What We Do Not Know about Fungal Cell Adhesion Molecules
Abstract
:1. Introduction: The Breadth of the Problem
1.1. Some Relevant Reviews
1.2. Genomics and the Taxonomy of Fungal Cell Adhesion
2. An Incomplete Catalog of Fungal Adhesins
2.1. Ascomycota
2.1.1. Saccharomycotina
2.1.2. Pezizomycotina
Adhesins from Pezizomycotina
Genomics Approaches in Pezizomycotina
2.1.3. Taphrinomycotina
2.2. Basidiomycota
2.3. Other Fungal Phyla
3. Structural Characteristics of Known Fungal Cell Adhesins
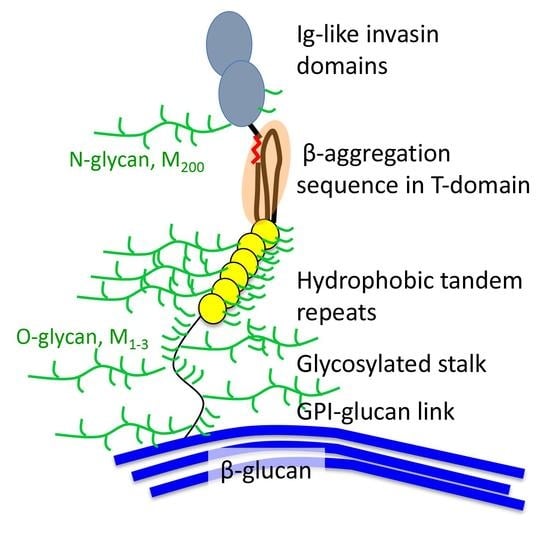
3.1. “Typical” Adhesins
3.2. Sequence Characteristics
3.2.1. N-Terminal Domains
3.2.2. Tandem Repeats
3.2.3. Cys-Rich Domains
3.3. Cell Surface Attachments
3.4. Ligand Specificities
3.5. Amyloid-Like Interactions in Fungal Adhesins
3.6. Cytoplasmic Proteins in Cell Adhesion
3.7. Are There Surface Molecules that Inhibit Adhesion?
3.8. Adhesion and Glycans
4. Bioinformatics and Discovery
4.1. Searching Databases for Adhesins
4.2. Is It Really an Adhesin? Some Approaches
5. What Do We Not Know?
5.1. Problems that Are Currently Soluble
5.1.1. How Many Types of Fungal Adhesins Are There, and How Many in Each Fungus?
5.1.2. How Widespread Are Amyloid-Based Clustering and Aggregation?
5.1.3. Are There More Roles for Cys-Rich Domains?
5.1.4. Is There Covalent Bonding between Cells?
5.1.5. What Are the Roles of Dibasic Sequences?
5.2. Problems that Are Not So Easily Approached
- Structure and function of surface glues that stick fungi to hosts are open questions. An extreme example would be lichen-associated fungi,-associatedle wo which stick to rocks and have a mutualistic interaction with algae. How are glues related to extracellular matrix components in fungal and mixed fungal-bacterial biofilms [6,45]?
- What are the functions of “anti-adhesins”, proteins whose deletion leads to increased activity of other adhesins [91]?
5.3. Why Do We Need to Know More?
5.3.1. Evolution
5.3.2. Biofilms
5.3.3. Pathogenesis
6. A Proposal
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Rauch, M.; Graef, H.; Rozenzhak, S.; Jones, S.; Bleckmann, C.; Kruger, R.; Naik, R.; Stone, M. Characterization of microbial contamination in United States Air Force aviation fuel tanks. J. Ind. Microbiol. Biotechnol. 2006, 33, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Krivushina, A.A.; Chekunova, L.N.; Mokeeva, V.L.; Polyakova, A.V. Micromycetes in fuel tanks of exploiting planes. Mikol. Fitopatol. 2016, 50, 108–114. [Google Scholar]
- Itah, A.Y.; Brooks, A.A.; Ogar, B.O.; Okure, A.B. Biodegradation of iInternational Jet A-1 aviation fuel by microorganisms isolated from aircraft tank and joint hydrant storage systems. Bull. Environ. Contam. Toxicol. 2009, 83, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Rola, K.; Osyczka, P.; Kafel, A. Different Heavy Metal Accumulation Strategies of Epilithic Lichens Colonising Artificial Post-Smelting Wastes. Arch. Environ. Contam. Toxicol. 2016, 70, 418–428. [Google Scholar] [CrossRef] [PubMed]
- De Groot, P.W.; Bader, O.; de Boer, A.D.; Weig, M.; Chauhan, N. Adhesins in human fungal pathogens: Glue with plenty of stick. Eukaryot. Cell 2013, 12, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Epstein, L.; Nicholson, R. Adhesion and adhesives of fungi and oomycetes. In Biological Adhesives, 2nd ed.; Springer: Cham, Switzerland, 2016; pp. 25–55. [Google Scholar]
- Tronchin, G.; Pihet, M.; Lopes-Bezerra, L.; Bouchara, J. Adherence mechanisms in human pathogenic fungi. Med. Mycol. 2008, 46, 749–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dranginis, A.M.; Rauceo, J.R.; Coronado, J.E.; Lipke, P.N. A biochemical guide to yeast adhesins: Glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 2007, 71, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Spatafora, J.W.; Aime, M.C.; Grigoriev, I.V.; Martin, F.; Stajich, J.E.; Blackwell, M. The fungal tree of life: From molecular systematics to genome-scale phylogenies. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Stajich, J.E. Fungal Genomes and Insights into the Evolution of the Kingdom. Microbiol. Spectr. 2017, 5, 2016. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, A.; Almen, M.S.; Fredriksson, R.; Schioth, H.B. The origin of GPCRs: Identification of mammalian like Rhodopsin, Adhesion, Glutamate and Frizzled GPCRs in fungi. PLoS ONE 2012, 7, e29817. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Rosenblum, E.B.; Poorten, T.J.; Joneson, S.; Settles, M. Substrate-specific gene expression in Batrachochytrium dendrobatidis, the chytrid pathogen of amphibians. PLoS ONE 2012, 7, e49924. [Google Scholar] [CrossRef] [PubMed]
