR-1275 | hsa-miR-601 |
| hsa-miR-126 | hsa-miR-940 | hsa-miR-1301 |
| hsa-miR-20b | hsa-miR-572 | hsa-miR-328 |
| hsa-let-7e | hsa-miR-29c | hsa-miR-30d |
| hsa-let-7c | hsa-miR-548q | hsa-let-7d |
| Dataset | Lung | Nasopharyngeal | Melanoma |
|---|---|---|---|
| Proposed algorithm | 0.1931 | 0.3257 | 0.1605 |
| 0.0159 | 0.0317 | 0.0179 | |
| MIFS-k-NN | 0.1822 − | 0.3741 + | 0.2173 + |
| 0.0121 | 0.0324 | 0.0198 | |
| JMI-k-NN | 0.2274 + | 0.3499 + | 0.2394 + |
| 0.0213 | 0.0327 | 0.0219 | |
| MRMR-k-NN | 0.2182 + | 0.3807 + | 0.2201 + |
| 0.0195 | 0.0351 | 0.0214 | |
| ICAP-k-NN | 0.2418 + | 0.3305 ≈ | 0.1994 + |
| 0.0269 | 0.0319 | 0.0201 | |
| CIFE-k-NN | 0.2362 + | 0.3677 + | 0.2319 + |
| 0.0275 | 0.0359 | 0.0237 | |
| DISR-k-NN | 0.2479 + | 0.3803 + | 0.2505 + |
| 0.0304 | 0.0381 | 0.0241 | |
| Better | 5 | 5 | 6 |
| Worse | 1 | 0 | 0 |
| Similar | 0 | 1 | 0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dowlatshahi, M.B.; Derhami, V.; Nezamabadi-pour, H. A Novel Three-Stage Filter-Wrapper Framework for miRNA Subset Selection in Cancer Classification. Informatics 2018, 5, 13. https://doi.org/10.3390/informatics5010013
Dowlatshahi MB, Derhami V, Nezamabadi-pour H. A Novel Three-Stage Filter-Wrapper Framework for miRNA Subset Selection in Cancer Classification. Informatics. 2018; 5(1):13. https://doi.org/10.3390/informatics5010013
Chicago/Turabian StyleDowlatshahi, Mohammad Bagher, Vali Derhami, and Hossein Nezamabadi-pour. 2018. "A Novel Three-Stage Filter-Wrapper Framework for miRNA Subset Selection in Cancer Classification" Informatics 5, no. 1: 13. https://doi.org/10.3390/informatics5010013