- Chaffin, W.L. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 2008, 72, 495–544. [Google Scholar] [CrossRef] [PubMed]
- Perez, A.; Pedros, B.; Murgui, A.; Casanova, M.; Lopez-Ribot, J.L.; Martinez, J.P. Biofilm formation by Candida albicans mutants for genes coding fungal proteins exhibiting the eight-cysteine-containing CFEM domain. FEMS Yeast Res. 2006, 6, 1074–1084. [Google Scholar] [CrossRef] [PubMed]
- Reuter, L.; Ritala, A.; Linder, M.; Joensuu, J. Novel Hydrophobin Fusion Tags for Plant-Produced Fusion Proteins. PLoS ONE 2016, 11, e0164032. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Kitaoka, H.; Park, P.; Ikeda, K. Novel aspects of hydrophobins in wheat isolate of Magnaporthe oryzae: Mpg1, but not Mhp1, is essential for adhesion and pathogenicity. J. Gen. Plant Pathol. 2016, 82, 18–28. [Google Scholar] [CrossRef]
- Sunde, M.; Pham, C.L.L.; Kwan, A.H. Molecular characteristics and biological functions of surface-active and surfactant proteins. Annu. Rev. Biochem. 2017, 86. [Google Scholar] [CrossRef] [PubMed]
- Prados-Rosales, R.; Luque-Garcia, J.L.; Martinez-Lopez, R.; Gil, C.; Di Pietro, A. The Fusarium oxysporum cell wall proteome under adhesion-inducing conditions. Proteomics 2009, 9, 4755–4769. [Google Scholar] [CrossRef] [PubMed]
- Ghamrawi, S.; Gastebois, A.; Zykwinska, A.; Vandeputte, P.; Marot, A.; Mabilleau, G.; Cuenot, S.; Bouchara, J.P. A Multifaceted Study of Scedosporium boydii Cell Wall Changes during Germination and Identification of GPI-Anchored Proteins. PLoS ONE 2015, 10, e0128680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhuri, R.; Ansari, F.A.; Raghunandanan, M.V.; Ramachandran, S. FungalRV: Adhesin prediction and immunoinformatics portal for human fungal pathogens. BMC Genom. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Wyrebek, M.; Bidochka, M.J. Variability in the Insect and Plant Adhesins, Mad1 and Mad2, within the Fungal Genus Metarhizium Suggest Plant Adaptation as an Evolutionary Force. PLoS ONE 2013, 8, e59357. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, Y.; Zhu, H.; Zhang, K.Q. Phylogenic analysis of adhesion related genes Mad1 revealed a positive selection for the evolution of trapping devices of nematode-trapping fungi. Sci. Rep. 2016, 6, 22609. [Google Scholar] [CrossRef] [PubMed]
- Brandhorst, T.T.; Wüthrich, M.; Warner, T.; Klein, B. Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis. J. Exp. Med. 1999, 189, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Brandhorst, T.T.; Roy, R.; Wüthrich, M.; Nanjappa, S.; Filutowicz, H.; Galles, K.; Tonelli, M.; McCaslin, D.R.; Satyshur, K.; Klein, B. Structure and Function of a Fungal Adhesin that Binds Heparin and Mimics Thrombospondin-1 by Blocking T Cell Activation and Effector Function. PLoS Pathog. 2013, 9, e1003464. [Google Scholar] [CrossRef] [PubMed]
- Beaussart, A.; Brandhorst, T.; Dufrene, Y.F.; Klein, B.S. Blastomyces virulence adhesin-1 protein binding to glycosaminoglycans is enhanced by protein disulfide isomerase. mBio 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Takahashi-Nakaguchi, A.; Sakai, K.; Takahashi, H.; Hagiwara, D.; Toyotome, T.; Chibana, H.; Watanabe, A.; Yaguchi, T.; Yamaguchi, M.; Kamei, K.; et al. Aspergillus fumigatus adhesion factors in dormant conidia revealed through comparative phenotypic and transcriptomic analyses. Cell. Microbiol. 2018, 20. [Google Scholar] [CrossRef] [PubMed]
- Lee, S. Identification of Fungal Adhesins for the Development of Potential Drug Targets in the Treatment of Aspergillosis; ProQuest Dissertations Publishing: New York, NY, USA, 2011. [Google Scholar]
- Matsuzawa, T.; Morita, T.; Tanaka, N.; Tohda, H.; Takegawa, K. Identification of a galactose-specific flocculin essential for non-sexual flocculation and filamentous growth in Schizosaccharomyces pombe. Mol. Microbiol. 2011, 82, 1531–1544. [Google Scholar] [CrossRef] [PubMed]
- Lo, W.S.; Dranginis, A.M. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by S. cerevisiae. Mol. Biol. Cell. 1998, 9, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Linder, T.; Gustafsson, C.M. Molecular phylogenetics of Ascomycotal adhesins-A novel family of putative cell-surface adhesive proteins in fission yeasts. Fungal Genet. Biol. 2007, 1, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Sharifmoghadam, M.R.; Valdivieso, M.H. The Schizosaccharomyces pombe Map4 adhesin is a glycoprotein that can be extracted from the cell wall with alkali but not with beta-glucanases and requires the C-terminal DIPSY domain for function. Mol. Microbiol. 2008, 69, 1476–1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ecker, M.; Deutzmann, R.; Lehle, L.; Mrsa, V.; Tanner, W. Pir proteins of Saccharomyces cerevisiae are attached to beta-1,3-glucan by a new protein-carbohydrate linkage. J. Biol. Chem. 2006, 281, 11523–11529. [Google Scholar] [CrossRef] [PubMed]
- Kottom, T.J.; Kennedy, C.C.; Limper, A.H. Pneumocystis PCINT1, a molecule with integrin-like features that mediates organism adhesion to fibronectin. Mol. Microbiol. 2008, 67, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Kutty, G.; England, K.J.; Kovacs, J.A. Expression of Pneumocystis jirovecii major surface glycoprotein in Saccharomyces cerevisiae. J. Infect. Dis. 2013, 208, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, P.A.; Penha, L.L.; Mendonca-Previato, L.; Previato, J.O. Mannoprotein MP84 mediates the adhesion of Cryptococcus neoformans to epithelial lung cells. Front. Cell. Infect. Microbiol. 2014, 4, 106. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhai, B.; Lin, X. The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog. 2012, 8, e1002765. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tian, X.; Gyawali, R.; Lin, X. Fungal adhesion protein guides community behaviors and autoinduction in a paracrine manner. Proc. Natl. Acad. Sci. USA 2013, 110, 11571–11576. [Google Scholar] [CrossRef] [PubMed]
- Ao, J.; Free, S.J. Genetic and biochemical characterization of the GH72 family of cell wall transglycosylases in Neurospora crassa. Fungal Genet. Biol. 2017, 101, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Ao, J.; Aldabbous, M.; Notaro, M.J.; Lojacono, M.; Free, S.J. A proteomic and genetic analysis of the Neurospora crassa conidia cell wall proteins identifies two glycosyl hydrolases involved in cell wall remodeling. Fungal Genet. Biol. 2016, 94, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Li, W.; Liu, X.; Che, J.; Wu, Y.; Lu, J. Distinct expression levels of ALS, LIP, and SAP genes in Candida tropicalis with diverse virulent activities. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Karkowska-Kuleta, J.; Zajac, D.; Bras, G.; Bochenska, O.; Rapala-Kozik, M.; Kozik, A. Binding of human plasminogen and high-molecular-mass kininogen by cell surface-exposed proteins of Candida parapsilosis. Acta Biochim. Pol. 2017, 64, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Cabral, V.; Znaidi, S.; Walker, L.A.; Martin-Yken, H.; Dague, E.; Legrand, M.; Lee, K.; Chauvel, M.; Firon, A.; Rossignol, T.; et al. Targeted Changes of the Cell Wall Proteome Influence Candida albicans Ability to Form Single- and Multi-strain Biofilms. PLoS Pathog. 2014, 10, e1004542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Younes, S.; Bahnan, W.; Dimassi, H.I.; Khalaf, R.A. The Candida albicans Hwp2 is necessary for proper adhesion, biofilm formation and oxidative stress tolerance. Microbiol. Res. 2011, 166, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.B.; Gulati, M.; Johnson, A.D.; Nobile, C.J. Development and regulation of single- and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 2018, 16, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Bennett, D.; Erdman, S.E. Maintenance of mating cell integrity requires the adhesin Fig2p. Eukaryot. Cell 2002, 1, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Coronado, J.E.; Attie, O.; Epstein, S.L.; Qiu, W.; Lipke, P.N. Composition-Modified Matrices Improve Identification of Homologs of Saccharomyces cerevisiae Low-Complexity Glycoproteins. Eukaryot. Cell. 2006, 5, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Escamilla, A.M.; Rousseau, F.; Schymkowitz, J.; Serrano, L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004, 22, 1302–1306. [Google Scholar] [CrossRef] [PubMed]
- Salgado, P.S.; Yan, R.; Taylor, J.D.; Burchell, L.; Jones, R.; Hoyer, L.L.; Matthews, S.J.; Simpson, P.J.; Cota, E. Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 2011, 108, 15775–15779. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Oh, S.H.; Jones, R.; Garnett, J.A.; Salgado, P.S.; Rusnakova, S.; Matthews, S.J.; Hoyer, L.L.; Cota, E. The peptide-binding cavity is essential for Als3-mediated adhesion of Candida albicans to human cells. J. Biol. Chem. 2014, 289, 18401–18412. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, D.C.; Yeaman, M.R.; Welch, W.H.; Phan, Q.T.; Fu, Y.; Ibrahim, A.S.; Filler, S.G.; Zhang, M.; Waring, A.J.; Edwards, E.J., Jr. Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 2004, 279, 30480–30489. [Google Scholar] [CrossRef] [PubMed]
- Kraushaar, T.; Bruckner, S.; Veelders, M.; Rhinow, D.; Schreiner, F.; Birke, R.; Pagenstecher, A.; Mosch, H.U.; Essen, L.O. Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-like Adhesin Domain Girdled by Aromatic Bands. Structure 2015, 23, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Maestre-Reyna, M.; Diderrich, R.; Veelders, M.S.; Eulenburg, G.; Kalugin, V.; Bruckner, S.; Keller, P.; Rupp, S.; Mosch, H.U.; Essen, L.O. Structural basis for promiscuity and specificity during Candida glabrata invasion of host epithelia. Proc. Natl. Acad. Sci. USA 2012, 109, 16864–16869. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.V.; Stassen, C.; Stals, I.; Donohue, D.S.; Devreese, B.; De Greve, H.; Willaert, R.G. The N-terminal domain of the Flo1 flocculation protein from Saccharomyces cerevisiae binds specifically to mannose carbohydrates. Eukaryot. Cell 2011, 10, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Coronado, J.E.; Epstein, S.L.; Qiu, W.; Lipke, P.N. Discovery of Recurrent Sequence Motifs in Saccharomyces cerevisiae Cell Wall Proteins. Match 2007, 58, 281–299. [Google Scholar] [PubMed]
- Fidalgo, M.; Barrales, R.R.; Ibeas, J.I.; Jimenez, J. Adaptive evolution by mutations in the FLO11 gene. Proc. Natl. Acad. Sci. USA 2006, 103, 11228–11233. [Google Scholar] [CrossRef] [PubMed]
- Frank, A.T.; Ramsook, C.B.; Otoo, H.N.; Tan, C.; Soybelman, G.; Rauceo, J.M.; Gaur, N.K.; Klotz, S.A.; Lipke, P.N. Structure and function of glycosylated tandem repeats from Candida albicans Als adhesins. Eukaryot. Cell 2010, 9, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.N.; Klotz, S.A.; Dufrene, Y.F.; Jackson, D.N.; Garcia-Sherman, M.C. Amyloid-Like beta-Aggregates as Force-Sensitive Switches in Fungal Biofilms and Infections. Microbiol. Mol. Biol. Rev. 2017, 82, 17. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.X.; El-Kirat-Chatel, S.; Joseph, I.G.; Jackson, D.N.; Ramsook, C.B.; Dufrene, Y.F.; Lipke, P.N. Force Sensitivity in Saccharomyces cerevisiae Flocculins. mSphere 2016, 1, e00128-16. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.X.; Lipke, P.N. Role of force-sensitive amyloid-like interactions in fungal catch bonding and biofilms. Eukaryot. Cell 2014, 13, 1136–1142. [Google Scholar] [CrossRef] [PubMed]
- El-Kirat-Chatel, S.; Beaussart, A.; Vincent, S.P.; Abellan Flos, M.; Hols, P.; Lipke, P.N.; Dufrene, Y.F. Forces in yeast flocculation. Nanoscale 2015, 7, 1760–1767. [Google Scholar] [CrossRef] [PubMed]
- Verstrepen, K.J.; Jansen, A.; Lewitter, F.; Fink, G.R. Intragenic tandem repeats generate functional variability. Nat. Genet. 2005, 37, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Rauceo, J.M.; De Armond, R.; Otoo, H.; Kahn, P.C.; Klotz, S.A.; Gaur, N.K.; Lipke, P.N. Threonine-Rich Repeats Increase Fibronectin Binding in the Candida albicans Adhesin Als5p. Eukaryot. Cell. 2006, 5, 1664–1673. [Google Scholar] [CrossRef] [PubMed]
- Heinisch, J.J.; Lipke, P.; Beaussart, A.; El Kirat Chatel, S.; Dupres, V.; Alsteens, D.; Dufrene, Y.F. Atomic force microscopy—Looking at mechanosensors on the cell surface. J. Cell Sci. 2012, 125, 4189–4195. [Google Scholar] [CrossRef] [PubMed]
- Alsteens, D.; Garcia, M.C.; Lipke, P.N.; Dufrene, Y.F. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc. Natl. Acad. Sci. USA 2010, 107, 20744–20749. [Google Scholar]
- Huang, G.; Dougherty, S.D.; Erdman, S.E. Conserved WCPL and CX4C domains mediate several mating adhesin interactions in Saccharomyces cerevisiae. Genetics 2009, 182, 173–189. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.N.; Kurjan, J. Sexual agglutination in budding yeasts: Structure, function, and regulation of adhesion glycoproteins. Microbiol. Rev. 1992, 56, 180–194. [Google Scholar] [PubMed]
- Shen, M.; Wang, L.; Pike, J.; Jue, C.K.; Zhao, H.; de Nobel, H.; Kurjan, J.; Lipke, P.N. Delineation of Functional Regions within the Subunits of the Saccharomyces cerevisiae Cell Adhesion Molecule a-Agglutinin. J. Biol. Chem. 2002, 276, 15768–15775. [Google Scholar]
- Tammi, M.; Ballou, L.; Taylor, A.; Ballou, C.E. Effect of glycosylation on yeast invertase oligomer stability. J. Biol. Chem. 1987, 262, 4395–4401. [Google Scholar] [PubMed]
- Li, F.; Palecek, S.P. Distinct domains of the Candida albicans adhesin EAP1 p mediate cell-cell and cell-substrate interactions. Microbiology 2008, 154, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Svarovsky, M.J.; Karlsson, A.J.; Wagner, J.P.; Marchillo, K.; Oshel, P.; Andes, D.; Palecek, S.P. Eap1p, an adhesin that mediates Candida albicans biofilm formation in vitro and in vivo. Eukaryot. Cell. 2007, 6, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Ielasi, F.S.; Alioscha-Perez, M.; Donohue, D.; Claes, S.; Sahli, H.; Schols, D.; Willaert, R.G. Lectin-glycan interaction network-based identification of host receptors of microbial pathogenic adhesins. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Rieg, G.; Fonzi, W.A.; Belanger, P.H.; Edwards, E.J., Jr.; Filler, S.G. Expression of the Candida albicans gene ALS1 in Saccharomyces cerevisiae induces adherence to endothelial and epithelial cells. Infect. Immun. 1998, 66, 1783–1786. [Google Scholar] [PubMed]
- Liu, Y.; Filler, S.G. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot. Cell 2011, 10, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.S.; Brunke, S.; Albrecht, A.; Thewes, S.; Laue, M.; Edwards, J.E.; Filler, S.G.; Hube, B. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 2008, 4, e1000217. [Google Scholar] [CrossRef] [PubMed]
- Klotz, S.A.; Gaur, N.K.; Lake, D.F.; V, C.; Rauceo, J.; Lipke, P.N. Degenerate peptide recognition by Candida albicans adhesins Als5p and Als1p. Infect. Immun. 2004, 72, 2029–2034. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.C.; Lee, J.T.; Ramsook, C.B.; Alsteens, D.; Dufrene, Y.F.; Lipke, P.N. A role for amyloid in cell aggregation and biofilm formation. PLoS ONE 2011, 6, e17632. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.N.; Garcia, M.C.; Alsteens, D.; Ramsook, C.B.; Klotz, S.A.; Dufrene, Y.F. Strengthening relationships: Amyloids create adhesion nanodomains in yeasts. Trends Microbiol. 2012, 20, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Palecek, S.P. Identification of Candida albicans genes that induce Saccharomyces cerevisiae cell adhesion and morphogenesis. Biotechnol. Prog. 2005, 21, 1601–1609. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Nett, J.E.; Andes, D.R.; Mitchell, A.P. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 2006, 5, 1604–1610. [Google Scholar] [CrossRef] [PubMed]
- Staab, J.F.; Bradway, S.D.; Fidel, P.L.; Sundstrom, P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 1999, 283, 1535–1538. [Google Scholar] [CrossRef] [PubMed]
- Staab, J.F.; Bahn, Y.S.; Tai, C.H.; Cook, P.F.; Sundstrom, P. Expression of transglutaminase substrate activity on Candida albicans germ tubes through a coiled, disulfide-bonded N-terminal domain of Hwp1 requires C-terminal glycosylphosphatidylinositol modification. J. Biol. Chem. 2004, 279, 40737–40747. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Halfmann, R.; King, O.; Kapila, A.; Lindquist, S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009, 137, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Orlean, P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 2012, 192, 775–818. [Google Scholar] [CrossRef] [PubMed]
- Gozalbo, D.; Gil-Navarro, I.; Azorin, I.; Renau-Piqueras, J.; Martinez, J.P.; Gil, M.L. The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect. Immun. 1998, 66, 2052–2059. [Google Scholar] [PubMed]
- Urban, C.; Sohn, K.; Lottspeich, F.; Brunner, H.; Rupp, S. Identification of cell surface determinants in Candida albicans reveals Tsa1p, a protein differentially localized in the cell. FEBS Lett. 2003, 544, 228–235. [Google Scholar] [CrossRef]
- Marcos, C.M.; de Fatima da Silva, J.; de Oliveira, H.C.; Moraes da Silva, R.A.; Mendes-Giannini, M.J.; Fusco-Almeida, A.M. Surface-expressed enolase contributes to the adhesion of Paracoccidioides brasiliensis to host cells. FEMS Yeast Res. 2012, 12, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Jue, C.K.; Lipke, P.N. Role of Fig2p in agglutination in Saccharomyces cerevisiae. Eukaryot. Cell 2002, 1, 843–845. [Google Scholar] [CrossRef] [PubMed]
- Granger, B.L. Accessibility and contribution to glucan masking of natural and genetically tagged versions of yeast wall protein 1 of Candida albicans. PLoS ONE 2018, 13, e0191194. [Google Scholar] [CrossRef] [PubMed]
- Granger, B.L. Insight into the antiadhesive effect of yeast wall protein 1 of Candida albicans. Eukaryot. Cell 2012, 11, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Oh, S.H.; Hoyer, L.L. Deletion of ALS5, ALS6 or ALS7 increases adhesion of Candida albicans to human vascular endothelial and buccal epithelial cells. Med. Mycol. 2007, 45, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S. Biophysics of biofilm infection. Pathog. Dis. 2014, 70, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.; Lincoln, L.; Marchillo, K.; Massey, R.; Holoyda, K.; Hoff, B.; VanHandel, M.; Andes, D. Putative role of β-1,3 glucans in Candida albicans biofilm resistance. Antimicrob. Agents Chemother. 2007, 51, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Ma, S.; Leonhard, M.; Moser, D.; Schneider-Stickler, B. β-1,3-glucanase disrupts biofilm formation and increases antifungal susceptibility of Candida albicans DAY185. Int. J. Biol. Macromol. 2018, 108, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Geller, A.M.; Bamford, N.C.; Liu, H.; Gravelat, F.N.; Snarr, B.D.; Le Mauff, F.; Chabot, J.; Ralph, B.; Ostapska, H.; et al. Deacetylation of fungal exopolysaccharide mediates adhesion and biofilm formation. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Te Riet, J.; Joosten, B.; Reinieren-Beeren, I.; Figdor, C.G.; Cambi, A. N-glycan mediated adhesion strengthening during pathogen-receptor binding revealed by cell-cell force spectroscopy. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.F.; Taff, H.T.; Cuevas, M.A.; Reinicke, E.L.; Sanchez, H.; Andes, D.R. Role of matrix beta-1,3 glucan in antifungal resistance of non-albicans Candida biofilms. Antimicrob. Agents Chemother. 2013, 57, 1918–1920. [Google Scholar] [CrossRef] [PubMed]
- Taff, H.T.; Nett, J.E.; Zarnowski, R.; Ross, K.M.; Sanchez, H.; Cain, M.T.; Hamaker, J.; Mitchell, A.P.; Andes, D.R. A Candida biofilm-induced pathway for matrix glucan delivery: Implications for drug resistance. PLoS Pathog. 2012, 8, e1002848. [Google Scholar] [CrossRef] [PubMed]
- Abeyratne-Perera, H.K.; Chandran, P.L. Mannose Surfaces Exhibit Self-Latching, Water Structuring, and Resilience to Chaotropes: Implications for Pathogen Virulence. Langmuir 2017, 33, 9178–9189. [Google Scholar] [CrossRef] [PubMed]
- Almeida, C.A.; de Campos-Takaki, G.M.; Portela, M.B.; Travassos, L.R.; Alviano, C.S.; Alviano, D.S. Sialoglycoproteins in morphological distinct stages of Mucor polymorphosporus and their influence on phagocytosis by human blood phagocytes. Mycopathologia 2013, 176, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Qiu, W.; Lipke, P. Accelerated and adaptive evolution of yeast sexual adhesins. Mol. Biol. Evol. 2011, 28, 3127–3137. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.V.; Ielasi, F.S.; Nookaew, I.; Stals, I.; Alonso-Sarduy, L.; Daenen, L.; Van Mulders, S.E.; Stassen, C.; van Eijsden, R.G.; Siewers, V.; et al. Molecular mechanism of flocculation self-recognition in yeast and its role in mating and survival. MBio 2015, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Mitchell, A.P. Microbial biofilms: E pluribus unum. Curr. Biol. 2007, 17, 349. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.N. Glycomics for microbes and microbiologists. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Gaur, N.K.; Klotz, S.A. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect. Immun. 1997, 65, 5289–5294. [Google Scholar] [PubMed]
- Gonzalez, M.; Goddard, N.; Hicks, C.; Ovalle, R.; Rauceo, J.M.; Jue, C.K.; Lipke, P.N. A screen for deficiencies in GPI-anchorage of wall glycoproteins in yeast. Yeast 2010, 27, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wang, T.; Zhang, J.; Liu, Q.; Wang, L.; Chen, P.; Wang, F.; Li, H.; Xiao, Y.; Zhao, X. Display of heterologous proteins on the Saccharomyces cerevisiae surface display system using a single constitutive expression vector. Biotechnol. Prog. 2014, 30, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Mathias, A.; Stavrou, S.; Neville, D.M., Jr. A new yeast display vector permitting free scFv amino termini can augment ligand binding affinities. Protein Eng. Des. Sel. 2005, 18, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Younger, D.; Berger, S.; Baker, D.; Klavins, E. High-throughput characterization of protein-protein interactions by reprogramming yeast mating. Proc. Natl. Acad. Sci. USA 2017, 114, 12166–12171. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, 344. [Google Scholar] [CrossRef] [PubMed]
- Coronado, J.E.; Mneimneh, S.; Epstein, S.L.; Qiu, W.G.; Lipke, P.N. Conserved processes and lineage-specific proteins in fungal cell wall evolution. Eukaryot. Cell 2007, 6, 2269–2277. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, F.; Schymkowitz, J.; Serrano, L. Protein aggregation and amyloidosis: Confusion of the kinds? Curr. Opin. Struct. Biol. 2006, 16, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt, L.; Teng, P.K.; Riek, R.; Eisenberg, D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. USA 2010, 107, 3487–3492. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sherman, M.C.; Lysak, N.; Filonenko, A.; Richards, H.; Sobonya, R.E.; Klotz, S.A.; Lipke, P.N. Peptide detection of fungal functional amyloids in infected tissue. PLoS ONE 2014, 9, e86067. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.N.; Ramsook, C.; Garcia-Sherman, M.C.; Jackson, D.N.; Chan, C.X.; Bois, M.; Klotz, S.A. Between Amyloids and Aggregation Lies a Connection with Strength and Adhesion. New J. Sci. 2014, 2014, 815102. [Google Scholar] [CrossRef] [PubMed]
- Gelis, S.; de Groot, P.W.; Castillo, L.; Moragues, M.D.; Sentandreu, R.; Gómez, M.M.; Valentín, E. Pga13 in Candida albicans is localized in the cell wall and influences cell surface properties, morphogenesis and virulence. Fungal Genet. Biol. 2012, 49, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Cappellaro, C.; Baldermann, C.; Rachel, R.; Tanner, W. Mating type-specific cell-cell recognition of Saccharomyces cerevisiae: Cell wall attachment and active sites of a- and alpha-agglutinin. EMBO J. 1994, 13, 4737–4744. [Google Scholar] [PubMed]
- Serra, D.O.; Richter, A.M.; Klauck, G.; Mika, F.; Hengge, R. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio 2013, 4, 103. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Price, J.E.; Burby, P.E.; Blanco, L.P.; Chamberlain, J.; Chapman, M.R. The Production of Curli Amyloid Fibers Is Deeply Integrated into the Biology of Escherichia coli. Biomolecules 2017, 7, 75. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, M.S. Differential adhesion in morphogenesis: A modern view. Curr. Opin. Genet. Dev. 2007, 17, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Watnick, P.; Kolter, R. Biofilm, city of microbes. J. Bacteriol. 2000, 182, 2675–2679. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Schneider, H.A.; Nett, J.E.; Sheppard, D.C.; Filler, S.G.; Andes, D.R.; Mitchell, A.P. Complementary adhesin function in C. albicans biofilm formation. Curr. Biol. 2008, 18, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Nobbs, A.H.; Vickerman, M.M.; Jenkinson, H.F. Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae Reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot. Cell 2010, 9, 1622–1634. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Andes, D.R.; Nett, J.E.; Smith, F.J.; Yue, F.; Phan, Q.T.; Edwards, J.E.; Filler, S.G.; Mitchell, A.P. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006, 2, e63. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sherman, M.C.; Lundgren, T.; Sobonya, R.; Lipke, P.N.; Klotz, S.A. A Unique Biofilm in Human Deep Mycoses: Fungal Amyloid is bound by Host Serum Amyloid P Component. NPJ Biofilms Microb. 2015, 1, 15009. [Google Scholar] [CrossRef] [PubMed]
- Klotz, S.A.; Sobonya, R.E.; Lipke, P.N.; Garcia-Sherman, M.C. Serum Amyloid P Component and Systemic Fungal Infection: Does It Protect the Host or Is It a Trojan Horse? In Open Forum Infectious Diseases; Oxford University Press: Oxford, UK, 2016. [Google Scholar]
- Schonherr, F.A.; Sparber, F.; Kirchner, F.R.; Guiducci, E.; Trautwein-Weidner, K.; Gladiator, A.; Sertour, N.; Hetzel, U.; Le, G.T.T.; Pavelka, N.; et al. The intraspecies diversity of C. albicans triggers qualitatively and temporally distinct host responses that determine the balance between commensalism and pathogenicity. Mucosal Immunol. 2017, 10, 1335–1350. [Google Scholar] [CrossRef] [PubMed]



| Figure 2 Model | Length (AA) | Signal | GPI | Cys-Rich Region | Tandem Repeats | Dibasic Sequence | TANGO >10% | N-glyco-sylation | QQQ | Ligands | Comments | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ascomycetes | ||||||||||||
| Saccharomycotina | ||||||||||||
| CaAls2 | A1 | 2362 | X | X | X | 1 | 11 | 22 | oral epithelium | |||
| CaAls5 | A1 | 1347 | X | X | X | 2 | 6 | 4 | peptides, Φ, epithelial cells | very broad ligand specificity | ||
| CaEap1 | A2 | 653 | X | X | X | X | 1 | 3 | 1 | Φ | ||
| CaHwp1 | A2 | 634 | X | X | X | X | 1 | 3 | 3 | X | Gln transaminase, buccal cells | |
| ScAga1 | A3 | 725 | X | X | X | X | - | 5 | - | - | SS bonded to Aga2 | |
| ScAga2 | A3 | 87 | X | - | 1 | - | Sc Sag1 | SS bonded to Aga1; sexual adhesin | ||||
| ScFig2 | A1 | 2127 | X | X | X | X | 3 | 16 | 14 | agar | secondary sexual adhesin | |
| ScFlo1 | A1 | 1537 | X | X | X | X | - | 22 | 5 | ⍺-man | ||
| ScFlo11 | A1 | 1367 | X | X | X | X | 2 | 4 | 3 | Sc Flo11 | homotypic binder | |
| Pezizomycotina | ||||||||||||
| AfHydrophobin | C | 155 | X | X | X | X | 1 | - | - | Φ | homopolymerizes | |
| AfRodA | C | 149 | X | X | - | - | - | Φ | homopolymerizes | |||
| AfRodB | C | 140 | X | X | X | - | 1 | - | Φ | homopolymerizes | ||
| Po Veg Hyd 2 | C | 112 | X | X | X | - | 2 | - | Φ | homopolymerizes | ||
| AoMad1 | A2? | 718 | X | X | X | X | 2 | 2 | 1 | invertebrate exoskeleton | ||
| BdBad1 | D | 1146 | X | CW | X | 1 | 2 | - | heparin | |||
| Vd Fas1-like | F | 469 | X | X | 3 | 15 | agar | |||||
| Vd Wsc1-like | A2? | 468 | X | X | X | X | 4 | 2 | - | agar | ||
| Taphrinomycotina | ||||||||||||
| Pj Int1 | ? | 1005 | X | X | 11 | 8 | 9 | fibronectin | signal missing, PH/Bud4 domain | |||
| Pj Msg | A2? | 1088 | X | X | X | 19 | 1 | 1 | epithelial cells | signal missing | ||
| SpGsf2 | A1 | 1563 | X | X | X | - | 13 | 23 | galactose | |||
| SpMam3 | E | 1082 | X | X | - | 10 | 9 | Sp Map4? | sexual adhesin | |||
| SpMap4 | E | 948 | X | X | X | 2 | 9 | 10 | Sp Mam3? | sexual adhesin | ||
| Basidiomycete | ||||||||||||
| CnCfl1 | ? | 309 | X | CW | X | 1 | - | - | X | C-terminal CAAX sequence | ||
| CnMP84 | F | 411 | X | X | 1 | 3 | 5 | epithelial cells | chitin de-acetylase domain | |||
| Entries in Table: 24 | 22 | 18 | 16 | 18 | 15 | 21 | 15 | 2 | ||||
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lipke, P.N. What We Do Not Know about Fungal Cell Adhesion Molecules. J. Fungi 2018, 4, 59. https://doi.org/10.3390/jof4020059
Lipke PN. What We Do Not Know about Fungal Cell Adhesion Molecules. Journal of Fungi. 2018; 4(2):59. https://doi.org/10.3390/jof4020059
Chicago/Turabian StyleLipke, Peter N. 2018. "What We Do Not Know about Fungal Cell Adhesion Molecules" Journal of Fungi 4, no. 2: 59. https://doi.org/10.3390/jof4020059